Key Points
-
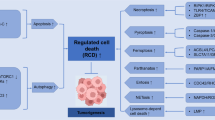
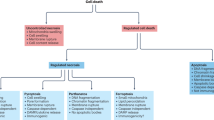
In response to cytotoxic insults, cells die by different subroutines that are mainly based on apoptosis and necrosis and often preceded by a global cytoprotective response centred on autophagy. Thus, cell-based tests that measure cell death-related phenomena must answer two critical questions: do the cells die and, if so, through which lethal pathway do they die?
-
Cells should be considered as dead when they fulfil at least one of the following criteria: the plasma membrane has lost its integrity, the cell has fragmented into apoptotic bodies, or the corpse or its fragments have been taken up by neighbouring cells.
-
Cell death assays fall into two major groups: assays that measure bona fide cell death and tests that quantify biochemical processes that are viewed as surrogate viability markers.
-
Apoptosis assays can be grossly subdivided into two categories: methods that detect events occurring in most (if not all) instances of apoptotic cell death and tests that measure pathway-specific processes, which depend on the apoptosis-initiating stimulus and the precise cellular context of lethal signalling.
-
Autophagy can be quantified by steady-state assessment of the subcellular morphology, long-lived molecule content and the lipidation status of the autophagosomal membrane protein LC3. In addition, flux measurements allow discrimination between efficient and stalled autophagic degradation. Efficient autophagic degradation generally exerts cytoprotective functions, whereas stalled autophagic degradation frequently represents a pathological event.
-
Necrotic cell death can be differentiated from other cell death subroutines by monitoring the kinetics of appearance of common markers, such as disintegration of the plasma membrane.
-
Sophisticated mitotic catastrophe assays are based on fluorescent biosensor cell lines that are assessed by robotized video microscopy and automated image analysis, both of which allow us to monitor several indicators of the nuclear status and establish cell fate profiles.
-
It can be anticipated that the combination of ever more sophisticated high-content and high-throughput microscopy methods with microfluidic devices or on-chip technologies will accelerate the cell-based identification of cell death-modulatory drugs.
Abstract
Cell death has an important role in many human diseases, and strategies aimed at modulating the associated pathways have been successfully applied to treat various disorders. Indeed, several clinically promising cytotoxic and cytoprotective agents with potential applications in cancer, ischaemic and neurodegenerative diseases have recently been identified by high-throughput screening (HTS), based on appropriate cell death assays. Given that different cell death modalities may be dysregulated in different diseases, it is becoming increasingly clear that such assays need to not only quantify the extent of cell death, but they must also be able to distinguish between the various pathways. Here, we systematically describe approaches to accurately quantify distinct cell death pathways, discuss their advantages and pitfalls, and focus on those techniques that are amenable to HTS.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Brown, J. M. & Attardi, L. D. The role of apoptosis in cancer development and treatment response. Nature Rev. Cancer 5, 231–237 (2005).
Degterev, A. et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nature Chem. Biol. 1, 112–119 (2005). Based on HTS of a small-molecule library, this article identifies necrostatin 1 as a specific inhibitor of necroptosis that works through the inhibition of RIPK1.
Kroemer, G. et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 16, 3–11 (2009). This article provides up-to-date guidelines for the use of cell death-related terminology in scientific publications, as provided by the Nomenclature Committee on Cell Death, an organization comprising reputed researchers in the field of cell death worldwide.
Vandenabeele, P., Galluzzi, L., Vanden Berghe, T. & Kroemer, G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nature Rev. Mol. Cell Biol. 11, 700–714 (2010). This review provides the first detailed analysis of the molecular mechanisms that underlie programmed necrosis and its pathophysiological implications.
Levine, B. & Kroemer, G. Autophagy in the pathogenesis of disease. Cell 132, 27–42 (2008).
Grumati, P. et al. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nature Med. 16, 1313–1320 (2010).
Obeid, M. et al. Calreticulin exposure dictates the immunogenicity of cancer cell death. Nature Med. 13, 54–61 (2007).
