Key Points
-
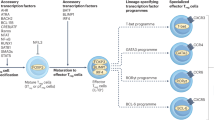
Forkhead box protein P3 (FOXP3) is a crucial regulator of regulatory T (Treg) cell gene expression that is responsible for much of the suppressive potential displayed by these cells.
-
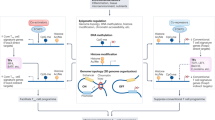
The regulation of FOXP3 expression in Treg cells occurs through the concerted action of transcription factors and extensive epigenetic control mechanisms; furthermore, post-translational modifications are also capable of modulating FOXP3 function.
-
These several layers of FOXP3 control are responsive to positive and negative regulation by factors in the tissue environment, including cytokines, inflammatory mediators and metabolic factors.
-
Modulating FOXP3 expression and Treg cell function by targeting newly discovered regulatory nodes may lead to the development of new immunotherapies for cancer and autoimmune diseases.
Abstract
The proper restraint of the destructive potential of the immune system is essential for maintaining health. Regulatory T (Treg) cells ensure immune homeostasis through their defining ability to suppress the activation and function of other leukocytes. The expression of the transcription factor forkhead box protein P3 (FOXP3) is a well-recognized characteristic of Treg cells, and FOXP3 is centrally involved in the establishment and maintenance of the Treg cell phenotype. In this Review, we summarize how the expression and activity of FOXP3 are regulated across multiple layers by diverse factors. The therapeutic implications of these topics for cancer and autoimmunity are also discussed.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Wang, J., Ioan-Facsinay, A., van der Voort, E. I., Huizinga, T. W. & Toes, R. E. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur. J. Immunol. 37, 129–138 (2007).
Gavin, M. A. et al. Single-cell analysis of normal and FOXP3-mutant human T cells: FOXP3 expression without regulatory T cell development. Proc. Natl Acad. Sci. USA 103, 6659–6664 (2006).
Sakaguchi, S., Wing, K. & Miyara, M. Regulatory T cells — a brief history and perspective. Eur. J. Immunol. 37 (Suppl. 1), S116–S123 (2007).
Vignali, D. A., Collison, L. W. & Workman, C. J. How regulatory T cells work. Nat. Rev. Immunol. 8, 523–532 (2008).
Shevach, E. M. Mechanisms of Foxp3+ T regulatory cell-mediated suppression. Immunity 30, 636–645 (2009).
Deaglio, S. et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J. Exp. Med. 204, 1257–1265 (2007).
Cao, X. et al. Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity 27, 635–646 (2007).
Ramsdell, F. & Ziegler, S. F. FOXP3 and scurfy: how it all began. Nat. Rev. Immunol. 14, 343–349 (2014).
Brunkow, M. E. et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27, 68–73 (2001).
Bennett, C. L. et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27, 20–21 (2001). References 9 and 10 describe the extremely negative consequences of FOXP3 mutation on immune regulation in mice and humans.
Zheng, Y. et al. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 445, 936–940 (2007).
Samstein, R. M. et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell 151, 153–166 (2012).
Marson, A. et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature 445, 931–935 (2007).
Morikawa, H. & Sakaguchi, S. Genetic and epigenetic basis of Treg cell development and function: from a FoxP3-centered view to an epigenome-defined view of natural Treg cells. Immunol. Rev. 259, 192–205 (2014).
Rudra, D. et al. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat. Immunol. 13, 1010–1019 (2012). This study reveals the extensive interactions of FOXP3 with other transcription factors and epigenetic-modifying enzymes.
Hori, S. The Foxp3 interactome: a network perspective of Treg cells. Nat. Immunol. 13, 943–945 (2012).
Wu, Y. et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell 126, 375–387 (2006).
Ono, M. et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature 446, 685–689 (2007).
Zheng, Y. et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control TH2 responses. Nature 458, 351–356 (2009).
Darce, J. et al. An N-terminal mutation of the Foxp3 transcription factor alleviates arthritis but exacerbates diabetes. Immunity 36, 731–741 (2012).
Zhang, F., Meng, G. & Strober, W. Interactions among the transcription factors Runx1, RORγt and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat. Immunol. 9, 1297–1306 (2008).
