Abstract
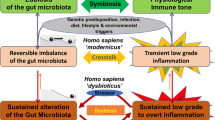
Spurred on by ever-evolving developments in analytical methodology, the microbiome, and the gut microbiome in particular, has become the hot topic in biomedical research. Ingenious experiments in animal models have revealed the extent to which the gut microbiota sustains health and how its disruption might contribute to disease pathogenesis. Not surprisingly, associations between the microbiota and disease states in humans have been the subject of considerable interest and many links proposed. However, with rare exceptions, the incrimination of an altered microbiota in disease pathogenesis seems premature at this time given our incomplete understanding of the composition of the gut microbiota in health and the effect of many confounding factors in the interpretation of supposedly abnormal microbial signatures. Future studies must account for these variables and the bidirectionality of host–microorganism interactions in health and disease. In this Perspectives, the status of microbiota signatures in the clinical arena (for facilitating diagnosis or refining prognosis) will be critically assessed and guidance toward future progress provided.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Blaser, M. J. The microbiome revolution. J. Clin. Invest. 124, 4162–4165 (2014).
Iqbal, S. & Quigley, E. M. M. Progress in our understanding of the gut microbiome: implications for the clinician. Curr. Gastroenterol. Rep. 18, 49 (2016).
O'Toole, P. W. & Flemer, B. From culture to high-throughput sequencing and beyond: a layperson's guide to the “omics” and diagnostic potential of the microbiome. Gastroenterol. Clin. North Am. 46, 9–17 (2017).
Fung, T. C., Olson, C. A. & Hsiao, E. Y. Interactions between the microbiota, immune and nervous systems in health and disease. Nat. Neurosci. 20, 145–155 (2017).
Parashar, A. & Udayabanu, M. Gut microbiota: implications in Parkinson's disease. Parkinsonism Relat. Disord. http://dx.doi.org/10.1016/j.parkreldis.2017.02.002 (2017).
Vuong, H. E. & Hsiao, E. Y. Emerging roles for the gut microbiome in autism spectrum disorder. Biol. Psychiatry 81, 411–423 (2017).
Paun, A., Yau, C. & Danska, J. S. The influence of the microbiome on type 1 diabetes. J. Immunol. 198, 590–595 (2017).
Stiemsma, L. T. & Turvey, S. E. Asthma and the microbiome: defining the critical window in early life. Allergy Asthma Clin. Immunol. 13, 3 (2017).
Marasco, G. et al. Gut microbiota and celiac disease. Dig. Dis. Sci. 61, 1461–1472 (2016).
Sender, R., Fuchs, S. & Milo, R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 164, 337–340 (2016).
Sekirov, I., Russell, S. L., Antunes, L. C. & Finlay, B. B. Gut microbiota in health and disease. Physiol. Rev. 90, 859–904 (2010).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Zhernakova, A. et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569 (2016).
Falony, G. et al. Population-level analysis of gut microbiome variation. Science 352, 560–564 (2016).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Marques, T. M. et al. Programming infant gut microbiota: influence of dietary and environmental factors. Curr. Opin. Biotechnol. 21, 149–156 (2010).
Dominguez-Bello, M. G. et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA 107, 11971–11975 (2010).
McGuire, M. K. & McGuire, M. A. Human milk: mother nature's prototypical probiotic food? Adv. Nutr. 6, 112–123 (2015).
Claesson, M. J. et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 488, 178–184 (2012).
Clarke, S. F. et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut 63, 1913–1920 (2014).
Engen, P. A., Green, S. J., Voigt, R. M., Forsyth, C. B. & Keshavarzian, A. The gastrointestinal microbiome: alcohol effects on the composition of intestinal microbiota. Alcohol Res. 7, 223–236 (2015).
Vangay, P., Ward, T., Gerber, J. S. & Knights, D. Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe. 17, 553–564 (2015).
Doré, J. & Blottière, H. The influence of diet on the gut microbiota and its consequences for health. Curr. Opin. Biotechnol. 32, 195–199 (2015).
Smith, M. I. et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339, 548–554 (2013).
Subramanian, S. et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510, 417–421 (2014).
Sonnenburg, E. D. & Sonnenburg, J. L. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. 20, 779–786 (2014).
McIntosh, K. et al. FODMAPs alter symptoms and the metabolome of patients with IBS: a randomised controlled trial. Gut http://dx.doi.org/10.1136/gutjnl-2015-311339 (2016).
Heinritz, S. N. et al. Intestinal microbiota and microbial metabolites are changed in a pig model fed a high-fat/low-fiber or a low-fat/high-fiber diet. PLoS ONE 11, e0154329 (2016).
Bonder, M. J. et al. The influence of a short-term gluten-free diet on the human gut microbiome. Genome Med. 8, 45 (2016).