Zitvogel, L., Kepp, O. & Kroemer, G. Decoding cell death signals in inflammation and immunity. Cell 140, 798–804 (2010). A comprehensive review summarizing the latest insights in the field of immunogenic cell death.
Guidicelli, G. et al. The necrotic signal induced by mycophenolic acid overcomes apoptosis-resistance in tumor cells. PLoS One 4, e5493 (2009).
Galluzzi, L. et al. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 16, 1093–1107 (2009). This review provides a comprehensive set of guidelines for assessing cell death in mammalian cells.
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4, 151–175 (2008). This review provides a comprehensive set of guidelines for measuring autophagy in mammalian cells.
Mizushima, N., Yoshimori, T. & Levine, B. Methods in mammalian autophagy research. Cell 140, 313–326 (2010).
Kerr, J. F. A histochemical study of hypertrophy and ischaemic injury of rat liver with special reference to changes in lysosomes. J. Pathol. Bacteriol. 90, 419–435 (1965).
Kerr, J. F., Wyllie, A. H. & Currie, A. R. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 26, 239–257 (1972).
Castedo, M. et al. Cell death by mitotic catastrophe: a molecular definition. Oncogene 23, 2825–2837 (2004).
Gilmore, A. P. Anoikis. Cell Death Differ. 12 (Suppl. 2), 1473–1477 (2005).
Overholtzer, M. et al. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell 131, 966–979 (2007).
Brennan, M. A. & Cookson, B. T. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol. Microbiol. 38, 31–40 (2000).
Andrabi, S. A., Dawson, T. M. & Dawson, V. L. Mitochondrial and nuclear cross talk in cell death: parthanatos. Ann. NY Acad. Sci. 1147, 233–241 (2008).
Gilloteaux, J. et al. Cancer cell necrosis by autoschizis: synergism of antitumor activity of vitamin C: vitamin K3 on human bladder carcinoma T24 cells. Scanning 20, 564–575 (1998).
Schweichel, J. U. & Merker, H. J. The morphology of various types of cell death in prenatal tissues. Teratology 7, 253–266 (1973).
Kroemer, G. & Levine, B. Autophagic cell death: the story of a misnomer. Nature Rev. Mol. Cell Biol. 9, 1004–1010 (2008).
Berry, D. L. & Baehrecke, E. H. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131, 1137–1148 (2007).
Lemasters, J. J. et al. The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy. Biochim. Biophys. Acta 1366, 177–196 (1998).
Berridge, M. V., Herst, P. M. & Tan, A. S. Tetrazolium dyes as tools in cell biology: new insights into their cellular reduction. Biotechnol. Annu. Rev. 11, 127–152 (2005).
Hitomi, J. et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 135, 1311–1323 (2008). This was the first time systems biology was applied to compare necroptosis to apoptosis in murine cells. Genome-wide RNAi-based screens coupled to in silico and in vitro analyses allowed the delineation of a signalling network that regulates the molecular bifurcation between necroptosis and apoptosis.
Arora, S. et al. RNAi phenotype profiling of kinases identifies potential therapeutic targets in Ewing's sarcoma. Mol. Cancer 9, 218 (2010).
Zeng, F. Y., Cui, J., Liu, L. & Chen, T. PAX3–FKHR sensitizes human alveolar rhabdomyosarcoma cells to camptothecin-mediated growth inhibition and apoptosis. Cancer Lett. 284, 157–164 (2009).
Atienza, J. M., Zhu, J., Wang, X., Xu, X. & Abassi, Y. Dynamic monitoring of cell adhesion and spreading on microelectronic sensor arrays. J. Biomol. Screen. 10, 795–805 (2005).
Solly, K., Wang, X., Xu, X., Strulovici, B. & Zheng, W. Application of real-time cell electronic sensing (RT-CES) technology to cell-based assays. Assay Drug Dev. Technol. 2, 363–372 (2004).
Xing, J. Z. et al. Dynamic monitoring of cytotoxicity on microelectronic sensors. Chem. Res. Toxicol. 18, 154–161 (2005).
