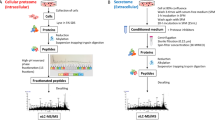
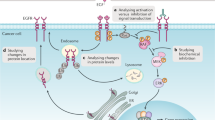
Abstract
Proteomics has revealed that many proteins are present in unexpected cellular locations. Moreover, it is increasingly recognized that proteins can translocate between intracellular and extracellular compartments in non-conventional ways. This increases gene pleiotrophy as the diverse functions of the protein that the gene encodes are dependent on the cellular location. Given that trafficking drug targets may exist in various forms — often with completely different functions — in multiple cellular compartments, careful interpretation of proteomics data is needed for an accurate understanding of gene function. This Perspective is intended to inspire the investigation of unusual protein localizations, rather than assuming that they are due to mislocalization or artefacts. Given a fair chance, proteomics could reveal novel and unforeseen biology with important ramifications for target validation in drug discovery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Walgren, J. L. & Thompson, D. C. Application of proteomic technologies in the drug development process. Toxicol. Lett. 149, 377–385 (2004).
Sleno, L. & Emili, A. Proteomic methods for drug target discovery. Curr. Opin. Chem. Biol. 12, 46–54 (2008).
Stelzl, U. et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell 122, 957–968 (2005).
Ideker, T., Galitski, T. & Hood, L. A new approach to decoding life: systems biology. Annu. Rev. Genomics Hum. Genet. 2, 343–372 (2001).
Butler, G. S., Dean, R. A., Tam, E. & Overall, C. M. Pharmacoproteomics of a metalloproteinase hydroxamate inhibitor in breast cancer cells: dynamics of matrix metalloproteinase-14 (MT1-MMP) mediated membrane protein shedding. Mol. Cell Biol. 28, 4896–4914 (2008).
Chen, G. et al. Proteomic analysis identifies protein targets responsible for depsipeptide sensitivity in tumor cells. J. Proteome Res. 7, 2733–2742 (2008).
Hu, W. et al. Proteomic identification of heat shock protein 70 as a candidate target for enhancing apoptosis induced by farnesyl transferase inhibitor. Proteomics 3, 1904–1911 (2003).
Verrills, N. M., Walsh, B. J., Cobon, G. S., Hains, P. G. & Kavallaris, M. Proteome analysis of vinca alkaloid response and resistance in acute lymphoblastic leukemia reveals novel cytoskeletal alterations. J. Biol. Chem. 278, 45082–45093 (2003).
MacKeigan, J. P. et al. Proteomic profiling drug-induced apoptosis in non-small cell lung carcinoma: identification of RS/DJ-1 and RhoGDIα. Cancer Res. 63, 6928–6934 (2003).
Ong, S. E. et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc. Natl Acad. Sci. USA 106, 4617–4622 (2009).
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Adams, M. D. et al. The genome sequence of Drosophila melanogaster. Science 287, 2185–2195 (2000).
Jeffery, C. J. Multifunctional proteins: examples of gene sharing. Ann. Med. 35, 28–35 (2003).
Piatigorsky, J. Gene sharing in lens and cornea: facts and implications. Prog. Retin. Eye Res. 17, 145–174 (1998).
Doucet, A., Butler, G. S., Rodriguez, D., Prudova, A. & Overall, C. M. Metadegradomics: toward in vivo quantitative degradomics of proteolytic post-translational modifications of the cancer proteome. Mol. Cell Proteomics 7, 1925–1951 (2008).
Roshy, S., Sloane, B. F. & Moin, K. Pericellular cathepsin B and malignant progression. Cancer Metastasis Rev. 22, 271–286 (2003).
Falsone, S. F., Gesslbauer, B., Rek, A. & Kungl, A. J. A proteomic approach towards the Hsp90-dependent ubiquitinylated proteome. Proteomics 7, 2375–2383 (2007).
Banerji, U. Heat shock protein 90 as a drug target: some like it hot. Clin. Cancer Res. 15, 9–14 (2009).
Solit, D. B. & Chiosis, G. Development and application of Hsp90 inhibitors. Drug Discov. Today 13, 38–43 (2008).
Li, W. et al. Extracellular heat shock protein-90α: linking hypoxia to skin cell motility and wound healing. EMBO J. 26, 1221–1233 (2007).
Patterson, S. D. & Aebersold, R. H. Proteomics: the first decade and beyond. Nature Genet. 33, S311–S323 (2003).
Pan, S. & Aebersold, R. Quantitative proteomics by stable isotope labeling and mass spectrometry. Methods Mol. Biol. 367, 209–218 (2007).
Rozanas, C. R. & Loyland, S. M. Capabilities using 2-D DIGE in proteomics research: the new gold standard for 2-D gel electrophoresis. Methods Mol. Biol. 441, 1–18 (2008).
Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003).
Eustace, B. K. et al. Functional proteomic screens reveal an essential extracellular role for HSP90α in cancer cell invasiveness. Nature Cell Biol. 6, 507–514 (2004).
Greenberg, Y. et al. The novel fragment of tyrosyl tRNA synthetase, mini-TyrRS, is secreted to induce an angiogenic response in endothelial cells. FASEB J. 22, 1597–1605 (2008).
Park, S. G. et al. Human lysyl-tRNA synthetase is secreted to trigger proinflammatory response. Proc. Natl Acad. Sci. USA 102, 6356–6361 (2005).
Wang, H. et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science 285, 248–251 (1999).
Fages, C., Nolo, R., Huttunen, H. J., Eskelinen, E. & Rauvala, H. Regulation of cell migration by amphoterin. J. Cell Sci. 113, 611–620 (2000).
Lee, S. W., Cho, B. H., Park, S. G. & Kim, S. Aminoacyl-tRNA synthetase complexes: beyond translation. J. Cell Sci. 117, 3725–3734 (2004).
Liu, J., Shue, E., Ewalt, K. L. & Schimmel, P. A new γ-interferon-inducible promoter and splice variants of an anti-angiogenic human tRNA synthetase. Nucleic Acids Res. 32, 719–727 (2004).
Westbrook, C. A., Gasson, J. C., Gerber, S. E., Selsted, M. E. & Golde, D. W. Purification and characterization of human T-lymphocyte-derived erythroid-potentiating activity. J. Biol. Chem. 259, 9992–9996 (1984).
