Key Points
-
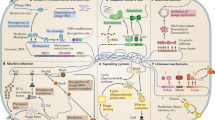
The Toll-like receptors (TLRs) and retinoic-acid-inducible gene I (RIG-I)-like receptors (RLRs) both participate in sensing viral infection, but do so in different cells and fill different 'niches'. Whereas the RLRs detect viral nucleic acids within the cytoplasm of infected cells and are therefore cell-autonomous sensors, the TLRs can allow non-infected cells to detect infection in other cells, perhaps even in cells at a considerable distance.
-
The biochemical pathways for TLR-induced and RLR-induced signalling have been largely deciphered, and are described here. These pathways are both capable of activating nuclear factor-κB (NF-κB) and interferon-regulatory factor 3 (IRF3) and/or IRF7, which are central regulators of inflammatory-cytokine induction and type I interferon (IFN) production, respectively.
-
A division of labour exists among immune cells, in that plasmacytoid dendritic cells (pDCs), conventional DCs, natural killer (NK) cells, lymphocytes and other cell types all mediate host resistance to viral infection, and each cell type has non-redundant functions in the containment of specific viral infections.
-
For a defined β-herpesvirus infection, multiple classes of DCs have a vital sensory role, whereas NK cells are of key importance as effectors. Numerous viral-evasion mechanisms have been identified, many mutations in the 'resistome' cause susceptibility to infection and many other mutations yet to be identified cause resistance to infection.
-
Studies using the invertebrate model Drosophila melanogaster show that insects sense viral infection and respond by inducing gene expression. Some of the genes that are induced by viral infection are controlled by the Janus kinase–signal transducer and activator of transcription (JAK–STAT) pathway, establishing a parallel with IFN signalling.
-
RNA interference is an intrinsic defence against viral infections in invertebrates. The RNaseIII enzyme Dicer-2 processes viral double-stranded RNAs, and generate small interfering RNAs that guide the RNaseH-like enzyme Argonaute-2 to viral RNA molecules, providing sequence-specific immunity.
Abstract
As machines that reprogramme eukaryotic cells to suit their own purposes, viruses present a difficult problem for multicellular hosts, and indeed, have become one of the central pre-occupations of the immune system. Unable to permanently outpace individual viruses in an evolutionary footrace, higher eukaryotes have evolved broadly active mechanisms with which to sense viruses and suppress their proliferation. These mechanisms have recently been elucidated by a combination of forward and reverse genetic methods. Some of these mechanisms are clearly ancient, whereas others are relatively new. All are remarkably adept at discriminating self from non-self, and allow the host to cope with what might seem an impossible predicament.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Morens, D. M., Folkers, G. K. & Fauci, A. S. The challenge of emerging and re-emerging infectious diseases. Nature 430, 242–249 (2004).
Kurt-Jones, E. A. et al. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nature Immunol. 1, 398–401 (2000).
Haynes, L. M. et al. Involvement of toll-like receptor 4 in innate immunity to respiratory syncytial virus. J. Virol. 75, 10730–10737 (2001).
Georgel, P. et al. Vesicular stomatitis virus glycoprotein G activates a specific antiviral Toll-like receptor 4-dependent pathway. Virology 362, 304–313 (2007).
Alexopoulou, L., Holt, A. C., Medzhitov, R. & Flavell, R. A. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413, 732–738 (2001).
Diebold, S. S., Kaisho, T., Hemmi, H., Akira, S. & Reis e Sousa, C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303, 1529–1531 (2004).
Heil, F. et al. Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science 303, 1481–1482 (2004).
Hemmi, H. et al. A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 (2000).
Schulz, O. et al. Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature 433, 887–892 (2005).
Tabeta, K. et al. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc. Natl Acad. Sci. USA 101, 3516–3521 (2004).
Wang, T. et al. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nature Med. 10, 1366–1373 (2004).
Cella, M. et al. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nature Med. 5, 919–923 (1999).
Blasius, A. et al. A cell surface molecule selectively expressed on murine natural interferon producing cells that blocks secretion of interferon-α. Blood 103, 4201–4206 (2003).
De Bouteiller, O. et al. Recognition of double stranded RNA by human toll like receptor 3 and downstream receptor signaling requires multimerisation and an acidic pH. J. Biol. Chem. 280, 38133–38145 (2005).
Rutz, M. et al. Toll-like receptor 9 binds single-stranded CpG-DNA in a sequence- and pH-dependent manner. Eur. J. Immunol. 34, 2541–2550 (2004).
MacFarlane, D. E. & Manzel, L. Antagonism of immunostimulatory CpG-oligodeoxynucleotides by quinacrine, chloroquine, and structurally related compounds. J. Immunol. 160, 1122–1131 (1998).
Lee, J. et al. Molecular basis for the immunostimulatory activity of guanine nucleoside analogs: activation of Toll-like receptor 7. Proc. Natl Acad. Sci. USA 100, 6646–6651 (2003).
