Key Points
-
As a result of ongoing high-risk behaviours, people who inject drugs (PWID) might commonly harbour mixed HCV infections and are at risk of reinfection following spontaneous or treatment-induced clearance
-
The sensitivity of assays used to screen for mixed infection in individual samples varies widely, particularly in their capacity to detect minor variants
-
Mixed HCV infection among populations of PWID ranges from 14% to 39% when sensitive methods are used for detection and characterization
-
Mixed infection has been shown to compromise interferon-based HCV therapy, but few studies are available to adequately assess how often this occurs
-
HCV reinfection rates after successful interferon-based treatment among PWID are 0–5 cases per 100 person-years, but studies are limited by small sample sizes and heterogeneity in injecting risk after treatment
-
Future studies are needed to evaluate the impact of mixed HCV infection and reinfection following treatment with direct acting antiviral HCV therapies among PWID
Abstract
The majority of new and existing cases of HCV infection in high-income countries occur among people who inject drugs (PWID). Ongoing high-risk behaviours can lead to HCV re-exposure, resulting in mixed HCV infection and reinfection. Assays used to screen for mixed infection vary widely in sensitivity, particularly with respect to their capacity for detecting minor variants (<20% of the viral population). The prevalence of mixed infection among PWID ranges from 14% to 39% when sensitive assays are used. Mixed infection compromises HCV treatment outcomes with interferon-based regimens. HCV reinfection can also occur after successful interferon-based treatment among PWID, but the rate of reinfection is low (0–5 cases per 100 person-years). A revolution in HCV therapeutic development has occurred in the past few years, with the advent of interferon-free, but still genotype-specific regiments based on direct acting antiviral agents. However, little is known about whether mixed infection and reinfection has an effect on HCV treatment outcomes in the setting of new direct-acting antiviral agents. This Review characterizes the epidemiology and natural history of mixed infection and reinfection among PWID, methodologies for detection, the potential implications for HCV treatment and considerations for the design of future studies.
Similar content being viewed by others
Introduction
People who inject drugs (PWID) comprise the majority of new and existing patients with HCV infection in many high-income countries.1,2,3 HCV transmission is driven by a high incidence of infection among PWID, particularly among young PWID.4,5,6 The majority of those infected fail to clear the virus and also fail to develop protective immunity.7,8,9,10 As such, PWID are likely to be exposed to multiple HCV infections as a result of ongoing high-risk behaviours and might commonly harbour mixed HCV infections (that is, infection with two or more distinct viruses).11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26 PWID are also at risk of reinfection (clearance of initial virus followed by infection with a second genetically distinct virus) after spontaneous7,23,24,25,27,28,29,30,31,32,33,34 or treatment-induced clearance.21,22,23,24,25,27,35,36,37,38,39,40,41,42,43,44
The characterization of multiple infections, including the prevalence, incidence and natural history, has implications for HCV immunopathogenesis, vaccine development and therapeutic strategies. Advances in HCV treatment with the development of simple, tolerable and highly effective direct-acting antiviral (DAA) oral regimens, provides the foundation for markedly broadened treatment access. However, high pricing has led to restrictions on access for PWID, owing to concerns of suboptimal treatment adherence and HCV reinfection risk related to ongoing high-risk behaviours. Underlying mixed HCV infection can contribute to nonresponse during therapy,45,46,47,48 which has implications for DAA regimens that are preferentially active against specific viral genotypes or subtypes.44 This Review characterizes the epidemiology and natural history of mixed HCV infection and reinfection among PWID, methodologies for detection, the potential implications for HCV treatment and important considerations for the design of future studies in this area of research.
HCV diversity and genotypes
HCV is a member of the Flaviviridae family. The single positive RNA strand is ∼10 kb and encodes a polyprotein of ∼3,000 amino acids. HCV is highly efficient at replication, with an estimated daily reproduction of 1012 new virions. The key enzyme for replication, the RNA-dependent RNA polymerase, which is encoded by NS5B, lacks a proofreading function,49 resulting in the introduction of at least one mutation per replicative cycle.50 This error-prone replicase leads to the development of a diverse and continuously evolving population of viruses with variations in the genome moulded by host and virus selective pressures.51 Recombination between divergent HCV strains (defined as viruses of distinct genotypes or subtypes)—which also contributes to viral diversity and confers the ability to rapidly adapt and escape the selection pressures of host immune responses and antiviral therapy—has also been demonstrated, albeit uncommonly.52,53,54,55,56,57 Similarly, across ethnically and geographically diverse populations, the constant evolution of HCV has led to considerable global genetic diversity.
The different HCV genotypes and subtypes demonstrate a complex global geographic distribution, reflecting transmission patterns and ethnic variability.58 HCV genotypes 1–7, each with multiple subtypes (for example 1a, 1b), have been identified on the basis of phylogenetic analyses of HCV RNA sequences.59 The diversity of HCV across the entire genome is considerable, with >30% nucleotide difference between HCV genotypes, 15–30% nucleotide difference among HCV subtypes and up to 13% of the viral population evolving over years to decades from a common ancestor within a single host.59 Worldwide, it is estimated that HCV genotype 1 is the most prevalent genotype (46% of all patients infected with HCV), followed by genotype 3 (30%), genotypes 2, 4 and 6 (23% collectively), and genotype 5 and 7 (<1%).60
Phylogenetic analyses examining evolutionary relatedness among viral sequences have been used to reconstruct the history of sampled viral strains.58 The majority of global infections comprise subtypes 1a or 1b, 2a or 2b and 3a.61,62 The transfusion of contaminated blood products is particularly associated with HCV genotype 1b and 2a infections61,63 and HCV genotypes 1a and 3a are most common among PWID.61,64,65,66,67,68,69,70,71,72,73 Blood product screening over the past two decades or more has led to a decrease in the prevalence of HCV genotypes 1b and 2a. During the same period, genotypes 1a and 3a have increased in prevalence, particularly among PWID.61,64,67,68,69,70,71,72,73 In addition, a group of endemic HCV strains exist, such as genotype 6 in southeast Asia and genotype 5 in Africa, which have a more restricted geographic distribution and are less prevalent than HCV genotypes 1, 2 or 3.62 Finally, the last major group of strains reflect local epidemics and are highly prevalent in specific locations and risk groups, such as genotype 4d, which >10% of the Egyptian population are infected with (initially via contaminated medical devices)74 and is also prevalent globally among HIV and HCV co-infected men who have sex with men.75
Defining mixed HCV infection
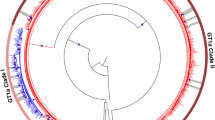
Given the general lack of protective immunity associated with primary infection, ongoing high-risk behaviours can lead to HCV re-exposure and multiple infection episodes.13,21,22,23,24,25 The detection of multiple genetically distinct HCV strains at a single time-point is often referred to as mixed HCV infection (Figure 1a, 2a, 2b). Mixed HCV infection can be the result of either HCV co-infection or superinfection. Co-infection refers to simultaneous infection with two or more distinct HCV strains. Superinfection refers to infection with an additional HCV strain following persistent infection with an initial single or mixed HCV infection (Figure 1a).22,76 Among individuals with mixed HCV infection detected at a single time-point, it is often difficult to distinguish between HCV superinfection and co-infection. The difficulty in making the distinction is owing to an absence of systematic screening for mixed HCV infection (particularly at an early time-point during infection) and often infrequent sampling intervals. Furthermore, the diagnostic genotyping assays and standard sequencing protocols used to detect mixed HCV infection have a poor level of sensitivity. Given that ongoing injection drug use has been associated with reinfection and superinfection,23 the likelihood of detecting mixed HCV infection is probably higher among those with ongoing injection drug use than those who do not participate in these high-risk behaviours. However, it is unknown whether mixed HCV infections can persist for long periods of time, as HCV does not integrate into the host genome and the virions have a short half-life. However, studies have demonstrated that therapy-resistant HCV variants can persist for up to 6 months following the cessation of therapy.77,78
a | Mixed HCV infection can be the result of either HCV co-infection (that is, simultaneous infection with ≥2 distinct HCV strains) or superinfection (that is, infection with a second HCV strain following persistent infection with an initial single HCV strain). Genotype-specific treatment might only supress the treatment-sensitive strains and result in persistence of a treatment-insensitive strain. Alternatively, treatment could clear all strains present. b | Reinfection, after treatment-induced clearance, is the detection of an HCV strain, which was heterologous to that detected before recurrence. Colours represent different viral strains, sizes represent relative abundance of the virus and grey areas represent treatment.
a | Viral persistence of treatment-insensitive strains in individuals with detectable mixed HCV infection. b | Viral persistence of minority, treatment-insensitive strains in individuals with undetectable mixed HCV infection. c | Relapse. d | Reinfection. Pie charts represent the proportion of each HCV strain to the total population. The dotted line represents the lowest concentration of virus that can be detected and the grey box indicates treatment.