Loizou, L., Andersen, K. G. & Betz, A. G. Foxp3 interacts with c-Rel to mediate NF-κB repression. PLoS ONE 6, e18670 (2011).
Chen, C., Rowell, E. A., Thomas, R. M., Hancock, W. W. & Wells, A. D. Transcriptional regulation by Foxp3 is associated with direct promoter occupancy and modulation of histone acetylation. J. Biol. Chem. 281, 36828–36834 (2006).
Li, B. et al. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc. Natl Acad. Sci. USA 104, 4571–4576 (2007).
Tao, R. et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 13, 1299–1307 (2007). References 24 and 25 are landmark studies demonstrating that FOXP3 is post-translationally modified by lysine acetylation (reference 24), and that preserving this modification through HDAC inhibition can augment T reg cell function in models of colitis and allograft transplantation.
Pan, F. et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science 325, 1142–1146 (2009).
Fu, W. et al. A multiply redundant genetic switch 'locks in' the transcriptional signature of regulatory T cells. Nat. Immunol. 13, 972–980 (2012). This landmark study demonstrates that FOXP3 functions together with other cofactors to activate the expression of most of the T reg cell signature.
Bettini, M. L. et al. Loss of epigenetic modification driven by the Foxp3 transcription factor leads to regulatory T cell insufficiency. Immunity 36, 717–730 (2012).
Xiao, Y. et al. Histone acetyltransferase mediated regulation of FOXP3 acetylation and Treg function. Curr. Opin. Immunol. 22, 583–591 (2010).
Liu, Y. et al. Inhibition of p300 impairs Foxp3+ T regulatory cell function and promotes antitumor immunity. Nat. Med. 19, 1173–1177 (2013).
Arvey, A. et al. Inflammation-induced repression of chromatin bound by the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 15, 580–587 (2014).
DuPage, M. et al. The chromatin-modifying enzyme Ezh2 is critical for the maintenance of regulatory T cell identity after activation. Immunity 42, 227–238 (2015).
Josefowicz, S. Z., Lu, L. F. & Rudensky, A. Y. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30, 531–564 (2012).
Ouyang, W. et al. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat. Immunol. 11, 618–627 (2010).
Kim, H. P. & Leonard, W. J. CREB/ATF-dependent T cell receptor-induced Foxp3 gene expression: a role for DNA methylation. J. Exp. Med. 204, 1543–1551 (2007).
Zheng, Y. et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812 (2010). This study demonstrates that the composition, size and maintenance of the T reg cell population are controlled by Foxp3 CNS elements that are engaged in response to distinct cell-extrinsic or cell-intrinsic cues.
Ruan, Q. et al. Development of Foxp3+ regulatory T cells is driven by the c-Rel enhanceosome. Immunity 31, 932–940 (2009).
Feng, Y. et al. A mechanism for expansion of regulatory T-cell repertoire and its role in self-tolerance. Nature 528, 132–136 (2015).
Toker, A. et al. Active demethylation of the Foxp3 locus leads to the generation of stable regulatory T cells within the thymus. J. Immunol. 190, 3180–3188 (2013).
Ohkura, N. et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity 37, 785–799 (2012).
Polansky, J. K. et al. DNA methylation controls Foxp3 gene expression. Eur. J. Immunol. 38, 1654–1663 (2008).
Wang, L. et al. Mbd2 promotes Foxp3 demethylation and T-regulatory-cell function. Mol. Cell. Biol. 33, 4106–4115 (2013).
Yue, X. et al. Control of Foxp3 stability through modulation of TET activity. J. Exp. Med. 213, 377–397 (2016).
Nair, V. S., Song, M. H., Ko, M. & Oh, K. I. DNA demethylation of the Foxp3 enhancer is maintained through modulation of ten-eleven-translocation and DNA methyltransferases. Mol. Cells 39, 888–897 (2016). References 43 and 44 provide insights into the mechanisms that are involved in regulating the methylation state of the CNS2 region of the Foxp3 gene.
Lu, L. et al. All-trans retinoic acid promotes TGF-β-induced Tregs via histone modification but not DNA demethylation on Foxp3 gene locus. PLoS ONE 6, e24590 (2011).