Gasson, J. C. et al. Molecular characterization and expression of the gene encoding human erythroid-potentiating activity. Nature 315, 768–771 (1985).
Docherty, A. J. et al. Sequence of human tissue inhibitor of metalloproteinases and its identity to erythroid-potentiating activity. Nature 318, 66–69 (1985).
Cawston, T. E., Galloway, W. A., Mercer, E., Murphy, G. & Reynolds, J. J. Purification of rabbit bone inhibitor of collagenase. Biochem. J. 195, 159–165 (1981).
Murphy, G., Cawston, T. E. & Reynolds, J. J. An inhibitor of collagenase from human amniotic fluid. Purification, characterization and action on metalloproteinases. Biochem. J. 195, 167–170 (1981).
Hayakawa, T., Yamashita, K., Kishi, J. & Harigaya, K. Tissue inhibitor of metalloproteinases from human bone marrow stromal cell line KM 102 has erythroid-potentiating activity, suggesting its possibly bifunctional role in the hematopoietic microenvironment. FEBS Lett. 268, 125–128 (1990).
Gurney, M. E., Heinrich, S. P., Lee, M. R. & Yin, H. S. Molecular cloning and expression of neuroleukin, a neurotrophic factor for spinal and sensory neurons. Science 234, 566–574 (1986).
Watanabe, H., Takehana, K., Date, M., Shinozaki, T. & Raz, A. Tumor cell autocrine motility factor is the neuroleukin/phosphohexose isomerase polypeptide. Cancer Res. 56, 2960–2963 (1996).
Yakirevich, E. & Naot, Y. Cloning of a glucose phosphate isomerase/neuroleukin-like sperm antigen involved in sperm agglutination. Biol. Reprod. 62, 1016–1023 (2000).
Faik, P., Walker, J. I., Redmill, A. A. & Morgan, M. J. Mouse glucose-6-phosphate isomerase and neuroleukin have identical 3′ sequences. Nature 332, 455–457 (1988).
Baumann, M. & Brand, K. Purification and characterization of phosphohexose isomerase from human gastrointestinal carcinoma and its potential relationship to neuroleukin. Cancer Res. 48, 7018–7021 (1988).
Goulet, B. et al. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol. Cell 14, 207–219 (2004).
Goulet, B. et al. Increased expression and activity of nuclear cathepsin L in cancer cells suggests a novel mechanism of cell transformation. Mol. Cancer Res. 5, 899–907 (2007).
Schulz, R. Intracellular targets of matrix metalloproteinase-2 in cardiac disease: rationale and therapeutic approaches. Annu. Rev. Pharmacol. Toxicol. 47, 211–242 (2007).
Strongin, A. Y. Mislocalization and unconventional functions of cellular MMPs in cancer. Cancer Metastasis Rev. 25, 87–98 (2006).
Ip, Y. C., Cheung, S. T. & Fan, S. T. Atypical localization of membrane type 1-matrix metalloproteinase in the nucleus is associated with aggressive features of hepatocellular carcinoma. Mol. Carcinog. 46, 225–230 (2007).
Si-Tayeb, K. et al. Matrix metalloproteinase 3 is present in the cell nucleus and is involved in apoptosis. Am. J. Pathol. 169, 1390–1401 (2006).
Eguchi, T. et al. Novel transcription-factor-like function of human matrix metalloproteinase 3 regulating the CTGF/CCN2 gene. Mol. Cell Biol. 28, 2391–2413 (2008).
Hashimoto, G. et al. Matrix metalloproteinases cleave connective tissue growth factor and reactivate angiogenic activity of vascular endothelial growth factor 165. J. Biol. Chem. 277, 36288–36295 (2002).
Tam, E. M., Morrison, C. J., Wu, Y. I., Stack, M. S. & Overall, C. M. Membrane protease proteomics: isotope-coded affinity tag MS identification of undescribed MT1-matrix metalloproteinase substrates. Proc. Natl Acad. Sci. USA 101, 6917–6922 (2004).
Dean, R. A. et al. Identification of candidate angiogenic inhibitors processed by matrix metalloproteinase 2 (MMP-2) in cell-based proteomic screens: disruption of vascular endothelial growth factor (VEGF)/heparin affin regulatory peptide (pleiotrophin) and VEGF/connective tissue growth factor angiogenic inhibitory complexes by MMP-2 proteolysis. Mol. Cell Biol. 27, 8454–8465 (2007).
Golubkov, V. S. et al. Membrane type-1 matrix metalloproteinase (MT1-MMP) exhibits an important intracellular cleavage function and causes chromosome instability. J. Biol. Chem. 280, 25079–25086 (2005).
McCulloch, D. R., Akl, P., Samaratunga, H., Herington, A. C. & Odorico, D. M. Expression of the disintegrin metalloprotease, ADAM-10, in prostate cancer and its regulation by dihydrotestosterone, insulin-like growth factor I, and epidermal growth factor in the prostate cancer cell model LNCaP. Clin. Cancer Res. 10, 314–323 (2004).
Arima, T. et al. Nuclear translocation of ADAM-10 contributes to the pathogenesis and progression of human prostate cancer. Cancer Sci. 98, 1720–1726 (2007).
Brix, K., Lemansky, P. & Herzog, V. Evidence for extracellularly acting cathepsins mediating thyroid hormone liberation in thyroid epithelial cells. Endocrinology 137, 1963–1974 (1996).
Bryant, D. M. & Stow, J. L. Nuclear translocation of cell-surface receptors: lessons from fibroblast growth factor. Traffic 6, 947–954 (2005).
Czupalla, C. et al. Proteomic analysis of lysosomal acid hydrolases secreted by osteoclasts. Implications for lytic enzyme transport and bone metabolism. Mol. Cell. Proteomics 5, 134–143 (2006).
Cardone, R. A., Casavola, V. & Reshkin, S. J. The role of disturbed pH dynamics and the Na+/H+ exchanger in metastasis. Nature Rev. Cancer 5, 786–795 (2005).
Wartha, F., Beiter, K., Normark, S. & Henriques-Normark, B. Neutrophil extracellular traps: casting the NET over pathogenesis. Curr. Opin. Microbiol. 10, 52–56 (2007).
Fuchs, T. A. et al. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 176, 231–241 (2007).
Clark, S. R. et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nature Med. 13, 463–469 (2007).