McGuire, M. K. & McGuire, M. A. Got bacteria? The astounding, yet not-so-surprising, microbiome of human milk. Curr. Opin. Biotechnol. 44, 63–68 (2016).
Kumar, H. et al. Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front. Microbiol. 7, 1619 (2016).
Blaser, M. J. Antibiotic use and its consequences for the normal microbiome. Science 352, 544–545 (2016).
Cho, I. et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488, 621–626 (2012).
Devkota, S. Prescription drugs obscure microbiome analyses. Science 351, 452–453 (2016).
Forslund, K. et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266 (2015).
Cox, L. M. et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158, 705–721 (2014).
Dabrowska, K. & Witkiewicz, W. Correlations of host genetics and gut microbiome composition. Front. Microbiol. 7, 1–7 (2016).
Elinav, E. et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757 (2011).
De Palma, G., Collins, S. M., Bercik, P. & Verdu, E. F. The microbiota–gut–brain axis in gastrointestinal disorders: stressed bugs, stressed brain or both? J. Physiol. 592, 2989–2997 (2014).
Clemente, J. C., Ursell, L. K., Parfrey, L. W. & Knight, R. The impact of the gut microbiota on human health: an integrative view. Cell 148, 1258–1270 (2012).
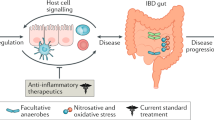
Dalal, S. R. & Chang, E. B. The microbial basis of inflammatory bowel diseases. J. Clin. Invest. 124, 4190–4196 (2014).
Surana, N. K. & Kasper, D. L. Deciphering the tête-à-tête between the microbiota and the immune system. J. Clin. Invest. 124, 4197–4203 (2014).
Carmody, R. N. & Turnbaugh, P. J. Host–microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J. Clin. Invest. 124, 4173–4181 (2014).
Mayer, E. A., Tillisch, K. & Gupta, A. Gut/brain axis and the microbiota. J. Clin. Invest. 125, 926–938 (2015).
Medzhitov, R. Origin and physiological roles of inflammation. Nature 454, 428–435 (2008).
Kelly, J. R. et al. Breaking down the barriers: the gut microbiome, intestinal permeability and stress-related psychiatric disorders. Front. Cell. Neurosci. 9, 392 (2015).
Seekatz, A. M. & Young, V. B. Clostridium difficile and the microbiota. J. Clin. Invest. 124, 4182–4819 (2014).
Chang, J. Y. et al. Decreased diversity of the fecal microbiome in recurrent clostridium difficile-associated diarrhea. J. Infect. Dis. 197, 435–438 (2008).
Buffie, C. G. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015).
Peek, R. M. Jr, Fiske, C. & Wilson, K. T. Role of innate immunity in Helicobacter pylori-induced gastric malignancy. Physiol. Rev. 90, 831–858 (2010).
Bianco, A. M., Girardelli, M. & Tommasini, A. Genetics of inflammatory bowel disease from multifactorial to monogenic forms. World J. Gastroenterol. 21, 12296–12310 (2015).
Villani, A. C. et al. Genetic risk factors for post-infectious irritable bowel syndrome following a waterborne outbreak of gastroenteritis. Gastroenterology 138, 1502–1513 (2010).
Quigley, E. M., Stanton, C. & Murphy, E. F. The gut microbiota and the liver. Pathophysiological and clinical implications. J. Hepatol. 58, 1020–1027 (2013).
Koutsounas, I., Kaltsa, G., Siakavellas, S. I. & Bamias, G. Markers of bacterial translocation in end-stage liver disease. World J. Hepatol. 7, 2264–2273 (2015).
Galipeau, H. J. & Verdu, E. F. The complex task of measuring intestinal permeability in basic and clinical science. Neurogastroenterol. Motil. 28, 957–965 (2016).
Weber, C. R. Dynamic properties of the tight junction barrier. Ann. NY Acad. Sci. 1257, 77–84 (2012).
Quigley, E. M. Leaky gut — concept or clinical entity? Curr. Opin. Gastroenterol. 32, 74–79 (2016).
Spadoni, I. et al. A gut-vascular barrier controls the systemic dissemination of bacteria. Science 350, 830–834 (2015).
Jarocki, P., Podles´ny, M., Glibowski, P. & Targon´ski, Z. A new insight into the physiological role of bile salt hydrolase among intestinal bacteria from the genus Bifidobacterium. PLoS ONE 9, e114379 (2014).
Li, T. & Chiang, J. Y. Bile acids as metabolic regulators. Curr. Opin. Gastroenterol. 31, 159–165 (2015).
Rao, A. S. et al. Chenodeoxycholate in females with irritable bowel syndrome-constipation: a pharmacodynamic and pharmacogenetic analysis. Gastroenterology 139, 1549–1558 (2010).