Lal, G. et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J. Immunol. 182, 259–273 (2009).
Li, C., Ebert, P. J. & Li, Q. J. T cell receptor (TCR) and transforming growth factor β (TGF-β) signaling converge on DNA (cytosine-5)-methyltransferase to control forkhead box protein 3 (Foxp3) locus methylation and inducible regulatory T cell differentiation. J. Biol. Chem. 288, 19127–19139 (2013).
Schlenner, S. M., Weigmann, B., Ruan, Q., Chen, Y. & von Boehmer, H. Smad3 binding to the Foxp3 enhancer is dispensable for the development of regulatory T cells with the exception of the gut. J. Exp. Med. 209, 1529–1535 (2012).
Josefowicz, S. Z. et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature 482, 395–399 (2012). This study demonstrates the importance of CNS1 for pT reg cell generation and the maintenance of immune homeostasis at barrier sites.
Samstein, R. M., Josefowicz, S. Z., Arvey, A., Treuting, P. M. & Rudensky, A. Y. Extrathymic generation of regulatory T cells in placental mammals mitigates maternal–fetal conflict. Cell 150, 29–38 (2012).
Benoist, C. & Mathis, D. Treg cells, life history, and diversity. Cold Spring Harb. Perspect. Biol. 4, a007021 (2012).
Tai, X., Cowan, M., Feigenbaum, L. & Singer, A. CD28 costimulation of developing thymocytes induces Foxp3 expression and regulatory T cell differentiation independently of interleukin 2. Nat. Immunol. 6, 152–162 (2005).
Salomon, B. et al. B7/CD28 costimulation is essential for the homeostasis of the CD4+CD25+ immunoregulatory T cells that control autoimmune diabetes. Immunity 12, 431–440 (2000).
Willoughby, J. E. et al. Raf signaling but not the ERK effector SAP-1 is required for regulatory T cell development. J. Immunol. 179, 6836–6844 (2007).
Huehn, J., Polansky, J. K. & Hamann, A. Epigenetic control of FOXP3 expression: the key to a stable regulatory T-cell lineage? Nat. Rev. Immunol. 9, 83–89 (2009).
Thornton, A. M., Donovan, E. E., Piccirillo, C. A. & Shevach, E. M. Cutting edge: IL-2 is critically required for the in vitro activation of CD4+CD25+ T cell suppressor function. J. Immunol. 172, 6519–6523 (2004).
Chen, Q., Kim, Y. C., Laurence, A., Punkosdy, G. A. & Shevach, E. M. IL-2 controls the stability of Foxp3 expression in TGF-β-induced Foxp3+ T cells in vivo. J. Immunol. 186, 6329–6337 (2011).
Miyara, M. et al. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity 30, 899–911 (2009). This paper describes the functional heterogeneity that is present within the human FOXP3+ T reg cell population, and the relationship between CD25 levels and suppressive potency.
Laurence, A. et al. STAT3 transcription factor promotes instability of nTreg cells and limits generation of iTreg cells during acute murine graft-versus-host disease. Immunity 37, 209–222 (2012).
Nguyen, D. X. & Ehrenstein, M. R. Anti-TNF drives regulatory T cell expansion by paradoxically promoting membrane TNF–TNF-RII binding in rheumatoid arthritis. J. Exp. Med. 213, 1241–1253 (2016).
Liu, Y. et al. A critical function for TGF-β signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat. Immunol. 9, 632–640 (2008).
Curotto de Lafaille, M. A. & Lafaille, J. J. Natural and adaptive Foxp3+ regulatory T cells: more of the same or a division of labor? Immunity 30, 626–635 (2009).
Ouyang, W., Beckett, O., Ma, Q. & Li, M. O. Transforming growth factor-β signaling curbs thymic negative selection promoting regulatory T cell development. Immunity 32, 642–653 (2010).
Floess, S. et al. Epigenetic control of the Foxp3 locus in regulatory T cells. PLoS Biol. 5, e38 (2007).
Hill, J. A. et al. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi cells. Immunity 29, 758–770 (2008).