Gupta, A. K., Hasler, P., Holzgreve, W. & Hahn, S. Neutrophil NETs: a novel contributor to preeclampsia-associated placental hypoxia? Semin. Immunopathol. 29, 163–167 (2007).
Taghbalout, A. & Rothfield, L. RNaseE and the other constituents of the RNA degradosome are components of the bacterial cytoskeleton. Proc. Natl Acad. Sci. USA 104, 1667–1672 (2007).
Knull, H. R. & Walsh, J. L. Association of glycolytic enzymes with the cytoskeleton. Curr. Top. Cell Regul. 33, 15–30 (1992).
Nickel, W. Unconventional secretory routes: direct protein export across the plasma membrane of mammalian cells. Traffic 6, 607–614 (2005).
Shin, B. K. et al. Global profiling of the cell surface proteome of cancer cells uncovers an abundance of proteins with chaperone function. J. Biol. Chem. 278, 7607–7616 (2003).
Eustace, B. K. & Jay, D. G. Extracellular roles for the molecular chaperone, hsp90. Cell Cycle 3, 1098–1100 (2004).
Pratt, W. B., Morishima, Y. & Osawa, Y. The hsp90 chaperone machinery regulates signaling by modulating ligand binding clefts. J. Biol. Chem. 283, 22885–22889 (2008).
Ray, I., Chauhan, A., Wegiel, J. & Chauhan, V. P. Gelsolin inhibits the fibrillization of amyloid beta-protein, and also defibrillizes its preformed fibrils. Brain Res. 853, 344–351 (2000).
Fishel, M. L. & Kelley, M. R. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol. Aspects Med. 28, 375–395 (2007).
Yang, S., Irani, K., Heffron, S. E., Jurnak, F. & Meyskens, F. L. Jr. Alterations in the expression of the apurinic/apyrimidinic endonuclease-1/redox factor-1 (APE/Ref-1) in human melanoma and identification of the therapeutic potential of resveratrol as an APE/Ref-1 inhibitor. Mol. Cancer Ther. 4, 1923–1935 (2005).
Overall, C. M. & Kleifeld, O. Validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nature Rev. Cancer 6, 227–239 (2006).
Mayer, M. P., Prodromou, C. & Frydman, J. The Hsp90 mosaic: a picture emerges. Nature Struct. Mol. Biol. 16, 2–6 (2009).
Mann, M. Functional and quantitative proteomics using SILAC. Nature Rev. Mol. Cell Biol. 7, 952–958 (2006).
McQuibban, G. A. et al. Inflammation dampened by gelatinase A cleavage of monocyte chemoattractant protein-3. Science 289, 1202–1206 (2000).
Vergote, D. et al. Proteolytic processing of SDF-1α reveals a change in receptor specificity mediating HIV-associated neurodegeneration. Proc. Natl Acad. Sci. USA 103, 19182–19187 (2006).
Autelitano, D. J. et al. The cryptome: a subset of the proteome, comprising cryptic peptides with distinct bioactivities. Drug Discov. Today 11, 306–314 (2006).
O'Reilly, M. S. et al. Angiostatin: a novel angiogenesis inhibitor that mediates the suppression of metastases by a Lewis lung carcinoma. Cell 79, 315–328 (1994).
Pike, S. E. et al. Vasostatin, a calreticulin fragment, inhibits angiogenesis and suppresses tumor growth. J. Exp. Med. 188, 2349–2356 (1998).
Ehlers, M. R. & Riordan, J. F. Membrane proteins with soluble counterparts: role of proteolysis in the release of transmembrane proteins. Biochemistry 30, 10065–10074 (1991).
Werb, Z. ECM and cell surface proteolysis: regulating cellular ecology. Cell 91, 439–442 (1997).
Noel, A. et al. Membrane associated proteases and their inhibitors in tumour angiogenesis. J. Clin. Pathol. 57, 577–584 (2004).
Cauwe, B., Steen, P. E. & Opdenakker, G. The biochemical, biological, and pathological kaleidoscope of cell surface substrates processed by matrix metalloproteinases. Crit. Rev. Biochem. Mol. Biol. 42, 113–185 (2007).
Carl-McGrath, S., Lendeckel, U., Ebert, M. & Rocken, C. Ectopeptidases in tumour biology: a review. Histol. Histopathol. 21, 1339–1353 (2006).
Roy., R., Zhang, B. & Moses, M. A. Making the cut: protease-mediated regulation of angiogenesis. Exp. Cell Res. 312, 608–622 (2006).
Wakasugi, K. & Schimmel, P. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science 284, 147–151 (1999).
Schluesener, H. Tyrosyl-tRNA synthetase: a housekeeping protein and an attractive harbinger of cellular death. Angew. Chem. Int. Ed. Engl. 38, 3635–3637 (1999).
Vong, L. et al. Annexin 1 cleavage in activated neutrophils: a pivotal role for proteinase 3. J. Biol. Chem. 282, 29998–30004 (2007).
Dean, R. A. & Overall, C. M. Proteomics discovery of metalloproteinase substrates in the cellular context by iTRAQ labeling reveals a diverse MMP-2 substrate degradome. Mol. Cell. Proteomics 6, 611–623 (2007).
Zolla, L. Proteomics studies reveal important information on small molecule therapeutics: a case study on plasma proteins. Drug Discov. Today 13, 1042–1051 (2008).
Kolch, W., Mischak, H. & Pitt, A. R. The molecular make-up of a tumour: proteomics in cancer research. Clin. Sci. (Lond.) 108, 369–383 (2005).
Witzmann, F. A. & Grant, R. A. Pharmacoproteomics in drug development. Pharmacogenomics J. 3, 69–76 (2003).
Bantscheff, M. et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nature Biotech. 25, 1035–1044 (2007).
Chapal, N. et al. Pharmacoproteomic approach to the study of drug mode of action, toxicity, and resistance: applications in diabetes and cancer. Fundam. Clin. Pharmacol. 18, 413–422 (2004).
Wang, Y., Dai, Z., Sadee, W. & Hancock, W. S. A pharmacoproteomics study of the cancer cell line EKVX using capillary-LC/MS/MS. Mol. Pharm. 3, 566–578 (2006).