Xia, M. et al. Compound cytotoxicity profiling using quantitative high-throughput screening. Environ. Health Perspect. 116, 284–291 (2008).
Metivier, D. et al. Cytofluorometric detection of mitochondrial alterations in early CD95/Fas/APO-1-triggered apoptosis of Jurkat T lymphoma cells. Comparison of seven mitochondrion-specific fluorochromes. Immunol. Lett. 61, 157–163 (1998).
Shiau, A. K., Massari, M. E. & Ozbal, C. C. Back to basics: label-free technologies for small molecule screening. Comb. Chem. High Throughput Screen. 11, 231–237 (2008).
Lipinski, M. M. et al. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev. Cell 18, 1041–1052 (2010). This screen identifies multiple starvation-independent autophagy-relevant genes. This paper is also a good example of the use of secondary HTS to characterize autophagy flux and upstream autophagic pathways.
Neumann, B. et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464, 721–727 (2010).
Buttner, S. et al. Why yeast cells can undergo apoptosis: death in times of peace, love, and war. J. Cell Biol. 175, 521–525 (2006).
Eisenberg, T. et al. Induction of autophagy by spermidine promotes longevity. Nature Cell Biol. 11, 1305–1314 (2009).
Chew, S. K. et al. Genome-wide silencing in Drosophila captures conserved apoptotic effectors. Nature 460, 123–127 (2009).
Stilwell, G. E., Saraswati, S., Littleton, J. T. & Chouinard, S. W. Development of a Drosophila seizure model for in vivo high-throughput drug screening. Eur. J. Neurosci. 24, 2211–2222 (2006).
Evensen, L., Link, W. & Lorens, J. B. Imaged-based high-throughput screening for anti-angiogenic drug discovery. Curr. Pharm. Des. 16, 3958–3963 (2010).
Maiuri, M. C., Zalckvar, E., Kimchi, A. & Kroemer, G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nature Rev. Mol. Cell Biol. 8, 741–752 (2007).
Galluzzi, L. et al. No death without life: vital functions of apoptotic effectors. Cell Death Differ. 15, 1113–1123 (2008).
Yi, C. H. & Yuan, J. The Jekyll and Hyde functions of caspases. Dev. Cell 16, 21–34 (2009).
Cen, H., Mao, F., Aronchik, I., Fuentes, R. J. & Firestone, G. L. DEVD-NucView488: a novel class of enzyme substrates for real-time detection of caspase-3 activity in live cells. FASEB J. 22, 2243–2252 (2008).
Tyas, L., Brophy, V. A., Pope, A., Rivett, A. J. & Tavare, J. M. Rapid caspase-3 activation during apoptosis revealed using fluorescence-resonance energy transfer. EMBO Rep. 1, 266–270 (2000).
Bouchier-Hayes, L. et al. Characterization of cytoplasmic caspase-2 activation by induced proximity. Mol. Cell 35, 830–840 (2009).
Yi, C. H. et al. A genome-wide RNAi screen reveals multiple regulators of caspase activation. J. Cell Biol. 179, 619–626 (2007).
Coleman, M. L. et al. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nature Cell Biol. 3, 339–345 (2001).
Chekeni, F. B. et al. Pannexin 1 channels mediate 'find-me' signal release and membrane permeability during apoptosis. Nature 467, 863–867 (2010).
Martin, S. J. et al. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J. Exp. Med. 182, 1545–1556 (1995).
Zwaal, R. F., Comfurius, P. & Bevers, E. M. Surface exposure of phosphatidylserine in pathological cells. Cell. Mol. Life Sci. 62, 971–988 (2005).
Qu, X. et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 128, 931–946 (2007).
Fischer, K. et al. Antigen recognition induces phosphatidylserine exposure on the cell surface of human CD8+ T cells. Blood 108, 4094–4101 (2006).
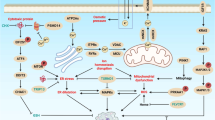
Kroemer, G., Galluzzi, L. & Brenner, C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 87, 99–163 (2007). This review provides a comprehensive analysis of the mitochondrial pathway of cell death and its multifaceted implications in human physiology and pathology.