Xiao, S. et al. Retinoic acid increases Foxp3+ regulatory T cells and inhibits development of Th17 cells by enhancing TGF-β-driven Smad3 signaling and inhibiting IL-6 and IL-23 receptor expression. J. Immunol. 181, 2277–2284 (2008).
Kang, S. G., Lim, H. W., Andrisani, O. M., Broxmeyer, H. E. & Kim, C. H. Vitamin A metabolites induce gut-homing FoxP3+ regulatory T cells. J. Immunol. 179, 3724–3733 (2007).
Zhou, X. et al. Cutting edge: all-trans retinoic acid sustains the stability and function of natural regulatory T cells in an inflammatory milieu. J. Immunol. 185, 2675–2679 (2010).
Kaur, G., Goodall, J. C., Jarvis, L. B. & Hill Gaston, J. S. Characterisation of Foxp3 splice variants in human CD4+ and CD8+ T cells — identification of Foxp3Δ7 in human regulatory T cells. Mol. Immunol. 48, 321–332 (2010).
Ryder, L. R. et al. FoxP3 mRNA splice forms in synovial CD4+ T cells in rheumatoid arthritis and psoriatic arthritis. APMIS 120, 387–396 (2012).
Smith, E. L., Finney, H. M., Nesbitt, A. M., Ramsdell, F. & Robinson, M. K. Splice variants of human FOXP3 are functional inhibitors of human CD4+ T-cell activation. Immunology 119, 203–211 (2006).
Aarts-Riemens, T., Emmelot, M. E., Verdonck, L. F. & Mutis, T. Forced overexpression of either of the two common human Foxp3 isoforms can induce regulatory T cells from CD4+CD25− cells. Eur. J. Immunol. 38, 1381–1390 (2008).
Mailer, R. K. et al. IL-1β promotes Th17 differentiation by inducing alternative splicing of FOXP3. Sci. Rep. 5, 14674 (2015).
Xie, X. et al. The regulatory T cell lineage factor Foxp3 regulates gene expression through several distinct mechanisms mostly independent of direct DNA binding. PLoS Genet. 11, e1005251 (2015).
Lopes, J. E. et al. Analysis of FOXP3 reveals multiple domains required for its function as a transcriptional repressor. J. Immunol. 177, 3133–3142 (2006).
Li, B. et al. FOXP3 is a homo-oligomer and a component of a supramolecular regulatory complex disabled in the human XLAAD/IPEX autoimmune disease. Int. Immunol. 19, 825–835 (2007). This study is noteworthy as it reveals that human FOXP3 forms a large molecular complex that is dysfunctional in T cells from patients with IPEX syndrome.
Deng, G. et al. Molecular and biological role of the FOXP3 N-terminal domain in immune regulation by T regulatory/suppressor cells. Exp. Mol. Pathol. 93, 334–338 (2012).
Hancock, W. W. & Ozkaynak, E. Three distinct domains contribute to nuclear transport of murine Foxp3. PLoS ONE 4, e7890 (2009).
van Loosdregt, J. et al. Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood 115, 965–974 (2010).
Samanta, A. et al. TGF-β and IL-6 signals modulate chromatin binding and promoter occupancy by acetylated FOXP3. Proc. Natl Acad. Sci. USA 105, 14023–14027 (2008). References 79 and 80 show that acetylation promotes the stability and DNA-binding ability of FOXP3.
Xiao, Y. et al. Dynamic interactions between TIP60 and p300 regulate FOXP3 function through a structural switch defined by a single lysine on TIP60. Cell Rep. 7, 1471–1480 (2014).
Wang, L. et al. Ubiquitin-specific protease-7 inhibition impairs Tip60-dependent Foxp3+ T-regulatory cell function and promotes antitumor immunity. EbioMedicine 13, 99–112 (2016).
Gao, Z. et al. Synergy between IL-6 and TGF-β signaling promotes FOXP3 degradation. Int. J. Clin. Exp. Pathol. 5, 626–633 (2012).
Wang, L., de Zoeten, E. F., Greene, M. I. & Hancock, W. W. Immunomodulatory effects of deacetylase inhibitors: therapeutic targeting of FOXP3+ regulatory T cells. Nat. Rev. Drug Discov. 8, 969–981 (2009).
van Loosdregt, J. et al. Rapid temporal control of Foxp3 protein degradation by sirtuin-1. PLoS ONE 6, e19047 (2011).