Sung, F. L. et al. Pharmacoproteomics study of cetuximab in nasopharyngeal carcinoma. J. Proteome Res. 5, 3260–3267 (2006).
Stockwin, L. H. et al. Proteomic analysis identifies oxidative stress induction by adaphostin. Clin. Cancer Res. 13, 3667–3681 (2007).
Milli, A. et al. A proteomic approach for evaluating the cell response to a novel histone deacetylase inhibitor in colon cancer cells. Biochim. Biophys. Acta 1784, 1702–1710 (2008).
Lin, Z., Crockett, D. K., Jenson, S. D., Lim, M. S. & Elenitoba-Johnson, K. S. Quantitative proteomic and transcriptional analysis of the response to the p38 mitogen-activated protein kinase inhibitor SB203580 in transformed follicular lymphoma cells. Mol. Cell Proteomics 3, 820–833 (2004).
Lee, S. J. & Wang, J. Y. Exploiting the promiscuity of imatinib. J. Biol. 8, 30 (2009).
Youn, J. H. & Shin, J. S. Nucleocytoplasmic shuttling of HMGB1 is regulated by phosphorylation that redirects it toward secretion. J. Immunol. 177, 7889–7897 (2006).
Oh, Y. J. et al. HMGB1 is phosphorylated by classical protein kinase C and is secreted by a calcium-dependent mechanism. J. Immunol. 182, 5800–5809 (2009).
Lopéz-Otín, C. & Overall, C. M. Protease degradomics: a new challenge for proteomics. Nature Rev. Mol. Cell Biol. 3, 509–519 (2002).
Schilling, O. & Overall, C. M. Proteomic discovery of protease substrates. Curr. Opin. Chem. Biol. 11, 36–45 (2007).
Doucet, A. & Overall, C. M. Protease proteomics: revealing protease in vivo functions using systems biology approaches. Mol. Aspects Med. 29, 339–358 (2008).
Cohen, A. A. et al. Dynamic proteomics of individual cancer cells in response to a drug. Science 322, 1511–1516 (2008).
Song, D. et al. Antitumor activity and molecular effects of the novel heat shock protein 90 inhibitor, IPI-504, in pancreatic cancer. Mol. Cancer Ther. 7, 3275–3284 (2008).
Maloney, A. et al. Gene and protein expression profiling of human ovarian cancer cells treated with the heat shock protein 90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 67, 3239–3253 (2007).
Schumacher, J. A., Crockett, D. K., Elenitoba-Johnson, K. S. & Lim, M. S. Proteome-wide changes induced by the Hsp90 inhibitor, geldanamycin in anaplastic large cell lymphoma cells. Proteomics 7, 2603–2616 (2007).
Clarke, P. A. et al. Gene expression profiling of human colon cancer cells following inhibition of signal transduction by 17-allylamino-17-demethoxygeldanamycin, an inhibitor of the hsp90 molecular chaperone. Oncogene 19, 4125–4133 (2000).
Liotta, L. A. et al. Metastatic potential correlates with enzymatic degradation of basement membrane collagen. Nature 284, 67–68 (1980).
Fingleton, B. in The Cancer Degradome: Proteases and Cancer Biology (eds Edwards, D., Hoyer-Hansen, G., Blasi, F. & Sloane, B. F.) 759–786 (Springer, New York, 2008).
Coussens, L. M., Fingleton, B. & Matrisian, L. M. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science 295, 2387–2392 (2002).
Fingleton, B. MMPs as therapeutic targets — still a viable option? Semin. Cell Dev. Biol. 19, 61–68 (2008).
Zucker, S., Cao, J. & Chen, W. T. Critical appraisal of the use of matrix metalloproteinase inhibitors in cancer treatment. Oncogene 19, 6642–6650 (2000).
McCawley, L. J. & Matrisian, L. M. Matrix metalloproteinases: they're not just for matrix anymore! Curr. Opin. Cell Biol. 13, 534–540 (2001).
Barbolina, M. V. & Stack, M. S. Membrane type 1-matrix metalloproteinase: substrate diversity in pericellular proteolysis. Semin. Cell Dev. Biol. 19, 24–33 (2007).
Overall, C. M. Dilating the degradome: matrix metalloproteinase 2 (MMP-2) cuts to the heart of the matter. Biochem. J. 383, e5–e7 (2004).
Page-McCaw, A., Ewald, A. J. & Werb, Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nature Rev. Mol. Cell Biol. 8, 221–233 (2007).
Egeblad, M. & Werb, Z. New functions for the matrix metalloproteinases in cancer progression. Nature Rev. Cancer 2, 161–174 (2002).
Overall, C. M. & Kleifeld, O. Towards third generation matrix metalloproteinase inhibitors for cancer therapy. Br. J. Cancer 94, 941–946 (2006).
Balbin, M. et al. Loss of collagenase-2 confers increased skin tumor susceptibility to male mice. Nature Genet. 35, 252–257 (2003).
Mantovani, A., Allavena, P., Sica, A. & Balkwill, F. Cancer-related inflammation. Nature 454, 436–444 (2008).
Cox, J. H. & Overall, C. M. in The Cancer Degradome: Proteases and Cancer Biology (eds Edwards, D., Hoyer-Hansen, G., Blasi, F. & Sloane, B. F.) 519–540 (Springer, New York, 2008).
Hautamaki, R. D., Kobayashi, D. K., Senior, R. M. & Shapiro, S. D. Requirement for macrophage elastase for cigarette smoke-induced emphysema in mice. Science 277, 2002–2004 (1997).
Houghton, A. M. et al. Macrophage elastase (matrix metalloproteinase-12) suppresses growth of lung metastases. Cancer Res. 66, 6149–6155 (2006).
Diarra, D. et al. Dickkopf-1 is a master regulator of joint remodeling. Nature Med. 13, 156–163 (2007).
Frazier, K., Williams, S., Kothapalli, D., Klapper, H. & Grotendorst, G. R. Stimulation of fibroblast cell growth, matrix production, and granulation tissue formation by connective tissue growth factor. J. Invest. Dermatol. 107, 404–411 (1996).
Chaqour, B. & Goppelt-Struebe, M. Mechanical regulation of the Cyr61/CCN1 and CTGF/CCN2 proteins. FEBS J. 273, 3639–3649 (2006).