Galluzzi, L. et al. Methods for the assessment of mitochondrial membrane permeabilization in apoptosis. Apoptosis 12, 803–813 (2007).
Zamzami, N. et al. Sequential reduction of mitochondrial transmembrane potential and generation of reactive oxygen species in early programmed cell death. J. Exp. Med. 182, 367–377 (1995).
Campalans, A., Amouroux, R., Bravard, A., Epe, B. & Radicella, J. P. UVA irradiation induces relocalisation of the DNA repair protein hOGG1 to nuclear speckles. J. Cell Sci. 120, 23–32 (2007).
Lipinski, M. M. et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer's disease. Proc. Natl Acad. Sci. USA 107, 14164–14169 (2010).
Goldstein, J. C., Waterhouse, N. J., Juin, P., Evan, G. I. & Green, D. R. The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nature Cell Biol. 2, 156–162 (2000).
Clarke, M. C., Savill, J., Jones, D. B., Noble, B. S. & Brown, S. B. Compartmentalized megakaryocyte death generates functional platelets committed to caspase-independent death. J. Cell Biol. 160, 577–587 (2003).
Campello, S. & Scorrano, L. Mitochondrial shape changes: orchestrating cell pathophysiology. EMBO Rep. 11, 678–684 (2010).
Jakobs, S. High resolution imaging of live mitochondria. Biochim. Biophys. Acta 1763, 561–575 (2006).
Zoratti, M. & Szabo, I. The mitochondrial permeability transition. Biochim. Biophys. Acta 1241, 139–176 (1995).
Stavrovskaya, I. G. et al. Clinically approved heterocyclics act on a mitochondrial target and reduce stroke-induced pathology. J. Exp. Med. 200, 211–222 (2004).
Lim, T. S., Davila, A., Wallace, D. C. & Burke, P. Assessment of mitochondrial membrane potential using an on-chip microelectrode in a microfluidic device. Lab. Chip 10, 1683–1688 (2010).
Deniaud, A. et al. Endoplasmic reticulum stress induces calcium-dependent permeability transition, mitochondrial outer membrane permeabilization and apoptosis. Oncogene 27, 285–299 (2008).
Liu, X., Kim, C. N., Yang, J., Jemmerson, R. & Wang, X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86, 147–157 (1996).
Zou, H., Henzel, W. J., Liu, X., Lutschg, A. & Wang, X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90, 405–413 (1997).
Hampton, M. B., Zhivotovsky, B., Slater, A. F., Burgess, D. H. & Orrenius, S. Importance of the redox state of cytochrome c during caspase activation in cytosolic extracts. Biochem. J. 329, 95–99 (1998).
Jiang, X. et al. Distinctive roles of PHAP proteins and prothymosin-α in a death regulatory pathway. Science 299, 223–226 (2003).
Kim, H. E., Jiang, X., Du, F. & Wang, X. PHAPI, CAS, and Hsp70 promote apoptosome formation by preventing Apaf-1 aggregation and enhancing nucleotide exchange on Apaf-1. Mol. Cell 30, 239–247 (2008).
Santamaria, B. et al. A nanoconjugate Apaf-1 inhibitor protects mesothelial cells from cytokine-induced injury. PLoS One 4, e6634 (2009).
Hoffman, G. R., Moerke, N. J., Hsia, M., Shamu, C. E. & Blenis, J. A high-throughput, cell-based screening method for siRNA and small molecule inhibitors of mTORC1 signaling using the in cell western technique. Assay Drug Dev. Technol. 8, 186–199 (2010).
MacDonald, M. L. et al. Identifying off-target effects and hidden phenotypes of drugs in human cells. Nature Chem. Biol. 2, 329–337 (2006).
Galluzzi, L. et al. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 70, 1793–1803 (2010).
Sirisoma, N. et al. Discovery of 2-chloro-N-(4-methoxyphenyl)-N-methylquinazolin-4-amine (EP128265, MPI-0441138) as a potent inducer of apoptosis with high in vivo activity. J. Med. Chem. 51, 4771–4779 (2008).