Beier, U. H. et al. Sirtuin-1 targeting promotes Foxp3+ T-regulatory cell function and prolongs allograft survival. Mol. Cell. Biol. 31, 1022–1029 (2011).
Kwon, H. S. et al. Three novel acetylation sites in the Foxp3 transcription factor regulate the suppressive activity of regulatory T cells. J. Immunol. 188, 2712–2721 (2012).
Li, J. et al. Mammalian sterile 20-like kinase 1 (Mst1) enhances the stability of forkhead box P3 (Foxp3) and the function of regulatory T cells by modulating Foxp3 acetylation. J. Biol. Chem. 290, 30762–30770 (2015).
Du, X. et al. Mst1/Mst2 regulate development and function of regulatory T cells through modulation of Foxo1/Foxo3 stability in autoimmune disease. J. Immunol. 192, 1525–1535 (2014).
Sivan, A. et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015).
Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013).
Arpaia, N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013). References 91 and 92 demonstrate that the microbial production of SCFAs can positively influence FOXP3 upregulation and the generation of T reg cells.
Nie, H. et al. Phosphorylation of FOXP3 controls regulatory T cell function and is inhibited by TNF-α in rheumatoid arthritis. Nat. Med. 19, 322–328 (2013). This study reveals a function-augmenting phosphorylation modification of FOXP3 that is adversely affected by TNF.
Morawski, P. A., Mehra, P., Chen, C., Bhatti, T. & Wells, A. D. Foxp3 protein stability is regulated by cyclin-dependent kinase 2. J. Biol. Chem. 288, 24494–24502 (2013).
Chunder, N., Wang, L., Chen, C., Hancock, W. W. & Wells, A. D. Cyclin-dependent kinase 2 controls peripheral immune tolerance. J. Immunol. 189, 5659–5666 (2012).
Li, Z. et al. PIM1 kinase phosphorylates the human transcription factor FOXP3 at serine 422 to negatively regulate its activity under inflammation. J. Biol. Chem. 289, 26872–26881 (2014).
Deng, G. et al. Pim-2 kinase influences regulatory T cell function and stability by mediating Foxp3 protein N-terminal phosphorylation. J. Biol. Chem. 290, 20211–20220 (2015). References 94, 96 and 97 demonstrate how the phosphorylation of FOXP3 can limit its regulatory activity and T reg cell function.
Basu, S., Golovina, T., Mikheeva, T., June, C. H. & Riley, J. L. Cutting edge: Foxp3-mediated induction of Pim 2 allows human T regulatory cells to preferentially expand in rapamycin. J. Immunol. 180, 5794–5798 (2008).
Nakahira, K., Morita, A., Kim, N. S. & Yanagihara, I. Phosphorylation of FOXP3 by LCK downregulates MMP9 expression and represses cell invasion. PLoS ONE 8, e77099 (2013).
Metzger, M. B., Hristova, V. A. & Weissman, A. M. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 125, 531–537 (2012).
Ben-Neriah, Y. Regulatory functions of ubiquitination in the immune system. Nat. Immunol. 3, 20–26 (2002).
Dang, E. V. et al. Control of TH17/Treg balance by hypoxia-inducible factor 1. Cell 146, 772–784 (2011). This study highlights the importance of metabolic cues in T cell fate determination and shows that FOXP3 levels in developing T reg cells are susceptible to ubiquitin-mediated degradation.
Chen, Z. et al. The ubiquitin ligase Stub1 negatively modulates regulatory T cell suppressive activity by promoting degradation of the transcription factor Foxp3. Immunity 39, 272–285 (2013). This paper demonstrates the crucial role of the stress-activated STUB1–HSP70 complex in promoting FOXP3 downregulation and T reg cell inactivation.
van Loosdregt, J. et al. Stabilization of the transcription factor Foxp3 by the deubiquitinase USP7 increases Treg-cell-suppressive capacity. Immunity 39, 259–271 (2013). This study reveals a molecular mechanism by which the rapid temporal control of FOXP3 expression in T reg cells can be regulated by USP7, thereby modulating T reg cell numbers and function.