Ongoing risk behaviours can also lead to reinfection after spontaneous7,23,24,25,27,28,29,30,31,32,33,34 or treatment-induced HCV clearance.21,22,23,24,25,27,35,36,37,38,39,40,41,42,43,44 The detection of HCV RNA following a period of aviraemia (defined by one or more negative test results for HCV RNA) is often referred to as viral recurrence. Among patients with recurrence, sequencing can be used to determine whether the recurrence is due to viral relapse, which is evidenced by the detection of an HCV strain similar (or homologous) to that detected before recurrence (Figures 1b, 2c), or reinfection, which is evidenced by the detection of an HCV strain substantially different (or heterologous) to that detected before recurrence (Figures 1b, 2d). However, among individuals with apparent viral relapse, excluding the possibility that reinfection has occurred from the same source as the initial infection is difficult. Furthermore, given the poor sensitivity of available methods to detect mixed HCV infections, excluding the possibility of viral relapse with a previously undetected, treatment-insensitive strain that was present before the period of aviraemia is difficult (Figure 2b).47
Methods of detection
Several methods are available for detecting mixed HCV infection, including: real-time subtype-specific nested reverse transcriptase PCR (nRT-PCR) assay sets; heteroduplex mobility assays (HMA); restriction fragment length polymorphism (RFLP) analysis; Sanger sequencing of nRT-PCR amplicons or molecular clones; and next-generation sequencing (NGS; Figure 3). Each of these methods have improved the ability to detect mixed HCV infections containing heterologous strains in comparison with the commercially available line-probe assay, which underestimates the true prevalence of mixed HCV infection and is often unable to detect multiple infections of the same subtype.11,26,79,80,81 The selection of a suitable mixed HCV infection screening methodology should consider the amplicon region, the genotype or subtype specificity, the capacity for quantitation and the lowest concentration of virus that can be detected.
All methods incorporate a prior RT-PCR step and hence are prone to failure of amplification, PCR errors and preferential amplification. a | Real-time nRT-PCR, a semi-quantitative method that is specific for genotypes 1a, 1b, 2a and 3a.25 b | HMA, a relatively insensitive, qualitative method relying on the formation of imperfect dimers between different oligonucleotide strands. HMA is specific for genotypes 1a, 1b, 2a/c, 2b, 3a, 3b, 4a and 6a.11,19 c | RFLP, a relatively insensitive, qualitative method that requires a priori knowledge of sequence variations and is specific for genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5 and 6.82,148 d | Sanger sequencing, a qualitative method, which is specific for genotypes 1a, 1b, 2a, 2b, 3a, 4a, 4d, 5a and 6a.15,48,46,107 e | Clonal Sanger sequencing, a quantitative method, which is specific for genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a.24,149 f | NGS, a quantitative method, which can detect all genotypes.47,150 Abbreviations: E, envelope; HMA, heteroduplex mobility assay; HVR, highly variable region; NGS, next-generation sequencing; nRT-PCR, nested reverse transcriptase PCR; NS, nonstructural protein; RFLP, restriction fragment length polymorphism; UTR, untranslated region.
Assays
Mixed HCV infection assays reported to date have targeted a variety of different regions of the genome, including: the 5′ untranslated region (5′UTR); the region encoding proteins that form the capsid (core); the region encoding the envelope glycoproteins, containing the highly variable region (E1-HVR); and the region encoding the RNA polymerase (NS5B). The diversity within each of these regions of the viral RNA genome influences the ability to detect minor variations between genotypes and subtypes.24,25,47,48,79,82 The high degree of genetic variability within the E1-HVR1 region, for example, provides sufficient diversity to distinguish between genotypes, subtypes and variants within subtypes.83 The more limited genetic variation in NS5B might be insufficient to distinguish subtype variants. The requirement for a qualitative or quantitative mixed HCV infection assay should be guided by the aim of the study. A qualitative assay that simply designates the presence or absence of mixed HCV infection might be sufficient for large studies assessing the prevalence of mixed infection. By contrast, a quantitative assay is more suitable for measuring the relative viral load dynamics over time in studies assessing the implications of mixed HCV infection and reinfection on treatment outcomes or disease natural history. However, even the most sensitive NGS methods for detecting mixed HCV infection might become technically impossible if the minor variant comprises <1% of the total viral population.
Limitations
In addition to the design features of each assay, inherent limitations are also important to consider. All current methods to detect mixed HCV infection start with PCR (usually nRT-PCR to generate an adequate signal) followed by hybridization of PCR products, real time nRT-PCR, HMA, RFLP, Sanger sequencing, molecular cloning, or NGS. Although RT-PCR has the benefit of being able to detect HCV at a low concentration, all RT-PCR methods are highly dependent on the primer choice during reverse transcription (unless random hexamers are used) and the initial PCR round—during which highly conserved regions should be targeted to enable amplification of all genotypes and subtypes. Therefore, current methods are all equally prone to failure of amplification (if the primers do not adequately match the viral target sequence [Figure 3]), PCR errors and preferential amplification of one viral strain. The latter might lead to the inadvertent selection of a nonrepresentative population of amplicons during the reverse transcription and/or PCR amplification processes.
Sequencing methods
Sequencing-based methods are increasingly used because they have the best ability to reliably differentiate between genetically distinct variants within HCV subtypes. Sanger sequencing the nRT-PCR amplicon (Figure 3d) offers the advantages of low cost, but has the key disadvantage of poor sensitivity in detection of minor variants84 and ambiguities are frequently evident in the nucleotide call of mixed bases within the composite sequence. Molecular cloning (Figure 3e) is highly labour intensive and hence costly, requiring the analysis of a large number of clones (generally >20) to provide adequate representation of minor variants.
NGS technologies (Figure 3f) offer high throughput analysis of a population of complex, highly variable sequences and thus a new opportunity to further characterize the dynamics of mixed HCV infection within an individual over time. Although NGS provides many potential advantages to clinical virology, including quantitation and the ability to detect low frequency variants, the absence of a standardized sample preparation and data analysis pipeline mandates careful consideration of many technical aspects to determine a sample's viral diversity.85,86,87 Multiple NGS platforms and chemistries are now available and the choice of workflow practices, including sample preparation and the genomic region analysed (targeted or whole genome), can influence the interpretation of results.88,89
In addition, a number of complex bioinformatics analysis algorithms are available, which are likely to influence the results.90,91 All algorithms use different strategies to filter, trim, error-correct, align and assemble sequences in order to estimate viral diversity and reconstruct individual haplotypes (or viral strains) within a patient sample.92 Given that the sequence reads are far shorter than the 9 kb genome, using the algorithms to reconstruct a haplotype (or viral strain) is an intrinsically probabilistic approach; therefore, strategies that can sequence viral populations without haplotype reconstruction (for example, full genome sequencing) should be sought.93 All algorithms require extensive validation to ensure data integrity, which is of course inherently difficult considering the rapidly emerging technology. Likewise, the capacity of NGS for high coverage and the ability to detect very low concentrations of virus requires well-designed procedures to minimize the risk of cross-contamination. The development of validated and robust NGS methodologies capable of automatically and accurately quantifying viral populations is required to determine the capacity for this technology in the analysis of mixed HCV infection and interpretation of its clinical importance. Full-length PCR amplification of the viral population, independent of the target sequences, accompanied by high throughput, deep-coverage NGS is the next frontier in the evolution of this technology.