Irshad, S., Mahul-Mellier, A. L., Kassouf, N., Lemarie, A. & Grimm, S. Isolation of ORCTL3 in a novel genetic screen for tumor-specific apoptosis inducers. Cell Death Differ. 16, 890–898 (2009).
Martins, I. et al. Restoration of the immunogenicity of cisplatin-induced cancer cell death by endoplasmic reticulum stress. Oncogene 13 Dec 2010 (doi:10.1038/onc.2010.500).
Madeo, F., Tavernarakis, N. & Kroemer, G. Can autophagy promote longevity? Nature. Cell Biol. 12, 842–846 (2010).
Amaravadi, R. K. et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J. Clin. Invest. 117, 326–336 (2007).
Ni, H. M. et al. Dissecting the dynamic turnover of GFP–LC3 in the autolysosome. Autophagy 7, 54–70 (2011).
Shintani, T. & Klionsky, D. J. Autophagy in health and disease: a double-edged sword. Science 306, 990–995 (2004).
Williams, A. et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nature Chem. Biol. 4, 295–305 (2008).
Zhang, L. et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc. Natl Acad. Sci. USA 104, 19023–19028 (2007).
Criollo, A. et al. The IKK complex contributes to the induction of autophagy. EMBO J. 29, 619–631 (2010).
Farkas, T., Hoyer-Hansen, M. & Jaattela, M. Identification of novel autophagy regulators by a luciferase-based assay for the kinetics of autophagic flux. Autophagy 5, 1018–1025 (2009).
Ju, J. S. et al. Quantitation of selective autophagic protein aggregate degradation in vitro and in vivo using luciferase reporters. Autophagy 5, 511–519 (2009).
Shvets, E., Fass, E. & Elazar, Z. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy 4, 621–628 (2008).
He, P. et al. High-throughput functional screening for autophagy-related genes and identification of TM9SF1 as an autophagosome-inducing gene. Autophagy 5, 52–60 (2009).
Balgi, A. D. et al. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One 4, e7124 (2009).
Sarkar, S. et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nature Chem. Biol. 3, 331–338 (2007). This paper describes a successful small-molecule screen for autophagy in yeast, which identified three conserved novel autophagy enhancers (from a library of 50,729 compounds) that act independently or downstream of the rapamycin target in human cells.
Kanki, T. et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol. Biol. Cell 20, 4730–4738 (2009).
Tian, Y. et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 141, 1042–1055 (2010).
Arsham, A. M. & Neufeld, T. P. A genetic screen in Drosophila reveals novel cytoprotective functions of the autophagy–lysosome pathway. PLoS One 4, e6068 (2009).
Morvan, J. et al. In vitro reconstitution of fusion between immature autophagosomes and endosomes. Autophagy 5, 676–689 (2009).
Koga, H., Kaushik, S. & Cuervo, A. M. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 24, 3052–3065 (2010).
Ichimura, Y. et al. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J. Biol. Chem. 279, 40584–40592 (2004).
Whitehurst, A. W. et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature 446, 815–819 (2007).
Bianchi, M. E., Beltrame, M. & Paonessa, G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science 243, 1056–1059 (1989).
Scaffidi, P., Misteli, T. & Bianchi, M. E. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 418, 191–195 (2002).
Bonaldi, T. et al. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 22, 5551–5560 (2003).
Bell, C. W., Jiang, W., Reich, C. F., & Pisetsky, D. S. The extracellular release of HMGB1 during apoptotic cell death. Am. J. Physiol. Cell Physiol. 291, C1318–C1325 (2006).
Christofferson, D. E. & Yuan, J. Cyclophilin A release as a biomarker of necrotic cell death. Cell Death Differ. 17, 1942–1943 (2010).
Handschumacher, R. E., Harding, M. W., Rice, J., Drugge, R. J. & Speicher, D. W. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science 226, 544–547 (1984).
Li, J. et al. Strategy for discovering chemical inhibitors of human cyclophilin A: focused library design, virtual screening, chemical synthesis and bioassay. J. Comb. Chem. 8, 326–337 (2006).