Chen, L. Wu, J., Pier, E., Zhao, Y. & Shen, Z. mTORC2–PKBα/Akt1 serine 473 phosphorylation axis is essential for regulation of FOXP3 stability by chemokine CCL3 in psoriasis. J. Invest. Dermatol. 133, 418–428 (2013).
Li, X. et al. CHIP promotes Runx2 degradation and negatively regulates osteoblast differentiation. J. Cell Biol. 181, 959–972 (2008).
Luo, W. et al. Hsp70 and CHIP selectively mediate ubiquitination and degradation of hypoxia-inducible factor (HIF)-1α but not HIF-2α. J. Biol. Chem. 285, 3651–3663 (2010).
Zhang, J. et al. Identification of the E3 deubiquitinase ubiquitin-specific peptidase 21 (USP21) as a positive regulator of the transcription factor GATA3. J. Biol. Chem. 288, 9373–9382 (2013).
Li, Y. et al. USP21 prevents the generation of T-helper-1-like Treg cells. Nat. Commun. 7, 13559 (2016).
Kirisako, T. et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25, 4877–4887 (2006).
Feuerer, M. et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med. 15, 930–939 (2009).
Burzyn, D. et al. A special population of regulatory T cells potentiates muscle repair. Cell 155, 1282–1295 (2013).
Wang, R. et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011).
Michalek, R. D. et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 186, 3299–3303 (2011). This study shows that distinct metabolic pathways are preferentially utilized by effector T cells and T reg cells, and that manipulating these pathways can alter T cell subset skewing and function.
Shi, L. Z. et al. HIF1α-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 208, 1367–1376 (2011).
Delgoffe, G. M. et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity 30, 832–844 (2009).
Beier, U. H. et al. Essential role of mitochondrial energy metabolism in Foxp3+ T-regulatory cell function and allograft survival. FASEB J. 29, 2315–2326 (2015).
Lee, J. H., Elly, C., Park, Y. & Liu, Y. C. E3 ubiquitin ligase VHL regulates hypoxia-inducible factor-1α to maintain regulatory T cell stability and suppressive capacity. Immunity 42, 1062–1074 (2015). This study shows that metabolic reprogramming in T reg cells can adversely affect their expression of FOXP3 and their suppressive function in an IFNγ-dependent manner.
Zeng, H. et al. mTORC1 couples immune signals and metabolic programming to establish Treg-cell function. Nature 499, 485–490 (2013).
De Rosa, V. et al. Glycolysis controls the induction of human regulatory T cells by modulating the expression of FOXP3 exon 2 splicing variants. Nat. Immunol. 16, 1174–1184 (2015). This paper introduces a possible connection between the metabolism of differentiating iT reg cells and the processes that are involved in the alternative splicing of the FOXP3 transcript.
Gerriets, V. A. et al. Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat. Immunol. 17, 1459–1466 (2016). This study demonstrates that the activation of T reg cells by TLR ligands can induce metabolic changes that disrupt their suppressive functions while promoting T reg cell population expansion; by contrast, FOXP3 directly regulates metabolic activity in T reg cells to restrict this TLR-dependent process.
Shrestha, S. et al. Treg cells require the phosphatase PTEN to restrain TH1 and TFH cell responses. Nat. Immunol. 16, 178–187 (2015).
Park, Y. et al. TSC1 regulates the balance between effector and regulatory T cells. J. Clin. Invest. 123, 5165–5178 (2013).
Ware, R. et al. Human CD8+ T lymphocyte clones specific for T cell receptor Vβ families expressed on autologous CD4+ T cells. Immunity 2, 177–184 (1995).
Kim, H. J. et al. CD8+ T regulatory cells express the Ly49 Class I MHC receptor and are defective in autoimmune prone B6-Yaa mice. Proc. Natl Acad. Sci. USA 108, 2010–2015 (2011).
Kiniwa, Y. et al. CD8+ Foxp3+ regulatory T cells mediate immunosuppression in prostate cancer. Clin. Cancer Res. 13, 6947–6958 (2007).