Chen, C. C., Chen, N. & Lau, L. F. The angiogenic factors Cyr61 and connective tissue growth factor induce adhesive signaling in primary human skin fibroblasts. J. Biol. Chem. 276, 10443–10452 (2001).
Overall, C. M. & Lopéz- Otín, C. Strategies for MMP inhibition in cancer: innovations for the post-trial era. Nature Rev. Cancer 2, 657–672 (2002).
Wang, H., Zhu, S., Zhou, R., Li, W. & Sama, A. E. Therapeutic potential of HMGB1-targeting agents in sepsis. Expert Rev. Mol. Med. 10, e32 (2008).
Vaughan, C. K., Piper, P. W., Pearl, L. H. & Prodromou, C. A common conformationally coupled ATPase mechanism for yeast and human cytoplasmic HSP90s. FEBS J. 276, 199–209 (2009).
Pearl, L. H., Prodromou, C. & Workman, P. The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem. J. 410, 439–453 (2008).
Chandarlapaty, S. et al. SNX2112, a synthetic heat shock protein 90 inhibitor, has potent antitumor activity against HER kinase-dependent cancers. Clin. Cancer Res. 14, 240–248 (2008).
Dickey, C. A. et al. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J. Clin. Invest. 117, 648–658 (2007).
Falsone, S. F., Gesslbauer, B., Tirk, F., Piccinini, A. M. & Kungl, A. J. A proteomic snapshot of the human heat shock protein 90 interactome. FEBS Lett. 579, 6350–6354 (2005).
Zhao, R. et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120, 715–727 (2005).
Millson, S. H. et al. A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p). Eukaryot. Cell 4, 849–860 (2005).
Toogun, O. A., Dezwaan, D. C. & Freeman, B. C. The hsp90 molecular chaperone modulates multiple telomerase activities. Mol. Cell Biol. 28, 457–467 (2008).
Meyer-Siegler, K. et al. A human nuclear uracil DNA glycosylase is the 37-kDa subunit of glyceraldehyde-3-phosphate dehydrogenase. Proc. Natl Acad. Sci. USA 88, 8460–8464 (1991).
Hegmans, J. P. et al. Proteomic analysis of exosomes secreted by human mesothelioma cells. Am. J. Pathol. 164, 1807–1815 (2004).
Kang, B. H. et al. Regulation of tumor cell mitochondrial homeostasis by an organelle-specific Hsp90 chaperone network. Cell 131, 257–270 (2007).
Chiosis, G., Vilenchik, M., Kim, J. & Solit, D. Hsp90: the vulnerable chaperone. Drug Discov. Today 9, 881–888 (2004).
Shooter, K. V., Goodwin, G. H. & Johns, E. W. Interactions of a purified non-histone chromosomal protein with DNA and histone. Eur. J. Biochem. 47, 263–270 (1974).
Parkkinen, J. & Rauvala, H. Interactions of plasminogen and tissue plasminogen activator (t-PA) with amphoterin. Enhancement of t-PA-catalyzed plasminogen activation by amphoterin. J. Biol. Chem. 266, 16730–16735 (1991).
Andersson, U. et al. High mobility group 1 protein (HMG-1) stimulates proinflammatory cytokine synthesis in human monocytes. J. Exp. Med. 192, 565–570 (2000).
Gong, Q. et al. Protective effect of antagonist of high-mobility group BOX 1 on lipopolysaccharide-induced acute lung injury in mice. Scand. J. Immunol. 69, 29–35 (2009).
Pisetsky, D. S., Erlandsson-Harris, H. & Andersson, U. High-mobility group box protein 1 (HMGB1): an alarmin mediating the pathogenesis of rheumatic disease. Arthritis Res. Ther. 10, 209 (2008).
Wang, H., Yang, H. & Tracey, K. J. Extracellular role of HMGB1 in inflammation and sepsis. J. Intern. Med. 255, 320–331 (2004).
Fossati, S. & Chiarugi, A. Relevance of high-mobility group protein BOX 1 to neurodegeneration. Int. Rev. Neurobiol. 82, 137–148 (2007).
Yang, H. et al. Reversing established sepsis with antagonists of endogenous high-mobility group BOX 1. Proc. Natl Acad. Sci. USA 101, 296–301 (2004).
Abraham, E., Arcaroli, J., Carmody, A., Wang, H. & Tracey, K. J. HMG-1 as a mediator of acute lung inflammation. J. Immunol. 165, 2950–2954 (2000).
Lange, S. S., Mitchell, D. L. & Vasquez, K. M. High mobility group protein B1 enhances DNA repair and chromatin modification after DNA damage. Proc. Natl Acad. Sci. USA 105, 10320–10325 (2008).
Stros, M., Bacikova, A., Polanska, E., Stokrova, J. & Strauss, F. HMGB1 interacts with human topoisomerase IIα and stimulates its catalytic activity. Nucleic Acids Res. 35, 5001–5013 (2007).
Mouri, F. et al. Intracellular HMGB1 transactivates the human IL1B gene promoter through association with an Ets transcription factor PU.1. Eur. J. Haematol. 80, 10–19 (2008).
Park, J. S. et al. High mobility group BOX 1 protein interacts with multiple Toll-like receptors. Am. J. Physiol. Cell Physiol. 290, C917–C924 (2006).
Yu, M. et al. HMGB1 signals through toll-like receptor (TLR) 4 and TLR2. Shock 26, 174–179 (2006).
Park, J. S. et al. Activation of gene expression in human neutrophils by high mobility group BOX 1 protein. Am. J. Physiol. Cell Physiol. 284, C870–C879 (2003).
Salmivirta, M., Rauvala, H., Elenius, K. & Jalkanen, M. Neurite growth-promoting protein (amphoterin, p30) binds syndecan. Exp. Cell Res. 200, 444–451 (1992).
Parkkinen, J. et al. Amphoterin, the 30-kDa protein in a family of HMG1-type polypeptides. Enhanced expression in transformed cells, leading edge localization, and interactions with plasminogen activation. J. Biol. Chem. 268, 19726–19738 (1993).
Huttunen, H. J., Fages, C., Kuja-Panula, J., Ridley, A. J. & Rauvala, H. Receptor for advanced glycation end products-binding COOH-terminal motif of amphoterin inhibits invasive migration and metastasis. Cancer Res. 62, 4805–4811 (2002).