The use of well validated and sensitive methods to detect mixed HCV infection influences the reliable identification of superinfection and reinfection. To conclude that an individual has demonstrated superinfection over the course of their follow-up, the absence of mixed HCV infection in previous samples needs to be confirmed to ensure the putatively observed superinfection is not the result of the emerging dominance of a pre-existing minor variant (Figure 2a). However, current methods can only reliably confirm the presence, and not the absence, of mixed HCV infection. A switch in the dominant strain has been demonstrated in many different populations of patients, including patients undergoing haemodialysis,94 blood transfusion recipients,95 PWID13,23 and patients with haemophilia.96,97 In the context of treatment, as Abdelrahman et al.47 demonstrated, a high degree of assay sensitivity is necessary to determine the cause of treatment failure and to classify a patient with viral recurrence as having relapsed owing to mixed HCV infection or a reinfection. In addition to high assay sensitivity, analysis of a region with adequate genetic diversity, such as found within the core-E2 region used by Pham et al.,25 is required to determine whether viral recurrence is due to relapse or reinfection.
Epidemiology
Mixed HCV infection
Mixed HCV infection has been reported in multiple case reports in humans95,98,99,100,101,102,103 and has been recapitulated experimentally in chimpanzees.104 Among PWID, the reported prevalence of mixed HCV infection ranges from 0% to 39%.11,14,15,16,18,19,24,25,105,106,107,108 Indirect evidence of mixed HCV infection has also been provided from the identification of genomic recombination of HCV in patient blood samples, which can confer drug-resistance, implying the coexistence of two strains within a single cell.54,55,56,57 Although genetic recombination seems to be rare, the rate of recombination might be underestimated, as sensitive detection methods that require amplification of at least two distant genomic regions are not commonly used. The considerable variation in the prevalence of mixed HCV infection between studies is probably related to a number of factors, including differences in study design (for example, frequency and duration of study follow-up), study populations (age, sex and high-risk behaviours such as drug injection) and the sensitivity of the laboratory methodologies used for detection. However, given the inadequate reporting of key data, comparing these studies is difficult (Table 1).
Initial studies using line-probe genotyping assays such as INNO-LiPA® (Fujirebio Europe, Ghent, Belgium) reported a prevalence of mixed HCV infection ranging from 1% to 5% among PWID populations (Table 1).11,16,18,105 However, given the poor sensitivity of this assay for the detection of minor variants11,109 and the inability of this assay to detect mixed HCV infections of the same subtype, it is clear these data underestimate the true prevalence of mixed HCV infection. For instance, Forns et al.79 compared the prevalence of HCV genotypes using multiple methods among people who had received blood transfusions, who had injected drugs or who had other unknown risk factors for exposure to HCV (n = 151). Overall, a 4% (6 of 151) prevalence of mixed HCV infection was found using line-probe genotyping, compared with 19% (28 of 151) using subtype-specific nRT-PCR.79
Subsequent studies have focused on the design of assays to improve their sensitivity for detecting mixed HCV infection, including nRT-PCR followed by Sanger sequencing of the amplicon. The prevalence of mixed HCV infection among PWID ranges from 14% to 39% when sensitive assays are used (Table 1).13,24,25 In one study of 25 PWID in San Francisco, USA, Herring et al.13 observed that 20% of participants had evidence of mixed HCV infection when using Sanger (consensus) sequencing of the E1-HVR1 region. However, a limitation of Sanger sequencing is that HCV strains present at a low prevalence (generally <20%) cannot be reliably detected. Molecular cloning followed by Sanger sequencing can be used to improve the successful sampling of viral populations present at low frequencies but is rarely used owing to cost and time constraints and the advent of high-throughput sequencing methods. For instance, van de Laar et al.24 used cloning and sequencing of NS5B region amplicons to assess 59 PWID in Amsterdam, Netherlands, and demonstrated a 39% prevalence of mixed HCV infection.
A sensitive, real-time based, subtype-specific, nRT-PCR assay was developed by Pham and colleagues to separately detect the presence of specific HCV subtypes, including 1a, 1b, 2a and 3a, by amplifying the core region.25 The protocol was shown to have a sensitivity of 100 copies per reaction with no crossreactivity detected, even in the presence of 106 copies of alternate serum-derived HCV subtype RNA molecules in the reaction.25 A group of 87 PWID recruited from prisons in New South Wales, Australia, demonstrated a mixed HCV infection prevalence of 25% using this assay.25
Although more data are required, the application of NGS after initial nRT-PCR amplification might improve reliability of subtype determination and increase the sensitivity for detecting mixed HCV infection. In a study of 114 clinical specimens from individuals with chronic HCV infection previously tested by line-probe assay (82 designated as genotype 1 and 32 with uninterpretable results), phylogenetic analysis of NGS reads matched subtype 1 determination by Sanger sequencing (69% genotype 1b, 31% genotype 1a) in 81 specimens and identified a mixed subtype infection (genotypes 1b, 3a and 1a) in one sample.81 Among the 32 previously indeterminate specimens, NGS genotype and subtype results were obtained in all samples, four of which had mixed HCV infection.81
Superinfection and co-infection
HCV superinfection is commonly seen in patients with thalassaemia who have had multiple blood transfusions and patients on renal dialysis.12,110,111,112,113 Several observational cohort studies of PWID with continuing risk behaviours for HCV exposure have demonstrated that the proportion of patients with HCV superinfection during follow-up is in the range of 16–24% (Table 2).13,23,24,25 However, no studies have reported the incidence of HCV superinfection. The comparison between studies is difficult because of the variation in the duration of follow-up (1–7 years) and the sensitivity of the different assays used (Table 2).
After HCV superinfection, elevations in alanine aminotransferase (ALT) levels and HCV RNA levels have been observed.23,100,101 In one study of people with recent HCV infection and superinfection from Australia, 83% of individuals (five of six) with superinfection demonstrated increases in ALT and HCV RNA levels.23 Although it is unknown whether elevations in ALT and/or HCV RNA following superinfection have an influence on the pathogenesis of HCV infection, regular monitoring of ALT and HCV RNA levels among participants at high-risk might provide a clinically useful tool for detecting superinfection.
Virological outcomes following HCV superinfection are heterogeneous, with some individuals demonstrating persistence of multiple infections, clearance of one infection (which might be represented by a 'switch' in HCV genotypes or subtypes) or clearance of all infections.13,23,24,25 Among individuals who demonstrate apparent clearance of one infection and persistence of a second infection, it is also possible that the persistent strain out-competes the other, but the latter remains within the viral population at very low levels. As such, it is important that sensitive detection methods are used for studies of superinfection.
Although several studies have identified potential patients with HCV co-infection,13,23,24,25 demonstrating true co-infection is difficult. The problem is that one cannot exclude the possibility that sequential infections at short time intervals before detection of the co-infection occurred. HCV co-infection identified early during acute HCV infection provides greater certainty that 'true' primary co-infection has been detected;23,25 however, the possibility of superinfection cannot be excluded given that the probability of transmission per injection event has been estimated at 0.6% (95% CI 0.3–1.1%) among PWID114 and more than once daily injection drug use is common.
HCV reinfection
Among patients infected with HCV, spontaneous clearance occurs in around 25%,29 with reduced clearance rates associated with male sex, unfavourable IFNL4 genotype, non-genotype 1 HCV infection and HIV co-infection.8,115 As reviewed elsewhere,8 HCV reinfection after spontaneous clearance can occur among PWID, with rates of HCV reinfection ranging between 1.8 and 46.7 cases per 100 person-years. Differences in study design, statistical analyses, variation in demographic factors, risk behaviours related to injection drug use and presence of viral co-infections make comparisons across studies difficult.8 These studies clearly show that HCV infection in humans generally does not confer full protective immunity against reinfection. However, the evidence also indicates that HCV RNA levels following reinfection are lower and generally transient in comparison with the initial infection in patients with spontaneous clearance; this evidence suggests that prior HCV infection with clearance alters the natural history of HCV infection following re-exposure, providing evidence for at least some protective HCV immunity.8,116
Treatment of HCV infection
For the past two decades, treatment of chronic HCV infection has been interferon-based.117 Innovations in drug discovery led first to the development of combination therapy with ribavirin,44 PEG-IFN118 and more recently DAAs (for example, telaprevir and boceprevir, which inhibit the NS3–NS4A protease),119,120 providing stepwise improvements in sustained virologic response (SVR; which equates with cure). However, the poor tolerability of PEG-IFN, ribavirin, telaprevir and boceprevir have limited global treatment uptake.121 A revolution in HCV therapeutic development has occurred in the past few years, with the advent of interferon-free, DAA-based regimens.122 Within a few more years, simple (single, daily dosing oral regimens), tolerable, short-duration (6–12 weeks) therapy with extremely high efficacy (cure rates >90%) across all genotypes is likely to be the norm.