Mayer, C. T. et al. CD8+ Foxp3+ T cells share developmental and phenotypic features with classical CD4+ Foxp3+ regulatory T cells but lack potent suppressive activity. Eur. J. Immunol. 41, 716–725 (2011).
Le, D. T. et al. CD8+ Foxp3+ tumor infiltrating lymphocytes accumulate in the context of an effective anti-tumor response. Int. J. Cancer 129, 636–647 (2011). References 127 and 128 suggest that FOXP3 expression is seen in some CD8+ T cells that have a suppressive function.
Bendelac, A., Savage, P. B. & Teyton, L. The biology of NKT cells. Annu. Rev. Immunol. 25, 297–336 (2007).
Monteiro, M. et al. Identification of regulatory Foxp3+ invariant NKT cells induced by TGF-β. J. Immunol. 185, 2157–2163 (2010).
Manrique, S. Z. et al. Foxp3-positive macrophages display immunosuppressive properties and promote tumor growth. J. Exp. Med. 208, 1485–1499 (2011).
Mayer, C. T., Kuhl, A. A., Loddenkemper, C. & Sparwasser, T. Lack of Foxp3+ macrophages in both untreated and B16 melanoma-bearing mice. Blood 119, 1314–1315 (2012).
Put, S. et al. Macrophages have no lineage history of Foxp3 expression. Blood 119, 1316–1318 (2012). References 132 and 133 show that, contrary to an initial study, FOXP3 is not expressed by several macrophage types.
Devaud, C. et al. Foxp3 expression in macrophages associated with RENCA tumors in mice. PLoS ONE 9, e108670 (2014).
Rosser, E. C. & Mauri, C. Regulatory B cells: origin, phenotype, and function. Immunity 42, 607–612 (2015).
Yang, M., Rui, K., Wang, S. & Lu, L. Regulatory B cells in autoimmune diseases. Cell. Mol. Immunol. 10, 122–132 (2013).
Olkhanud, P. B. et al. Tumor-evoked regulatory B cells promote breast cancer metastasis by converting resting CD4+ T cells to T-regulatory cells. Cancer Res. 71, 3505–3515 (2011).
Noh, J., Noh, G., Kim, H. S., Kim, A. R. & Choi, W. S. Allergen-specific responses of CD19+CD5+Foxp3+ regulatory B cells (Bregs) and CD4+Foxp3+ regulatory T cell (Tregs) in immune tolerance of cow milk allergy of late eczematous reactions. Cell. Immunol. 274, 109–114 (2012).
Vadasz, Z. et al. The expansion of CD25highIL-10highFoxP3high B regulatory cells is in association with SLE disease activity. J. Immunol. Res. 2015, 254245 (2015).
de Andres, C. et al. New regulatory CD19+CD25+ B-cell subset in clinically isolated syndrome and multiple sclerosis relapse. Changes after glucocorticoids. J. Neuroimmunol. 270, 37–44 (2014).
Zuo, T. et al. FOXP3 is an X-linked breast cancer suppressor gene and an important repressor of the HER-2/ErbB2 oncogene. Cell 129, 1275–1286 (2007).
Wang, L. et al. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer Cell 16, 336–346 (2009).
Buckner, J. H. Mechanisms of impaired regulation by CD4+CD25+FOXP3+ regulatory T cells in human autoimmune diseases. Nat. Rev. Immunol. 10, 849–859 (2010).
Sarkar, S. & Fox, D. A. Regulatory T cell defects in rheumatoid arthritis. Arthritis Rheum. 56, 710–713 (2007).
Talaat, R. M., Mohamed, S. F., Bassyouni, I. H. & Raouf, A. A. Th1/Th2/Th17/Treg cytokine imbalance in systemic lupus erythematosus (SLE) patients: correlation with disease activity. Cytokine 72, 146–153 (2015).
Long, S. A. et al. Defects in IL-2R signaling contribute to diminished maintenance of FOXP3 expression in CD4+CD25+ regulatory T-cells of type 1 diabetic subjects. Diabetes 59, 407–415 (2010).
Di Ianni, M. et al. Tregs prevent GVHD and promote immune reconstitution in HLA-haploidentical transplantation. Blood 117, 3921–3928 (2011).