Taguchi, A. et al. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature 405, 354–360 (2000).
Pedrazzi, M. et al. Selective proinflammatory activation of astrocytes by high-mobility group BOX 1 protein signaling. J. Immunol. 179, 8525–8532 (2007).
Sgarra, R. et al. Interaction proteomics of the HMGA chromatin architectural factors. Proteomics 8, 4721–4732 (2008).
Pierantoni, G. M. et al. Identification of new high mobility group A1 associated proteins. Proteomics 7, 3735–3742 (2007).
Yang, H., Wang, H., Czura, C. J. & Tracey, K. J. The cytokine activity of HMGB1. J. Leukoc. Biol. 78, 1–8 (2005).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, Research0034 (2002).
Mansur, N. R., Meyer-Siegler, K., Wurzer, J. C. & Sirover, M. A. Cell cycle regulation of the glyceraldehyde-3-phosphate dehydrogenase/uracil DNA glycosylase gene in normal human cells. Nucleic Acids Res. 21, 993–998 (1993).
Ishitani, R. et al. Proapoptotic protein glyceraldehyde-3-phosphate dehydrogenase: a possible site of action of antiapoptotic drugs. Prog. Neuropsychopharmacol Biol. Psychiatry 27, 291–301 (2003).
Park, S. G., Ewalt, K. L. & Kim, S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem. Sci. 30, 569–574 (2005).
Prudovsky, I. et al. Secretion without Golgi. J. Cell Biochem. 103, 1327–1343 (2007).
Keller, S., Sanderson, M. P., Stoeck, A. & Altevogt, P. Exosomes: from biogenesis and secretion to biological function. Immunol. Lett. 107, 102–108 (2006).
Fevrier, B. & Raposo, G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 16, 415–421 (2004).
Blott, E. J. & Griffiths, G. M. Secretory lysosomes. Nature Rev. Mol. Cell Biol. 3, 122–131 (2002).
Hugel, B., Martinez, M. C., Kunzelmann, C. & Freyssinet, J. M. Membrane microparticles: two sides of the coin. Physiology (Bethesda) 20, 22–27 (2005).
Cleves, A. E. & Kelly, R. B. Rehearsing the ABCs. Protein translocation. Curr. Biol. 6, 276–278 (1996).
Munro, S. & Pelham, H. R. A C-terminal signal prevents secretion of luminal ER proteins. Cell 48, 899–907 (1987).
von Heijne, G. Signal sequences. The limits of variation. J. Mol. Biol. 184, 99–105 (1985).
Dingwall, C. & Laskey, R. A. Nuclear targeting sequences — a consensus? Trends Biochem. Sci. 16, 478–481 (1991).
Kosugi, S., Hasebe, M., Tomita, M. & Yanagawa, H. Nuclear export signal consensus sequences defined using a localization-based yeast selection system. Traffic 9, 2053–2062 (2008).
Omura, T. Mitochondria-targeting sequence, a multi-role sorting sequence recognized at all steps of protein import into mitochondria. J. Biochem. 123, 1010–1016 (1998).
Subramani, S., Koller, A. & Snyder, W. B. Import of peroxisomal matrix and membrane proteins. Annu. Rev. Biochem. 69, 399–418 (2000).
Kwiatkowski, D. J., Mehl, R. & Yin, H. L. Genomic organization and biosynthesis of secreted and cytoplasmic forms of gelsolin. J. Cell Biol. 106, 375–384 (1988).
McCaw, B. J. et al. Identification and characterization of mErk5-T, a novel Erk5/Bmk1 splice variant. Gene 345, 183–190 (2005).
Raynal, P., Kuijpers, G., Rojas, E. & Pollard, H. B. A rise in nuclear calcium translocates annexins IV and V to the nuclear envelope. FEBS Lett. 392, 263–268 (1996).
Wiest, D. L., Burgess, W. H., McKean, D., Kearse, K. P. & Singer, A. The molecular chaperone calnexin is expressed on the surface of immature thymocytes in association with clonotype-independent CD3 complexes. EMBO J. 14, 3425–3433 (1995).
Godde, N. J., D'Abaco, G. M., Paradiso, L. & Novak, U. Efficient ADAM22 surface expression is mediated by phosphorylation-dependent interaction with 14-3-3 protein family members. J. Cell Sci. 119, 3296–3305 (2006).
Ashery, U., Yizhar, O., Rotblat, B. & Kloog, Y. Nonconventional trafficking of Ras associated with Ras signal organization. Traffic 7, 119–126 (2006).
Paukku, K. & Silvennoinen, O. STATs as critical mediators of signal transduction and transcription: lessons learned from STAT5. Cytokine Growth Factor Rev. 15, 435–455 (2004).
Constantinides, S. M. & Deal, W. C. Jr. Reversible dissociation of tetrameric rabbit muscle glyceraldehyde 3-phosphate dehydrogenase into dimers or monomers by adenosine triphosphate. J. Biol. Chem. 244, 5695–5702 (1969).
Hirano, W., Gotoh, I., Uekita, T. & Seiki, M. Membrane-type 1 matrix metalloproteinase cytoplasmic tail binding protein-1 (MTCBP-1) acts as an eukaryotic aci-reductone dioxygenase (ARD) in the methionine salvage pathway. Genes Cells 10, 565–574 (2005).
Uekita, T. et al. Membrane-type 1 matrix metalloproteinase cytoplasmic tail-binding protein-1 is a new member of the Cupin superfamily. A possible multifunctional protein acting as an invasion suppressor down-regulated in tumors. J. Biol. Chem. 279, 12734–12743 (2004).
Parrott, A. M., Walsh, M. R., Reichman, T. W. & Mathews, M. B. RNA binding and phosphorylation determine the intracellular distribution of nuclear factors 90 and 110. J. Mol. Biol. 348, 281–293 (2005).
Coppolino, M. G. & Dedhar, S. Ligand-specific, transient interaction between integrins and calreticulin during cell adhesion to extracellular matrix proteins is dependent upon phosphorylation/dephosphorylation events. Biochem. J. 340 (Pt 1), 41–50 (1999).
Deora, A. B., Kreitzer, G., Jacovina, A. T. & Hajjar, K. A. An annexin 2 phosphorylation switch mediates p11-dependent translocation of annexin 2 to the cell surface. J. Biol. Chem. 279, 43411–43418 (2004).