Although HCV treatment is improving, many currently available DAA agents are fairly genotype-specific and/or subtype-specific. Given that individuals with HCV genotype 1 demonstrate poor responses to interferon-based therapies, rational drug design has focused on the development of agents with potent activity against this genotype. Although this strategy has led to SVR rates of >90% among patients with genotype 1,123,124,125,126,127,128,129 depending on the agent, many DAA-based therapies have reduced activity against other genotypes.44,119,120,130,131,132,133,134,135,136 As >50% of the global population with HCV infection is infected with non-genotype 1 HCV, this reduced efficacy is problematic.60 For example, HCV genotype 3a infection is highly prevalent among populations of PWID globally and its prevalence has continued to increase over time.64,65,67,68,69,70,71,72,73
Interferon-free, pan-genotypic DAA combinations in development look very promising.122,137,138,139 The co-formulated sofosbuvir and GS-5816 single daily tablet, 12 week regimen (a nucleotide analogue and NS5A inhibitor, respectively), which is in phase II studies, has demonstrated a SVR rate of >90% for HCV genotypes 1, 2, 3, 4, and 6.137,138 Responses of >90% have been observed among treatment-naive patients receiving 12 weeks of sofosbuvir plus daclatasvir.139 However, the reality is that many HCV treatments will still be genotype-specific for the next 1–2 years. In addition, even within the genotype-specific DAA regimens, response rates vary between subtypes; for instance, the efficacy of the protease inhibitor simeprevir might be reduced in the presence of the naturally occurring Q80K resistance-association variation, which is found in subtype 1a.140 As such, the presence of mixed HCV infection before treatment initiation could be of concern. Furthermore, as HCV therapy is broadened to populations at increased risk of HCV re-exposure (for example, active PWID and HIV co-infected men who have sex with men), the risk of reinfection could increase. As such, it is important to consider both mixed infection and reinfection in the context of HCV treatment in the era of new DAA therapies.
Implications of mixed HCV infection for treatment
Data on the effect of mixed HCV infection on treatment outcomes are limited. In a study by Grebely et al.,23 one person who injected drugs who had acute HCV genotype 2b infection demonstrated HCV superinfection with genotype 1a during PEG-IFN monotherapy. The genotype 2a infection was cleared but the genotype 1a HCV persisted during and after therapy (Figure 4).23 Following the first course of unsuccessful therapy, this individual had a second superinfection with HCV genotype 3a before re-treatment with PEG-IFN plus ribavirin, after which the HCV genotype 3a infection was cleared but the genotype 1a infection persisted.23 In another report by Schröter et al.,46 an individual with HCV genotype 3a infection diagnosed by line-probe assay received interferon-based treatment, which led to the successful clearance of the infection; however, a previously unsuspected mixed infection with genotype 1a at baseline was found to have persisted.46 Another study included men who have sex with men, who had HIV and HCV co-infection and did not respond to PEG-IFN plus ribavirin therapy for acute HCV infection (n = 15). In this study, 100% of participants had evidence of mixed HCV infection in the pretreatment sample when NGS was performed, including mixed genotype 1a infections (15 of 15) and mixed subtype or genotype infections (6 of 15).47 Given the high prevalence of mixed HCV infection among PWID, further data are needed to assess the effect of mixed infection on response to HCV treatment among PWID.
The patient was first confirmed as having a genotype 2b HCV infection. After commencement of PEG-IFN therapy, genotype 2b was eradicated, but the patient had a persistent genotype 1a infection. Between consecutive treatments (firstly PEG-IFN then PEG-IFN plus RBV with ∼2 years between treatments), the individual became superinfected with genotype 3a and retained the mixed genotype 1a and 3a infection until PEG-IFN plus RBV therapy, at which time genotype 3a was cleared but genotype 1a remained persistent. Blue squares represent ALT levels and green circles represent total HCV RNA levels. The grey boxes represent treatments. Abbreviations: ALT, alanine aminotransferase; RBV, ribavirin. Permission obtained from John Wiley and Sons © Grebely, J. et al. Hepatology 55, 1058–1069 (2012).
Implications of HCV reinfection following treatment
Ongoing high-risk behaviours following HCV treatment-induced clearance might lead to reinfection and compromised treatment outcomes.21,22,23,24,25,27,35,36,37,38,39,40,41,42,43,44 Among patients with a treatment-induced period of aviraemia followed by HCV recurrence, nRT-PCR and sequencing can be used to determine whether the recurring virus is due to viral relapse (Figure 2c) or reinfection (Figure 2d).
Reinfection after treatment-induced HCV clearance was first described in case studies among PWID.141,142,143 To date, the reported incidence of HCV reinfection following successful treatment-induced HCV clearance among PWID is low, ranging from 0.0 to 5.3 cases per 100 person-years (Table 3).21,22,23,24,25,27,35,36,37,38,39,40,41,144 However, among individuals reporting ongoing injection drug use after successful treatment, the incidence of reinfection is higher, ranging from 1.8 to 33.0 cases per 100 person-years (Table 3).21,22,23,24,25,27,35,36,38,39,40,41,144 In a systematic review and meta-analysis of HCV reinfection among PWID, the pooled estimate of reinfection was 2.2 cases per 100 person-years (95% CI, 0.9–6.1) overall and 6.4 cases per 100 person-years (95% CI, 2.5–16.7) among individuals who reported injection drug use after treatment-induced HCV clearance.145 In a further meta-analysis performed by Hill et al.,146 the proportion of patients with HCV reinfection after 5 years was 1% in low-risk populations, 8% in PWID or prisoners and 24% in those with HIV co-infection.
Studies of reinfection after successful HCV treatment are limited by small sample sizes, retrospective study designs, incomplete longitudinal follow-up and a lack of sensitive methods to detect low levels of coexisting variants in pretreatment samples. As DAA-based therapy with improved efficacy and tolerability is expanded to populations at increased risk of re-exposure, further data are needed to assess reinfection following successful treatment among PWID. Reinfection leads to a reduced proportion of the PWID population with overall cure; therefore, it is incredibly important to understand how often HCV reinfection occurs. HCV reinfection has important cost implications with regards treatment and would affect our ability to control HCV infection among the PWID population; the rate at which chronic infections are eliminated through treatment would be reduced if people are becoming reinfected soon after successful treatment.
Clinical implications and recommendations
Among people with a history of injection drug use, HCV genotyping should be performed before the initiation of HCV treatment using the most sensitive clinical genotyping assay available (most often second generation line-probe assays, for example, INNO-LiPA® HCV 2.0 or Sanger sequencing). Although it is acknowledged that line-probe and Sanger sequencing assays might not be sensitive enough for the detection of low-level mixed viral variants, this drawback must be balanced against the practicality that more sensitive methods might not be available for use in clinical situations (for example, cloning or NGS).
In people with evidence of mixed HCV infection at baseline, genotype-specific treatment regimens should be avoided owing to potential inactivity against other strains and pan-genotype regimens should be considered in these patients. Among patients with a history of injection drug use and null-response to treatment, HCV genotyping at the end of therapy could be used to assess whether mixed infection at baseline, or superinfection during therapy, contributed to this treatment outcome (evidenced by a genotype switch from baseline compared to the end of therapy). Among patients with a history of injection drug use and viral breakthrough during therapy, HCV genotyping should be performed to assess whether viral breakthrough was likely to be due to viral relapse or HCV reinfection (useful in instances of reinfection with a different genotype or subtype). In the case of relapse with the same subtype as initial infection, Sanger sequencing (preferably of the E1-HVR1 region) would be required to distinguish relapse from reinfection; however, Sanger sequencing cannot exclude the possibility of reinfection from the same partner in the case of viral relapse. More sophisticated methods for distinguishing relapse from reinfection in a research setting could also include a comparison between the rate of viral evolution in the host and the presumed source.147
After successful treatment (undetectable HCV RNA at 12 weeks or 24 weeks after treatment cessation), patients should be tested for HCV RNA using a sensitive commercial assay. If HCV RNA is detectable, HCV genotyping should be performed to assess whether viral recurrence was probably due to viral relapse or HCV reinfection. In patients with a relapse from the same subtype as the initial infection, Sanger sequencing is likely to be required to distinguish relapse from reinfection. Among people with ongoing risk behaviours for HCV exposure and a successful treatment response, HCV RNA testing should be performed annually to assess for HCV reinfection.