Martelli, M. F. et al. HLA-haploidentical transplantation with regulatory and conventional T-cell adoptive immunotherapy prevents acute leukemia relapse. Blood 124, 638–644 (2014).
Masteller, E. L. et al. Expansion of functional endogenous antigen-specific CD4+CD25+ regulatory T cells from nonobese diabetic mice. J. Immunol. 175, 3053–3059 (2005).
Roncarolo, M. G. & Battaglia, M. Regulatory T-cell immunotherapy for tolerance to self antigens and alloantigens in humans. Nat. Rev. Immunol. 7, 585–598 (2007).
Bluestone, J. A. et al. Type 1 diabetes immunotherapy using polyclonal regulatory T cells. Sci. Transl Med. 7, 315ra189 (2015).
Marek-Trzonkowska, N. et al. Therapy of type 1 diabetes with CD4+CD25highCD127-regulatory T cells prolongs survival of pancreatic islets — results of one year follow-up. Clin. Immunol. 153, 23–30 (2014). References 151 and 152 report encouraging results from trials using the adoptive transfer of T reg cells as a therapy in patients with diabetes; the data suggest that ex vivo -expanded T reg cells can be used to treat autoimmune diseases.
Desreumaux, P. et al. Safety and efficacy of antigen-specific regulatory T-cell therapy for patients with refractory Crohn's disease. Gastroenterology 143, 1207–1217 (2012).
Nishikawa, H. & Sakaguchi, S. Regulatory T cells in cancer immunotherapy. Curr. Opin. Immunol. 27, 1–7 (2014).
Liu, C., Workman, C. J. & Vignali, D. A. Targeting regulatory T cells in tumors. FEBS J. 283, 2731–2748 (2016).
Rech, A. J. et al. CD25 blockade depletes and selectively reprograms regulatory T cells in concert with immunotherapy in cancer patients. Sci. Transl Med. 4, 134ra162 (2012). This study shows the potential benefits of targeting T reg cells in patients with cancer as a means to augment antitumour immunity.
Sugiyama, D. et al. Anti-CCR4 mAb selectively depletes effector-type FoxP3+CD4+ regulatory T cells, evoking antitumor immune responses in humans. Proc. Natl Acad. Sci. USA 110, 17945–17950 (2013).
Acknowledgements
The laboratory of F.P. is supported by grants from the Bloomberg–Kimmel Institute of Johns Hopkins University (Maryland, USA), the US National Institutes of Health (grants RO1AI099300 and RO1AI089830) and the US Department of Defense (grant PC130767). F.P. is the recipient of an Established Investigator Award from the Melanoma Research Alliance (Washington, USA). The research of J.B. is supported by a grant from the Roswell Park Alliance Foundation and by the US National Cancer Institute (grant P30CA016056). The laboratory of L.L. is supported by the National Natural Science Fund of China (grants 81571564 and 81522020), the 863 Young Scientists Special Fund (grant SS2015AA020932) and the Natural Science Foundation of China (grant 91442117). The authors thank S. Newman, A. Lebid, X. Ni, P. Wei and A. Ramaswamy for help with preparing the figures and for critically reviewing the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Lu, L., Barbi, J. & Pan, F. The regulation of immune tolerance by FOXP3. Nat Rev Immunol 17, 703–717 (2017). https://doi.org/10.1038/nri.2017.75
Published:
Issue Date:
DOI: https://doi.org/10.1038/nri.2017.75
This article is cited by
-
Loss of chromosome Y in regulatory T cells
BMC Genomics (2024)
-
The regulation and differentiation of regulatory T cells and their dysfunction in autoimmune diseases
Nature Reviews Immunology (2024)
-
Immune cell profiles of idiopathic inflammatory myopathy patients expressed anti-aminoacyl tRNA synthetase or anti-melanoma differentiation-associated gene 5 autoantibodies
BMC Immunology (2023)
-
Identification of key transcription factors and their functional role involved in Salmonella typhimurium infection in chicken using integrated transcriptome analysis and bioinformatics approach
BMC Genomics (2023)
-
CTLA4+CD4+CXCR5−FOXP3+ T cells associate with unfavorable outcome in patients with chronic HBV infection
BMC Immunology (2023)