Salmena, L. & Pandolfi, P. P. Changing venues for tumour suppression: balancing destruction and localization by monoubiquitylation. Nature Rev. Cancer 7, 409–413 (2007).
Brown, M. S. & Goldstein, J. L. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl Acad. Sci. USA 96, 11041–11048 (1999).
Schroeter, E. H., Kisslinger, J. A. & Kopan, R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 (1998).
Cao, X. & Sudhof, T. C. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 293, 115–120 (2001).
De Strooper, B. et al. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 398, 518–522 (1999).
Struhl, G. & Adachi, A. Nuclear access and action of notch in vivo. Cell 93, 649–660 (1998).
Garton, K. J. et al. Tumor necrosis factor-α-converting enzyme (ADAM17) mediates the cleavage and shedding of fractalkine (CX3CL1). J. Biol. Chem. 276, 37993–38001 (2001).
Hoyer-Hansen, G. et al. Urokinase plasminogen activator cleaves its cell surface receptor releasing the ligand-binding domain. J. Biol. Chem. 267, 18224–18229 (1992).
Andolfo, A. et al. Metalloproteases cleave the urokinase-type plasminogen activator receptor in the D1-D2 linker region and expose epitopes not present in the intact soluble receptor. Thromb. Haemost. 88, 298–306 (2002).
Giri, J. G. et al. Elevated levels of shed type II IL-1 receptor in sepsis. Potential role for type II receptor in regulation of IL-1 responses. J. Immunol. 153, 5802–5809 (1994).
Crowe, P. D. et al. A metalloprotease inhibitor blocks shedding of the 80-kD TNF receptor and TNF processing in T lymphocytes. J. Exp. Med. 181, 1205–1210 (1995).
Lum, L. et al. Evidence for a role of a tumor necrosis factor-α (TNF-α)-converting enzyme-like protease in shedding of TRANCE, a TNF family member involved in osteoclastogenesis and dendritic cell survival. J. Biol. Chem. 274, 13613–13618 (1999).
Yamashita, R. et al. Extracellular proteome of human hepatoma cell, HepG2 analyzed using two-dimensional liquid chromatography coupled with tandem mass spectrometry. Mol. Cell Biochem. 298, 83–92 (2007).
Ahram, M., Adkins, J. N., Auberry, D. L., Wunschel, D. S. & Springer, D. L. A proteomic approach to characterize protein shedding. Proteomics 5, 123–131 (2005).
Mbeunkui, F., Metge, B. J., Shevde, L. A. & Pannell, L. K. Identification of differentially secreted biomarkers using LC-MS/MS in isogenic cell lines representing a progression of breast cancer. J. Proteome Res. 6, 2993–3002 (2007).
Anderson, N. L. et al. The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol. Cell Proteomics 3, 311–326 (2004).
Pardo, M. et al. The characterization of the invasion phenotype of uveal melanoma tumour cells shows the presence of MUC18 and HMG-1 metastasis markers and leads to the identification of DJ-1 as a potential serum biomarker. Int. J. Cancer 119, 1014–1022 (2006).
Dacheux, J. L., Belghazi, M., Lanson, Y. & Dacheux, F. Human epididymal secretome and proteome. Mol. Cell Endocrinol. 250, 36–42 (2006).
Dupont, A. et al. The proteome and secretome of human arterial smooth muscle cells. Proteomics 5, 585–596 (2005).
Coppinger, J. A. et al. Characterization of the proteins released from activated platelets leads to localization of novel platelet proteins in human atherosclerotic lesions. Blood 103, 2096–2104 (2004).
Chevallet, M., Diemer, H., Van Dorssealer, A., Villiers, C. & Rabilloud, T. Toward a better analysis of secreted proteins: the example of the myeloid cells secretome. Proteomics 7, 1757–1770 (2007).
Dupont, A. et al. Two-dimensional maps and databases of the human macrophage proteome and secretome. Proteomics 4, 1761–1778 (2004).
Lafon-Cazal, M. et al. Proteomic analysis of astrocytic secretion in the mouse. Comparison with the cerebrospinal fluid proteome. J. Biol. Chem. 278, 24438–24448 (2003).
Wang, P. et al. Profiling of the secreted proteins during 3T3-L1 adipocyte differentiation leads to the identification of novel adipokines. Cell. Mol. Life Sci. 61, 2405–2417 (2004).
Wu, C. C. et al. Cancer cell-secreted proteomes as a basis for searching potential tumor markers: nasopharyngeal carcinoma as a model. Proteomics 5, 3173–3182 (2005).
Thery, C. et al. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 166, 7309–7318 (2001).
Gasteiger, E. et al. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 (2003).
Jacob, R. J. & Cramer, R. PIGOK: linking protein identity to gene ontology and function. J. Proteome Res. 5, 3429–3432 (2006).
Emans, N. et al. Annexin II is a major component of fusogenic endosomal vesicles. J. Cell Biol. 120, 1357–1369 (1993).
Mickleburgh, I. et al. Annexin A2 binds to the localization signal in the 3′ untranslated region of c-myc mRNA. FEBS J. 272, 413–421 (2005).
Filipenko, N. R., MacLeod, T. J., Yoon, C. S. & Waisman, D. M. Annexin A2 is a novel RNA-binding protein. J. Biol. Chem. 279, 8723–8731 (2004).
Boyko, V. et al. A major cellular substrate for protein kinases, annexin II, is a DNA-binding protein. FEBS Lett. 345, 139–142 (1994).
Liu, J. & Vishwanatha, J. K. Regulation of nucleo-cytoplasmic shuttling of human annexin A2: a proposed mechanism. Mol. Cell Biochem. 303, 211–220 (2007).
Eberhard, D. A., Karns, L. R., VandenBerg, S. R. & Creutz, C. E. Control of the nuclear-cytoplasmic partitioning of annexin II by a nuclear export signal and by p11 binding. J. Cell Sci. 114, 3155–3166 (2001).
Siever, D. A. & Erickson, H. P. Extracellular annexin II. Int. J. Biochem. Cell Biol. 29, 1219–1223 (1997).
Keller, M., Ruegg, A., Werner, S. & Beer, H. D. Active caspase-1 is a regulator of unconventional protein secretion. Cell 132, 818–831 (2008).