Future studies
Investigations into the prevalence, incidence and natural history of mixed infection (both superinfection and co-infection) and reinfection, including their effect on HCV treatment, could provide crucial insights into HCV immunopathogenesis, vaccine development and therapeutic strategies for PWID infected with HCV. Understanding the effect of mixed infection on treatment response and the long-term effect of HCV reinfection among PWID will be crucial, given the expanded access to treatment in this population.
Recommendations for the ideal study to improve understanding of mixed HCV infection, superinfection and reinfection during and after HCV treatment should include people with a history of injection drug use (particularly those with recent drug use). All patients should undergo HCV RNA testing to confirm chronic infection. Sensitive assays to detect and characterize mixed HCV infection, for example, genotype-specific nRT-PCR or pan-genotypic nRT-PCR with NGS, would be performed at baseline and subsequently on all viraemic samples taken longitudinally. Ideally, the most sensitive assays, such as NGS, should be used to effectively assess the dynamics of persistent infection with multiple strains. Detailed information about historical, recent and ongoing HCV risk behaviours should be collected at regular intervals. Follow-up should be extended for 1–2 years following an end-of-treatment response (or SVR) to evaluate the rate of HCV reinfection. Individuals with reinfection should be followed up for a long period after treatment to determine the course and outcome of reinfection events. Finally, mixed HCV infection, superinfection and reinfection events should be confirmed through phylogenetic characterization of the various viral strains identified.
Conclusions
Data from human studies suggest that mixed HCV infection and superinfection are common events among PWID (particularly those with ongoing injection drug use), and mixed HCV infection could have an effect on HCV treatment outcomes. Furthermore, HCV reinfection among PWID after treatment-induced HCV clearance can occur, but the rates of HCV reinfection reported to date have been low.21,22,23,24,25,27,35,36,37,38,39,40,41,144,145
However, in the setting of DAA-based HCV therapy, very few studies have reported on mixed infection, superinfection and reinfection. Given that the majority of people infected with HCV are recent or former PWID, the expansion of DAA-based HCV treatment in these populations will be crucial to reduce the global morbidity and mortality associated with HCV infection. However, as >50% of people infected with HCV worldwide are infected with a non-genotype 1 HCV strains and the prevalence of mixed HCV infection among PWID is high, it will be important to evaluate the effect of mixed HCV infection in the DAA era given that many of these agents are still genotype-specific. Furthermore, given the potential of increased HCV treatment access among PWID who have a greater risk of HCV re-exposure, it will be important to evaluate the rate of HCV reinfection after successful DAA treatment.
Review criteria
Data for this Review were collected by searching Medline with the following search criteria, “hepatitis C” AND “superinfection” OR “superinfections” OR “mixed infect” OR “multiple infect” OR “reinfect” OR “re-infect”. The references of identified articles were manually searched for further relevant papers. Key abstracts at international meetings were also considered. Only English language papers were included.
References
Nelson, P. K. et al. Global epidemiology of hepatitis B and hepatitis C in people who inject drugs: results of systematic reviews. Lancet 378, 571–583 (2011).
Grebely, J. & Dore, G. J. Can hepatitis C virus infection be eradicated in people who inject drugs? Antiviral Res. 104, 62–72 (2014).
Hajarizadeh, B., Grebely, J. & Dore, G. J. Epidemiology and natural history of HCV infection. Nat. Rev. Gastroenterol. Hepatol. 10, 553–562 (2013).
Grebely, J. et al. Declining incidence of hepatitis C virus infection among people who inject drugs in a Canadian setting, 1996–2012. PLoS ONE 9, e97726 (2014).
Hadland, S. E. et al. Prescription opioid injection and risk of hepatitis C in relation to traditional drugs of misuse in a prospective cohort of street youth. BMJ Open 4, e005419 (2014).
Page, K., Morris, M. D., Hahn, J. A., Maher, L. & Prins, M. Injection drug use and hepatitis C virus infection in young adult injectors: using evidence to inform comprehensive prevention. Clin. Infect. Dis. 57 (Suppl. 2), S32–S38 (2013).
Osburn, W. O. et al. Spontaneous control of primary hepatitis C virus infection and immunity against persistent reinfection. Gastroenterology 138, 315–324 (2010).
Grebely, J. et al. Hepatitis C virus clearance, reinfection, and persistence, with insights from studies of injecting drug users: towards a vaccine. Lancet Infect. Dis. 12, 408–414 (2012).
Elliot, L. N., Lloyd, A. R., Ziegler, J. B. & Ffrench, R. A. Protective immunity against hepatitis C virus infection. Immunol. Cell Biol. 84, 239–249 (2006).
Abdel-Hakeem, M. S. & Shoukry, N. H. Protective immunity against hepatitis C: many shades of gray. Front. Immunol. 5, 274 (2014).
Bowden, S., McCaw, R., White, P. A., Crofts, N. & Aitken, C. K. Detection of multiple hepatitis C virus genotypes in a cohort of injecting drug users. J. Viral Hep. 12, 322–324 (2005).
Diamantis, I. et al. Genotype distribution of hepatitis C virus infection in Greece: correlation with different risk factors and response to interferon therapy. Eur. J. Gastroenterol. Hepatol. 10, 75–79 (1998).
Herring, B. L., Page-Shafer, K., Tobler, L. H. & Delwart, E. L. Frequent hepatitis C virus superinfection in injection drug users. J. Infect. Dis. 190, 1396–1403 (2004).
Ye, Y. et al. Molecular epidemiology of hepatitis C virus among different groups of people in the province of Fujian, China. Arch. Virol. 158, 611–618 (2013).
Lee, Y. M. et al. Molecular epidemiology of HCV genotypes among injection drug users in Taiwan: full-length sequences of two new subtype 6w strains and a recombinant form_2b6w. J. Med. Virol. 82, 57–68 (2010).
Micalessi, M. I. et al. Distribution of hepatitis C virus genotypes among injecting drug users in contact with treatment centers in Belgium, 2004–2005. J. Med. Virol. 80, 640–645 (2008).
Rios-Olivares, E. et al. HCV genotype analysis in HCV-HIV-co-infected Puerto Ricans who are injecting drug users: undetermined and mixed infections. Cell. Mol. Biol. (Noisy-le-Grand, France) 47, 1017–1024 (2001).
Sereno, S., Perinelli, P. & Laghi, V. Changes in the prevalence of hepatitis C virus genotype among Italian injection drug users-relation to period of injection started. J. Clin. Virol. 45, 354–357 (2009).
White, P. A., Li, Z., Zhai, X., Marinos, G. & Rawlinson, W. D. Mixed viral infection identified using heteroduplex mobility analysis (HMA). Virology 271, 382–389 (2000).
Chen, B. F. et al. High prevalence of mixed genotype infections in hepatitis B virus infected intravenous drug users. J. Med. Virol. 74, 536–542 (2004).
Blackard, J. T. HCV superinfection and reinfection. Antivir. Ther. 17, 1443–1448 (2012).
Blackard, J. T. & Sherman, K. E. Hepatitis C virus coinfection and superinfection. J. Infect. Dis. 195, 519–524 (2007).
Grebely, J. et al. Hepatitis C virus reinfection and superinfection among treated and untreated participants with recent infection. Hepatology 55, 1058–1069 (2012).
van de Laar, T. J. et al. Frequent HCV reinfection and superinfection in a cohort of injecting drug users in Amsterdam. J. Hepatol. 51, 667–674 (2009).
Pham, S. T. et al. Frequent multiple hepatitis C virus infections among injection drug users in a prison setting. Hepatology 52, 1564–1572 (2010).