Ni, S. et al. Discovering potent small molecule inhibitors of cyclophilin A using de novo drug design approach. J. Med. Chem. 52, 5295–5298 (2009).
Kullertz, G., Luthe, S. & Fischer, G. Semiautomated microtiter plate assay for monitoring peptidylprolyl cis/trans isomerase activity in normal and pathological human sera. Clin. Chem. 44, 502–508 (1998).
Mori, T. et al. Use of a real-time fluorescence monitoring system for high-throughput screening for prolyl isomerase inhibitors. J. Biomol. Screen. 14, 419–424 (2009).
Jin, Z. G. et al. Cyclophilin A is a proinflammatory cytokine that activates endothelial cells. Arterioscler. Thromb. Vasc. Biol. 24, 1186–1191 (2004).
Degterev, A. et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nature Chem. Biol. 4, 313–321 (2008).
Christofferson, D. E. & Yuan, J. Necroptosis as an alternative form of programmed cell death. Curr. Opin. Cell Biol. 22, 263–268 (2010).
Kirkegaard, T. et al. Hsp70 stabilizes lysosomes and reverts Niemann-Pick disease-associated lysosomal pathology. Nature 463, 549–553 (2010).
Vakifahmetoglu, H., Olsson, M. & Zhivotovsky, B. Death through a tragedy: mitotic catastrophe. Cell Death Differ. 15, 1153–1162 (2008).
Vitale, I. et al. Multipolar mitosis of tetraploid cells: inhibition by p53 and dependency on Mos. EMBO J. 29, 1272–1284 (2010).
Weaver, B. A. & Cleveland, D. W. Decoding the links between mitosis, cancer, and chemotherapy: the mitotic checkpoint, adaptation, and cell death. Cancer Cell 8, 7–12 (2005).
Olsson, M. et al. DISC-mediated activation of caspase-2 in DNA damage-induced apoptosis. Oncogene 28, 1949–1959 (2009).
Tu, S. et al. In situ trapping of activated initiator caspases reveals a role for caspase-2 in heat shock-induced apoptosis. Nature Cell Biol. 8, 72–77 (2006).
Vakifahmetoglu, H. et al. DNA damage induces two distinct modes of cell death in ovarian carcinomas. Cell Death Differ. 15, 555–566 (2008).
Rello-Varona, S. et al. An automated fluorescence videomicroscopy assay for the detection of mitotic catastrophe. Cell Death Dis. 1, e25 (2010). This was the first approach towards an automated high-throughput image-based assessment of mitotic catastrophe by means of fluorescent biosensors utilized in robotized microscopy.
Hopkins, A. L. & Groom, C. R. The druggable genome. Nature Rev. Drug Discov. 1, 727–730 (2002).
Duan, Z., Choy, E. & Hornicek, F. J. NSC23925, identified in a high-throughput cell-based screen, reverses multidrug resistance. PLoS One 4, e7415 (2009).
Yoon, I. S., Au, Q., Barber, J. R., Ng, S. C. & Zhang, B. Development of a high-throughput screening assay for cytoprotective agents in rotenone-induced cell death. Anal. Biochem. 407, 205–210 (2010).
Muzzey, D. & van Oudenaarden, A. Quantitative time-lapse fluorescence microscopy in single cells. Annu. Rev. Cell Dev. Biol. 25, 301–327 (2009).
Conrad, C. & Gerlich, D. W. Automated microscopy for high-content RNAi screening. J. Cell Biol. 188, 453–461 (2010).
Zhao, L. et al. Analysis of nonadherent apoptotic cells by a quantum dots probe in a microfluidic device for drug screening. Anal. Chem. 81, 7075–7080 (2009).
Malo, N., Hanley, J. A., Cerquozzi, S., Pelletier, J. & Nadon, R. Statistical practice in high-throughput screening data analysis. Nature Biotech. 24, 167–175 (2006).
Shimizu, S. et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nature Cell Biol. 6, 1221–1228 (2004).