Coppolino, M. G. & Dedhar, S. Calreticulin. Int. J. Biochem. Cell Biol. 30, 553–558 (1998).
Johnson, S., Michalak, M., Opas, M. & Eggleton, P. The ins and outs of calreticulin: from the ER lumen to the extracellular space. Trends Cell Biol. 11, 122–129 (2001).
Funasaka, T. & Raz, A. The role of autocrine motility factor in tumor and tumor microenvironment. Cancer Metastasis Rev. 26, 725–735 (2007).
Ghiran, I., Klickstein, L. B. & Nicholson-Weller, A. Calreticulin is at the surface of circulating neutrophils and uses CD59 as an adaptor molecule. J. Biol. Chem. 278, 21024–21031 (2003).
Orr, A. W. et al. Low density lipoprotein receptor-related protein is a calreticulin coreceptor that signals focal adhesion disassembly. J. Cell Biol. 161, 1179–1189 (2003).
Pike, S. E. et al. Calreticulin and calreticulin fragments are endothelial cell inhibitors that suppress tumor growth. Blood 94, 2461–2468 (1999).
Dupuis, M., Schaerer, E., Krause, K. H. & Tschopp, J. The calcium-binding protein calreticulin is a major constituent of lytic granules in cytolytic T lymphocytes. J. Exp. Med. 177, 1–7 (1993).
Mitola, S. et al. Cutting edge: extracellular high mobility group box-1 protein is a proangiogenic cytokine. J. Immunol. 176, 12–15 (2006).
Scaffidi, P., Misteli, T. & Bianchi, M. E. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 418, 191–195 (2002).
Taniguchi, N. et al. Stage-specific secretion of HMGB1 in cartilage regulates endochondral ossification. Mol. Cell Biol. 27, 5650–5663 (2007).
Gardella, S. et al. The nuclear protein HMGB1 is secreted by monocytes via a non-classical, vesicle-mediated secretory pathway. EMBO Rep. 3, 995–1001 (2002).
Jin, Z. G. et al. Cyclophilin A is a proinflammatory cytokine that activates endothelial cells. Arterioscler. Thromb. Vasc. Biol. 24, 1186–1191 (2004).
Dhar-Chowdhury, P. et al. The glycolytic enzymes, glyceraldehyde-3-phosphate dehydrogenase, triose-phosphate isomerase, and pyruvate kinase are components of the KATP channel macromolecular complex and regulate its function. J. Biol. Chem. 280, 38464–38470 (2005).
Liu, Q. Y., Corjay, M., Feuerstein, G. Z. & Nambi, P. Identification and characterization of triosephosphate isomerase that specifically interacts with the integrin αIIb cytoplasmic domain. Biochem. Pharmacol. 72, 551–557 (2006).
Estey, T., Piatigorsky, J., Lassen, N. & Vasiliou, V. ALDH3A1: a corneal crystallin with diverse functions. Exp. Eye Res. 84, 3–12 (2007).
Butler, G. S. & Overall, C. M. Updated biological roles for matrix metalloproteinases and new “intracellular” substrates revealed by degradomics. Biochemistry (9 Oct 2009) (doi:10.1021/bi901656f).
Acknowledgements
C.M.O. is supported by a Canada Research Chair in Metalloproteinase Proteomics and Systems Biology. This research was supported by grants from the Canadian Institutes of Health Research, the National Cancer Institute of Canada (with funds raised by the Canadian Cancer Association), a Special Program Grant from the Canadian Breast Cancer Research Alliance Metastasis, and a Centre Grant from the Michael Smith Research Foundation to the University of British Columbia Centre for Blood Research.
Author information
Authors and Affiliations
Related links
Related links
FURTHER INFORMATION
The Overall laboratory homepage
Glossary
- Drug target
-
A molecule that is known to be involved in disease against which a drug is designed to block these pathological actions.
- Pharmacoproteomics
-
The proteomic characterization of the effects of a drug on a system.
- Anti-target
-
A molecule that must be therapeutically spared owing to host-protective effects in a disease, or the blockade of which produces unacceptable side effects.
- Counter-target
-
A molecule that might reduce the efficiency of a drug.
- Off-target
-
A molecule that interacts with a drug but is not the intended target of the drug.
- Shotgun proteomics
-
High-throughput proteomics using high-performance liquid chromatography and mass spectrometry to identify peptides generated from a proteome — for example, by trypsin — from which the protein components are identified.
- Chemical proteomics
-
The identification of small-molecule interactors in a complex sample using affinity capture.
- Target refinement
-
By mapping the network in which a drug target lies, alternative targets at different points in the pathway, which have the desired effect but with greater specificity, may be selected.
- Degradomics
-
The identification of all proteases, their substrates and inhibitors by genomics and proteomics techniques.
- Secretome
-
The portion of the proteome that is secreted.
- Sheddase
-
A protease that releases or 'sheds' all or a portion of a protein to the extracellular milieu.
- Neoproteins
-
Functional proteins that are released from a larger protein by specific proteolysis.
Rights and permissions
About this article
Cite this article
Butler, G., Overall, C. Proteomic identification of multitasking proteins in unexpected locations complicates drug targeting. Nat Rev Drug Discov 8, 935–948 (2009). https://doi.org/10.1038/nrd2945
Issue Date:
DOI: https://doi.org/10.1038/nrd2945
This article is cited by
-
Extracellular vimentin mimics VEGF and is a target for anti-angiogenic immunotherapy
Nature Communications (2022)
-
Monocyte Chemoattractant Protein-1 promotes cancer cell migration via c-Raf/MAPK/AP-1 pathway and MMP-9 production in osteosarcoma
Journal of Experimental & Clinical Cancer Research (2020)
-
TRPV1 translocated to astrocytic membrane to promote migration and inflammatory infiltration thus promotes epilepsy after hypoxic ischemia in immature brain
Journal of Neuroinflammation (2019)
-
Matrix Metalloproteinase-2 Single Nucleotide Polymorphism in Egyptian Non-Hodgkin Lymphoma Patients: Correlation with Clinicopathological Characteristics and Outcome
Indian Journal of Hematology and Blood Transfusion (2019)
-
Secretome profiling of heterotypic spheroids suggests a role of fibroblasts in HIF-1 pathway modulation and colorectal cancer photodynamic resistance
Cellular Oncology (2019)