Giannini, C. et al. Prevalence of mixed infection by different hepatitis C virus genotypes in patients with hepatitis C virus-related chronic liver disease. J. Lab. Clin. Med. 134, 68–73 (1999).
Currie, S. L. et al. A prospective study to examine persistent HCV reinfection in injection drug users who have previously cleared the virus. Drug Alcohol Depend. 93, 148–54 (2008).
Mehta, S. H. et al. Protection against persistence of hepatitis C. Lancet 359, 1478–1483 (2002).
Micallef, J. M., Kaldor, J. M. & Dore, G. J. Spontaneous viral clearance following acute hepatitis C infection: a systematic review of longitudinal studies. J. Viral Hep. 13, 34–41 (2006).
Grebely, J. et al. Hepatitis C virus reinfection in injection drug users. Hepatology 44, 1139–1145 (2006).
Aitken, C. K. et al. High incidence of hepatitis C virus reinfection in a cohort of injecting drug users. Hepatology 48, 1746–1752 (2008).
Dove, L. et al. Viral evolution of hepatitis C in injection drug users. J. Viral Hep. 12, 574–583 (2005).
Micallef, J. M. et al. High incidence of hepatitis C virus reinfection within a cohort of injecting drug users. J. Viral Hep. 14, 413–418 (2007).
Page, K. et al. Acute hepatitis C virus infection in young adult injection drug users: a prospective study of incident infection, resolution, and reinfection. J. Infect. Dis. 200, 1216–1226 (2009).
Backmund, M., Meyer, K. & Edlin, B. R. Infrequent reinfection after successful treatment for hepatitis C virus infection in injection drug users. Clin. Infect. Dis. 39, 1540–1543 (2004).
Dalgard, O. Follow-up studies of treatment for hepatitis C virus infection among injection drug users. Clin. Infect. Dis. 40 (Suppl. 5), S336–338 (2005).
Bate, J. P., Colman, A. J., Frost, P. J., Shaw, D. R. & Harley, H. A. High prevalence of late relapse and reinfection in prisoners treated for chronic hepatitis C. J. Gastroenterol. Hepatol. 25, 1276–1280 (2010).
Grady, B. P. et al. Low incidence of reinfection with the hepatitis C virus following treatment in active drug users in Amsterdam. Eur. J. Gastroenterol. Hepatol. 24, 1302–1307 (2012).
Grebely, J. et al. Reinfection with hepatitis C virus following sustained virological response in injection drug users. J. Gastroenterol. Hepatol. 25, 1281–1284 (2010).
Marco, A. et al. Hepatitis C virus reinfection among prisoners with sustained virological response after treatment for chronic hepatitis C. J. Hepatol. 59, 45–51 (2013).
Hilsden, R. J., Macphail, G., Grebely, J., Conway, B. & Lee, S. S. Directly observed pegylated interferon plus self-administered ribavirin for the treatment of hepatitis C virus infection in people actively using drugs: a randomized controlled trial. Clin. Infect. Dis. 57 (Suppl. 2), S90–96 (2013).
McHutchison, J. G. et al. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N. Engl. J. Med. 360, 1827–1838 (2009).
Sherman, K. E. et al. Response-guided telaprevir combination treatment for hepatitis C virus infection. N. Engl. J. Med. 365, 1014–1024 (2011).
Kohli, A., Shaffer, A., Sherman, A. & Kottilil, S. Treatment of hepatitis C: a systematic review. JAMA 312, 631–640 (2014).
Loulergue, P., Mir, O. & Sogni, P. Super-infection with genotype 4 hepatitis C virus in a patient treated for genotype 3 acute hepatitis C. AIDS 26, 655–656 (2012).
Schroter, M., Feucht, H. H., Zollner, B., Schafer, P. & Laufs, R. Multiple infections with different HCV genotypes: prevalence and clinical impact. J. Clin. Virol. 27, 200–204 (2003).
Abdelrahman, T. et al. Next-generation sequencing sheds light on the natural history of hepatitis C infection in patients who fail treatment. Hepatology 61, 88–97 (2014).
McNaughton, A. L., Thomson, E. C., Templeton, K., Gunson, R. N. & Leitch, E. C. Mixed genotype hepatitis C infections and implications for treatment. Hepatology 59, 1209 (2014).
Bartenschlager, R. & Lohmann, V. Replication of hepatitis C virus. J. Gen. Virol. 81, 1631–1648 (2000).
Duffy, S., Shackelton, L. A. & Holmes, E. C. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9, 267–276 (2008).
Jackowiak, P. et al. Phylogeny and molecular evolution of the hepatitis C virus. Infect. Genet. Evol. 21, 67–82 (2014).
Galli, A. & Bukh, J. Comparative analysis of the molecular mechanisms of recombination in hepatitis C virus. Trends Microbiol. 22, 354–364 (2014).
Gonzalez-Candelas, F., Lopez-Labrador, F. X. & Bracho, M. A. Recombination in hepatitis C virus. Viruses 3, 2006–2024 (2011).
Hedskog, C. et al. Characterization of hepatitis C virus intergenotypic recombinant strains and associated virological response to sofosbuvir/ribavirin. Hepatology 61, 471–480 (2014).
Kalinina, O., Norder, H., Mukomolov, S. & Magnius, L. O. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 76, 4034–4043 (2002).
Palmer, B. A. et al. Analysis of the evolution and structure of a complex intrahost viral population in chronic hepatitis C virus mapped by ultradeep pyrosequencing. J. Virol. 88, 13709–13721 (2014).
Shi, W. et al. Recombination in hepatitis C virus: identification of four novel naturally occurring inter-subtype recombinants. PLoS ONE 7, e41997 (2012).
Stumpf, M. P. & Pybus, O. G. Genetic diversity and models of viral evolution for the hepatitis C virus. FEMS Microbiol. Lett. 214, 143–152 (2002).
Smith, D. B. et al. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59, 318–327 (2014).
Messina, J. P. et al. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 61, 77–87 (2014).
Mondelli, M. U. & Silini, E. Clinical significance of hepatitis C virus genotypes. J. Hepatol. 31 (Suppl. 1), 65–70 (1999).
Smith, D. B. et al. The origin of hepatitis C virus genotypes. J. Gen. Virol. 78, 321–328 (1997).
Pybus, O. G. et al. The epidemic behavior of the hepatitis C virus. Science 292, 2323–2325 (2001).
Kalinina, O. et al. Shift in predominating subtype of HCV from 1b to 3a in St. Petersburg mediated by increase in injecting drug use. J. Med. Virol. 65, 517–524 (2001).
van Asten, L. et al. Spread of hepatitis C virus among European injection drug users infected with HIV: a phylogenetic analysis. J. Infect. Dis. 189, 292–302 (2004).
Jacka, B. et al. Phylogenetic clustering of hepatitis C virus among people who inject drugs in Vancouver, Canada. Hepatology 60, 1571–1580 (2014).
Romano, C. M. et al. Social networks shape the transmission dynamics of hepatitis C virus. PLoS ONE 5, e11170 (2010).
Dias, P. T. et al. Temporal changes in HCV genotype distribution in three different high risk populations in San Francisco, California. BMC Infect. Dis. 11, 208 (2011).
Pybus, O. G., Cochrane, A., Holmes, E. C. & Simmonds, P. The hepatitis C virus epidemic among injecting drug users. Infect. Genet. Evol. 5, 131–139 (2005).
Roman, F. et al. Hepatitis C virus genotypes distribution and transmission risk factors in Luxembourg from 1991 to 2006. World J. Gastroenterol. 14, 1237–1243 (2008).
Salehi Moghadam, F. et al. Phylogenetic analysis of hepatitis C virus strains and risk factors associated with infection and viral subtypes among Iranian patients. J. Med. Virol. 86, 1342–1349 (2014).
Samimi-Rad, K. et al. Molecular epidemiology of hepatitis C virus among injection drug users in Iran: a slight change in prevalence of HCV genotypes over time. Arch. Virol. 157, 1959–1965 (2012).
Morice, Y. et al. Molecular epidemiology of hepatitis C virus subtype 3a in injecting drug users. J. Med. Virol. 78, 1296–1303 (2006).