Apetoh, L. et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nature Med. 13, 1050–1059 (2007).
Ghiringhelli, F. et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1β-dependent adaptive immunity against tumors. Nature Med. 15, 1170–1178 (2009).
Juhasz, G. & Neufeld, T. P. Autophagy: a forty-year search for a missing membrane source. PLoS Biol. 4, e36 (2006).
Lum, J. J., DeBerardinis, R. J. & Thompson, C. B. Autophagy in metazoans: cell survival in the land of plenty. Nature Rev. Mol. Cell Biol. 6, 439–448 (2005).
Acknowledgements
G.K. is supported by the Ligue Nationale contre le Cancer (Equipes labellisée), the Agence Nationale pour la Recherche (ANR), the European Commission (Active p53, Apo-Sys, ChemoRes, ApopTrain and ArtForce), the Fondation pour la Recherche Médicale (FRM), the Institut National du Cancer (INCa), Cancéropôle Ile-de-France and the AXA Chair for Longevity Research. J.Y. is supported by a US National Institutes of Health Director's Pioneer Award, grants from the National Institute of Aging (USA) and the National Cancer Institute (USA). We are indebted to S. Shen for providing fluorescence microscopy images.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information Table S1
Recent successful high-throughput cell-based assays for the detection of cell death/survival-related variables (PDF 435 kb)
Related links
Glossary
- Apoptotic bodies
-
Membrane-surrounded vesicles that are shed from dying cells during the late stages of apoptosis, and that may include portions of the nucleus and/or seemingly normal organelles.
- Autophagic cell death
-
For a long time, this term has erroneously been used to indicate cell death that manifests with autophagic vacuolization; thus implying that cell death is mediated by rather than accompanied by autophagy.
- Caspases
-
Cysteine proteases that cleave their substrate after an aspartic acid residue. Caspases have a critical role in both the initiation (caspase 2, caspase 8, caspase 9 and caspase10) and execution (caspase 3, caspase 6 and caspase 7) of apoptosis.
- Necroptosis
-
A regulated form of necrosis that requires the catalytic activity of receptor-interacting protein kinase 1 (RIPK1) and RIPK3.
- Anoikis
-
A particular form of apoptosis that is triggered by the detachment of cells from the extracellular matrix.
- Entosis
-
A recently discovered and debated cell death mode in which one cell engulfs one of its live neighbours, which may then die within the phagosome.
- Pyroptosis
-
A pyrogenic cell death modality that is accompanied by the activation of caspase 1 and the secretion of the pro-inflammatory cytokine interleukin-1β. Pyroptosis can manifest with features of apoptosis or necrosis.
- Parthanatos
-
Caspase-independent cell death induced by poly(ADP-ribose) polymerase 1 and mediated by apoptosis-inducing factor.
- Autoschizis
-
A cell death modality that is initiated in certain cancer cells by combinatorial treatment with vitamins C and K3, and leads to the self-excision of cytoplasmic portions.
- Intrinsic apoptosis
-
Intracellular stress is sensed by mitochondria, which undergo mitochondrial membrane permeabilization and hence activate caspase-dependent and caspase-independent cell death executioner mechanisms.
- Extrinsic apoptosis
-
Apoptosis can be initiated by extracellular molecules that bind to 'death' receptors (for example, FAS), in turn activating the caspase 8–caspase 3 cascade.
- Necrostatin 1
-
The tryptophan-based molecule 5-(1H-indol-3-ylmethyl)-3-methyl-2-thioxo-4-imidazolidi-none that was first identified as a specific and potent inhibitor of necroptosis.
- Micronucleation and multinucleation
-
The presence of multiple nuclei often derives from mitotic problems — for instance, from chromosomes or chromosomal fragments that have not been evenly distributed at anaphase — or from deficient cytokinesis.
Rights and permissions
About this article
Cite this article
Kepp, O., Galluzzi, L., Lipinski, M. et al. Cell death assays for drug discovery. Nat Rev Drug Discov 10, 221–237 (2011). https://doi.org/10.1038/nrd3373
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd3373