Alrefai, W. A. et al. Taurodeoxycholate modulates apical Cl−/OH− exchange activity in Caco2 cells. Dig. Dis. Sci. 52, 1270–1278 (2007).
Guo, C., Chen, W.-D. & Wang, Y.-D. TGR5, not only a metabolic regulator. Front. Physiol. 7, 646 (2016).
Soldavini, J. & Kaunitz, J. D. Pathobiology and potential therapeutic value of intestinal short-chain fatty acids in gut inflammation and obesity. Dig. Dis. Sci. 58, 2756–2766 (2013).
Cushing, K., Alvarado, D. M. & Ciorba, M. A. Butyrate and mucosal inflammation: new scientific evidence supports clinical observation. Clin. Transl Gastroenterol. 6, e108 (2015).
Nayfach, S. & Pollard, K. S. Toward accurate and quantitative comparative metagenomics. Cell 166, 1103–1116 (2016).
Vernocchi, P., Del Chierico, F. & Putignani, L. Gut microbiota profiling: metabolomics based approach to unravel compounds affecting human health. Front. Microbiol. 7, 1144 (2016).
Altmann, F. et al. Genome analysis and characterisation of the exopolysaccharide produced by Bifidobacterium longum subsp. longum 35624™. PLoS ONE 11, e0162983 (2016).
Schiavi, E. et al. The surface-associated exopolysaccharide of Bifidobacterium longum 35624 plays an essential role in dampening host proinflammatory responses and repressing local TH17 responses. Appl. Environ. Microbiol. 82, 7185–7196 (2016).
Phear, E. A. & Ruebner, B. The in vitro production of ammonium and amines by intestinal bacteria in relation to nitrogen toxicity as a factor in hepatic coma. Br. J. Exp. Pathol. 37, 253–262 (1956).
Bajaj, J. S. et al. Modulation of the metabiome by rifaximin in patients with cirrhosis and minimal hepatic encephalopathy. PLoS ONE 8, e60042 (2013).
Browne, H. P. et al. Culturing of 'unculturable' human microbiota reveals novel taxa and extensive sporulation. Nature 533, 543–546 (2016).
Simpson, H. L. & Campbell, B. J. Review article: dietary fibre-microbiota interactions. Aliment. Pharmacol. Ther. 42, 158–179 (2015).
Donaldson, G. P., Lee, S. M. & Mazmanian, S. K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32 (2016).
Codling, C., O'Mahony, L., Shanahan, F., Quigley, E. M. & Marchesi, J. R. A molecular analysis of fecal and mucosal bacterial communities in irritable bowel syndrome. Dig. Dis. Sci. 55, 392–397 (2010).
Carroll, I. M. et al. Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am. J. Physiol. Gastrointest. Liver Physiol. 301, G799–G807 (2011).
Ringel, Y. et al. High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut Microbes 6, 173–181 (2015).
Bagdasarian, N., Rao, K. & Malani, P. N. Diagnosis and treatment of Clostridium difficile in adults: a systematic review. JAMA 313, 398–408 (2015).
Sonnenburg, E. D. et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016).
Kashyap, P. C. et al. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology 144, 967–977 (2013).
Nagao-Kitamoto, H. et al. Functional characterization of inflammatory bowel disease-associated gut dysbiosis in gnotobiotic mice. Cell. Mol. Gastroenterol. Hepatol. 2, 468–481 (2016).
Kelly, J. R. et al. Transferring the blues: depression-associated gut microbiota induces neurobehavioural changes in the rat. J. Psychiatr. Res. 82, 109–118 (2016).
Sampson, T. R. et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell 167, 1469–1480 (2016).
Shen, T. C. et al. Engineering the gut microbiota to treat hyperammonemia. J. Clin. Invest. 125, 2841–2850 (2015).
Anderson, E. L. et al. A robust ambient temperature collection and stabilization strategy: enabling worldwide functional studies of the human microbiome. Sci. Rep. 6, 31731 (2016).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Rights and permissions
About this article
Cite this article
Quigley, E. Gut microbiome as a clinical tool in gastrointestinal disease management: are we there yet?. Nat Rev Gastroenterol Hepatol 14, 315–320 (2017). https://doi.org/10.1038/nrgastro.2017.29
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrgastro.2017.29
This article is cited by
-
The intersection of undernutrition, microbiome, and child development in the first years of life
Nature Communications (2023)
-
Review of the Health Benefits of Cereals and Pseudocereals on Human Gut Microbiota
Food and Bioprocess Technology (2023)
-
Irritable bowel syndrome and microbiome; Switching from conventional diagnosis and therapies to personalized interventions
Journal of Translational Medicine (2022)
-
General anesthesia bullies the gut: a toxic relationship with dysbiosis and cognitive dysfunction
Psychopharmacology (2022)
-
Süßstoffe - Modulatoren des Darmmikrobioms?
Info Diabetologie (2022)