Frank, C. et al. The role of parenteral antischistosomal therapy in the spread of hepatitis C virus in Egypt. Lancet 355, 887–891 (2000).
van de Laar, T. et al. Evidence of a large, international network of HCV transmission in HIV-positive men who have sex with men. Gastroenterology 136, 1609–1617 (2009).
Smith, D. M., Richman, D. D. & Little, S. J. HIV superinfection. J. Infect. Dis. 192, 438–444 (2005).
Sarrazin, C. et al. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132, 1767–1777 (2007).
Wang, C. et al. Persistence of resistant variants in hepatitis C virus-infected patients treated with the NS5A replication complex inhibitor daclatasvir. Antimicrob. Agents Chemother. 57, 2054–2065 (2013).
Forns, X. et al. Comparative study of three methods for genotyping hepatitis C virus strains in samples from Spanish patients. J. Clin. Microbiol. 34, 2516–2521 (1996).
Hu, Y. W. et al. Comparison and application of a novel genotyping method, semiautomated primer-specific and mispair extension analysis, and four other genotyping assays for detection of hepatitis C virus mixed-genotype infections. J. Clin. Microbiol. 38, 2807–2813 (2000).
Quer, J. et al. High-resolution hepatitis C virus (HCV) subtyping, using NS5B deep sequencing and phylogeny, an alternative to current methods. J. Clin. Microbiol. 53, 219–226 (2014).
Davidson, F. et al. Survey of major genotypes and subtypes of hepatitis C virus using RFLP of sequences amplified from the 5′ non-coding region. J. Gen. Virol. 76, 1197–1204 (1995).
Jacka, B. et al. Sequencing of the hepatitis C virus: a systematic review. PLoS ONE 8, e67073 (2013).
Tsiatis, A. C. et al. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J. Mol. Diagn. 12, 425–432 (2010).
Barzon, L. et al. Next-generation sequencing technologies in diagnostic virology. J. Clin. Virol. 58, 346–350 (2013).
Buermans, H. P. & den Dunnen, J. T. Next generation sequencing technology: advances and applications. Biochim. Biophys. Acta 1842, 1932–1941 (2014).
Donaldson, E. F., Harrington, P. R., O'Rear, J. J. & Naeger, L. K. Clinical evidence and bioinformatics characterization of potential hepatitis C virus resistance pathways for sofosbuvir. Hepatology 61, 56–65 (2015).
Liu, L. et al. Comparison of next-generation sequencing systems. J. Biomed. Biotech. 2012, 251364 (2012).
Quinones-Mateu, M. E., Avila, S., Reyes-Teran, G. & Martinez, M. A. Deep sequencing: becoming a critical tool in clinical virology. J. Clin. Virol. 61, 9–19 (2014).
Fonseca, N. A., Rung, J., Brazma, A. & Marioni, J. C. Tools for mapping high-throughput sequencing data. Bioinformatics 28, 3169–3177 (2012).
Marz, M. et al. Challenges in RNA virus bioinformatics. Bioinformatics 30, 1793–1799 (2014).
McElroy, K., Thomas, T. & Luciani, F. Deep sequencing of evolving pathogen populations: applications, errors, and bioinformatic solutions. Microb. Inform. Exp. 4, 1 (2014).
Nelson, C. W. & Hughes, A. L. Within-host nucleotide diversity of virus populations: insights from next-generation sequencing. Infect. Genet. Evol. 30, 1–7 (2014).
Pujol, F. H., Devesa, M., Loureiro, C. L., Capriles, F. & Liprandi, F. Turnover of hepatitis C virus genotypes in hemodialysis patients. Arch. Virol. 143, 823–827 (1998).
Laskus, T. et al. Exposure of hepatitis C virus (HCV) RNA-positive recipients to HCV RNA-positive blood donors results in rapid predominance of a single donor strain and exclusion and/or suppression of the recipient strain. J. Virol. 75, 2059–2066 (2001).
Eyster, M. E., Sherman, K. E., Goedert, J. J., Katsoulidou, A. & Hatzakis, A. Prevalence and changes in hepatitis C virus genotypes among multitransfused persons with hemophilia. The Multicenter Hemophilia Cohort Study. J. Infect. Dis. 179, 1062–1069 (1999).
Jarvis, L. M. et al. Frequent reinfection and reactivation of hepatitis C virus genotypes in multitransfused hemophiliacs. J. Infect. Dis. 170, 1018–1022 (1994).
Kao, J. H., Chen, P. J., Lai, M. Y. & Chen, D. S. Superinfection of heterologous hepatitis C virus in a patient with chronic type C hepatitis. Gastroenterology 105, 583–587 (1993).
Lai, M. E. et al. Hepatitis C virus in multiple episodes of acute hepatitis in polytransfused thalassaemic children. Lancet 343, 388–390 (1994).
Widell, A. et al. Hepatitis C superinfection in hepatitis C virus (HCV)-infected patients transplanted with an HCV-infected kidney. Transplantation 60, 642–647 (1995).
Kao, J. H. et al. Superinfection by homotypic virus in hepatitis C virus carriers: studies on patients with post-transfusion hepatitis. J. Med. Virol. 50, 303–308 (1996).
Accapezzato, D., Fravolini, F., Casciaro, M. A. & Paroli, M. Hepatitis C flare due to superinfection by genotype 4 in an HCV genotype 1b chronic carrier. Eur. J. Gastroenterol. Hepatol. 14, 879–881 (2002).
Ghosn, J. et al. Sexually transmitted hepatitis C virus superinfection in HIV/hepatitis C virus co-infected men who have sex with men. AIDS 22, 658–661 (2008).
Okamoto, H. et al. Superinfection of chimpanzees carrying hepatitis C virus of genotype II/1b with that of genotype III/2a or I/1a. Hepatology 20, 1131–1136 (1994).
Lau, J. Y. et al. Distribution of hepatitis C virus genotypes determined by line probe assay in patients with chronic hepatitis C seen at tertiary referral centers in the United States. Hepatitis Interventional Therapy Group. Ann. Intern. Med. 124, 868–876 (1996).
van Asten, L. & Prins, M. Infection with concurrent multiple hepatitis C virus genotypes is associated with faster HIV disease progression. AIDS 18, 2319–2324 (2004).
Viazov, S., Widell, A. & Nordenfelt, E. Mixed infection with two types of hepatitis C virus is probably a rare event. Infection 28, 21–25 (2000).
Stamenkovic, G. et al. Distribution of HCV genotypes among risk groups in Serbia. Eur. J. Epidemiol. 16, 949–954 (2000).
Aitken, C. et al. Change in hepatitis C virus genotype in injecting drug users. J. Med. Virol. 74, 543–545 (2004).
Antipa, C., Ruta, S. & Cernescu, C. Serological profile assessment of the infection with hepatitis C virus (HCV) in haemophiliacs and thalassemic patients. Rom. J. Virol. 47, 3–11 (1996).
Natov, S. N. et al. Serologic and virologic profiles of hepatitis C infection in renal transplant candidates. New England Organ Bank Hepatitis C Study Group. Am. J. Kidney Dis. 31, 920–927 (1998).
Wu, J. S. et al. Genotype distribution of hepatitis C virus infection in Taiwan. J. Med. Virol. 44, 74–79 (1994).
Qian, K. P., Natov, S. N., Pereira, B. J. & Lau, J. Y. Hepatitis C virus mixed genotype infection in patients on haemodialysis. J. Viral Hep. 7, 153–160 (2000).
Boelen, L. et al. Per-event probability of hepatitis C infection during sharing of injecting equipment. PLoS ONE 9, e100749 (2014).
Grebely, J. et al. The effects of female sex, viral genotype, and IL28B genotype on spontaneous clearance of acute hepatitis C virus infection. Hepatology 59, 109–120 (2014).
Osburn, W. O. et al. Spontaneous control of primary hepatitis C virus infection and immunity against persistent reinfection. Gastroenterology 138, 315–324 (2009).
McHutchison, J. G. et al. Interferon α-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N. Engl. J. Med. 339, 1485–1492 (1998).
Manns, M. P. et al. Peginterferon α-2b plus ribavirin compared with interferon α-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358, 958–965 (2001).
Jacobson, I. M. et al. Telaprevir for previously untreated chronic hepatitis C virus infection. N. Engl. J. Med. 364, 2405–2416 (2011).
Poordad, F. et al. Boceprevir for untreated chronic HCV genotype 1 infection. N. Engl. J. Med. 364, 1195–1206 (2011).
Razavi, H., Estes, C., Pasini, K., Gower, E. & Hindman, S. HCV treatment rate in select European countries in 2004–2010. J. Hepatol. 58, S22–S23 (2013).
Liang, T. J. & Ghany, M. G. Therapy of hepatitis C—back to the future. N. Engl. J. Med. 370, 2043–2047 (2014).
Ferenci, P. et al. ABT-450/r-ombitasvir and dasabuvir with or without ribavirin for HCV. N. Engl. J. Med. 370, 1983–1992 (2014).
Feld, J. J. et al. Treatment of HCV with ABT-450/r-ombitasvir and dasabuvir with ribavirin. N. Engl. J. Med. 370, 1594–1603 (2014).
Poordad, F. et al. ABT-450/r-ombitasvir and dasabuvir with ribavirin for hepatitis C with cirrhosis. N. Engl. J. Med. 370, 1973–1982 (2014).
Everson, G. T. et al. Efficacy of an interferon- and ribavirin-free regimen of daclatasvir, asunaprevir, and BMS-791325 in treatment-naive patients with HCV genotype 1 infection. Gastroenterology 146, 420–429 (2014).
Afdhal, N. et al. Ledipasvir and sofosbuvir for untreated HCV genotype 1 infection. N. Engl. J. Med. 370, 1889–1898 (2014).
Afdhal, N. et al. Ledipasvir and sofosbuvir for previously treated HCV genotype 1 infection. N. Engl. J. Med. 370, 1483–1493 (2014).
Lawitz, E. et al. Efficacy and safety of MK-5172 and MK-8742 ± ribavirin in hepatitis C genotype 1 infected patients with cirrhosis or previous null response: the C-WORTHY Study. [abstract 196]. Hepatology 60 (Suppl. 1), S25–S26 (2014).
Gane, E. J. et al. Nucleotide polymerase inhibitor sofosbuvir plus ribavirin for hepatitis C. N. Engl. J. Med. 368, 34–44 (2013).
Zeuzem, S. et al. Sofosbuvir and ribavirin in HCV genotypes 2 and 3. N. Engl. J. Med. 370, 1993–2001 (2014).
McPhee, F. et al. Preclinical profile and characterization of the hepatitis C virus NS3 protease inhibitor asunaprevir (BMS-650032). Antimicrob. Agents Chemother. 56, 5387–5396 (2012).
Lawitz, E. et al. Sofosbuvir for previously untreated chronic hepatitis C infection. N. Engl. J. Med. 368, 1878–1887 (2013).
Link, J. O. et al. Discovery of ledipasvir (GS-5885): a potent, once-daily oral NS5A inhibitor for the treatment of hepatitis C virus infection. J. Med. Chem. 57, 2033–2046 (2014).
Lawitz, E. et al. Exploratory trial of ombitasvir and ABT-450/r with or without ribavirin for HCV genotype 1, 2, and 3 infection. J. Infect. 70, 197–205 (2014).
Summa, V. et al. MK-5172, a selective inhibitor of hepatitis C virus NS3/4a protease with broad activity across genotypes and resistant variants. Antimicrob. Agents Chemother. 56, 4161–4167 (2012).
Everson, G. T. et al. Safety and efficacy of treatment with interferon-free, ribavirin-free combination of sofosbuvir + GS-5816 for 12 weeks in treatment naïve patients with genotype 1–6 HCV infection [abstract 0111]. J. Hepatol. 60, S46 (2014).
Tran, T. T. et al. Safety and efficacy of treatment with sofosbuvir + GS-5816 ± ribavirin for 8 or 12 weeks in treatment naive patients with genotype 1–6 HCV infection. [abstract 80]. Hepatology 60 (Suppl. 1), 237A (2014).
Nelson, D. R., Cooper, J. N., Lalezari, J. P. & Lawitz, E. All-oral 12-week combination treatment with daclatasvir (DCV) and sofosbuvir (SOF) in patients infected with HCV genotype (GT) 3: ALLY-3 phase 3 study. [abstract LB3]. Hepatology 60 (Suppl. 1) (2014).
Izquierdo, L. et al. Simeprevir for the treatment of hepatitis C virus infection. Pharmacogenomics Pers. Med. 7, 241–249 (2014).
Kao, J. H., Lai, M. Y., Chen, P. J. & Chen, D. S. Probable reinfection with hepatitis C virus in a chronic hepatitis C patient with a sustained response to combination therapy. J. Formos. Med. Assoc. 100, 824–828 (2001).
Akuta, N. et al. Interferon-alpha for reinfection with hepatitis C virus in two patients with chronic hepatitis C who had responded to previous therapies. Dig. Dis. Sci. 48, 1654–1657 (2003).
Asselah, T. et al. Second infection with a different hepatitis C virus genotype in a intravenous drug user during interferon therapy. Gut 52, 900–902 (2003).
Backmund, M., Meyer, K., Von Zielonka, M. & Eichenlaub, D. Treatment of hepatitis C infection in injection drug users. Hepatology 34, 188–193 (2001).
Aspinall, E. J. et al. Treatment of hepatitis C virus infection among people who are actively injecting drugs: a systematic review and meta-analysis. Clin. Infect. Dis. 57 (Suppl 2), S80–S89 (2013).
Hill, A. M., Saleem, J., Heath, K. A. & Simmons, B. Effects of sustained virological response (SVR) on the risk of liver transplant, hepatocellular carcinoma, death and re-infection: meta-analysis of 129 studies in 23,309 patients with hepatitis C infection. [abstract 44]. Hepatology 60 (Suppl. 1), 218A (2014).
Bretaña, N. A. et al. Hepatitis C virus transmission dynamics in a high-risk incarcerated population. Emerg. Infect. Dis. (in press).
Parodi, C. et al. Evidence of occult HCV genotypes in haemophilic individuals with unapparent HCV mixed infections. Haemophilia 14, 816–822 (2008).
Thomson, E. C. et al. Predicting spontaneous clearance of acute hepatitis C virus in a large cohort of HIV-1-infected men. Gut 60, 837–845 (2011).
Wu, S. et al. Prevalence of hepatitis C virus subgenotypes 1a and 1b in Japanese patients: ultra-deep sequencing analysis of HCV NS5B genotype-specific region. PLoS ONE 8, e73615 (2013).
Acknowledgements
E.B.C. is supported by a National CIHR Research Training Program in Hepatitis C Scholarship. J.G. is supported by a National Health and Medical Research Council Career Development Fellowship. A.R.L. and G.J.D. are supported by National Health and Medical Research Council Practitioner Research Fellowships.
Author information
Authors and Affiliations
Contributions
E.B.C. and J.G. contributed equally to researching data for the article. All authors contributed equally to the discussion of content, writing and reviewing and editing the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
J.G. is a consultant, advisor and has received research grants from Abbvie, Bristol Myers Squibb, Gilead Sciences and Merck. G.D. is a consultant, advisor and has received research grants from Abbvie, Bristol Myers Squibb, Gilead Sciences, Janssen, Merck and Roche.
Rights and permissions
About this article
Cite this article
Cunningham, E., Applegate, T., Lloyd, A. et al. Mixed HCV infection and reinfection in people who inject drugs—impact on therapy. Nat Rev Gastroenterol Hepatol 12, 218–230 (2015). https://doi.org/10.1038/nrgastro.2015.36
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrgastro.2015.36
This article is cited by
-
Country versus pharmaceutical company interests for hepatitis C treatment
Health Care Management Science (2022)
-
An update on hepatitis C virus genotype distribution in Jordan: a 12-year retrospective study from a tertiary care teaching hospital in Amman
BMC Infectious Diseases (2020)
-
Infection with multiple hepatitis C virus genotypes detected using commercial tests should be confirmed using next generation sequencing
Scientific Reports (2019)
-
Reliable resolution of ambiguous hepatitis C virus genotype 1 results with the Abbott HCV Genotype Plus RUO assay
Scientific Reports (2019)
-
Hepatitis C virus infection status and associated factors among a multi-site sample of people who used illicit drugs in the Amazon region
BMC Infectious Diseases (2019)