Tabeta, K. et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nature Immunol. 7, 156–164 (2006).
Casrouge, A. et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science 314, 308–312 (2006).
Brinkmann, M. M. et al. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J. Cell Biol. 177, 265–275 (2007).
Hoebe, K. et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature 424, 743–748 (2003).
Yamamoto, M. et al. Role of adapter TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science 301, 640–643 (2003).
Meylan, E. et al. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-κB activation. Nature Immunol. 5, 503–507 (2004).
Sato, S. et al. Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-κB and IFN-regulatory factor-3, in the Toll-like receptor signaling. J. Immunol. 171, 4304–4310 (2003).
Beutler, B. et al. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu. Rev. Immunol. 24, 353–389 (2006).
Hacker, H. et al. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 439, 204–207 (2006).
Oganesyan, G. et al. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 439, 208–211 (2006).
Honda, K. & Taniguchi, T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nature Rev. Immunol. 6, 644–658 (2006).
Honda, K. et al. Role of a transductional–transcriptional processor complex involving MyD88 and IRF-7 in Toll-like receptor signaling. Proc. Natl Acad. Sci. USA 101, 15416–15421 (2004).
Hoshino, K. et al. IκB kinase-α is critical for interferon-α production induced by Toll-like receptors 7 and 9. Nature 440, 949–953 (2006).
Kawai, T. et al. Interferon-α induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nature Immunol. 5, 1061–1068 (2004).
Matsui, K. et al. Cutting edge: Role of TANK-binding kinase 1 and inducible IκB kinase in IFN responses against viruses in innate immune cells. J. Immunol. 177, 5785–5789 (2006).
Shinohara, M. L. et al. Osteopontin expression is essential for interferon-α production by plasmacytoid dendritic cells. Nature Immunol. 7, 498–506 (2006).
Faust, M., Hastings, K. E. & Millward, S. m7G5′ppp5′GmptcpUp at the 5′ terminus of reovirus messenger RNA. Nucleic Acids Res. 2, 1329–1343 (1975).
Rose, J. K. Heterogneeous 5′-terminal structures occur on vesicular stomatitis virus mRNAs. J. Biol. Chem. 250, 8098–8104 (1975).
Moss, B., Keith, J. M., Gershowitz, A., Ritchey, M. B. & Palese, P. Common sequence at the 5′ ends of the segmented RNA genomes of influenza A and B viruses. J. Virol. 25, 312–318 (1978).
Yoneyama, M. et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nature Immunol. 5, 730–737 (2004). This paper describes the initial identification of RIG-I as an intracellular viral dsRNA detector, the activation of which leads to the production of type I IFNs.
Kang, D. C. et al. mda-5: an interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl Acad. Sci. USA 99, 637–642 (2002).
Yoneyama, M. et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175, 2851–2858 (2005).
Rothenfusser, S. et al. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 175, 5260–5268 (2005).
Kato, H. et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23, 19–28 (2005).
Kato, H. et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105 (2006). In this paper, the authors generated mice that lacked RIG-I and MDA5, and showed that these two helicases recognize different RNA viruses.
Gitlin, L. et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl Acad. Sci. USA 103, 8459–8464 (2006).
Hornung, V. et al. 5′-triphosphate RNA is the ligand for RIG-I. Science 314, 994–997 (2006).
Pichlmair, A. et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314, 997–1001 (2006).
Marques, J. T. et al. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nature Biotechnol. 24, 559–565 (2006).
Gack, M. U. et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920 (2007).
Venkataraman, T. et al. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 178, 6444–6455 (2007).
Kawai, T. et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nature Immunol. 6, 981–988 (2005).
Meylan, E. et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437, 1167–1172 (2005).
Seth, R. B., Sun, L., Ea, C. K. & Chen, Z. J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell 122, 669–682 (2005).
Xu, L. G. et al. VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol. Cell 19, 727–740 (2005). References 49–52 identify a novel CARD-containing protein that functions as an adaptor for RIG-I- and MDA5-mediated signalling.
Kumar, H. et al. Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med. 203, 1795–1803 (2006).
Sun, Q. et al. The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24, 633–642 (2006).
Balachandran, S., Thomas, E. & Barber, G. N. A FADD-dependent innate immune mechanism in mammalian cells. Nature 432, 401–405 (2004).
Takahashi, K. et al. Roles of caspase-8 and caspase-10 in innate immune responses to double-stranded RNA. J. Immunol. 176, 4520–4524 (2006).
Li, K. et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl Acad. Sci. USA 102, 2992–2997 (2005).
Loo, Y. M. et al. Viral and therapeutic control of IFN-β promoter stimulator 1 during hepatitis C virus infection. Proc. Natl Acad. Sci. USA 103, 6001–6006 (2006).
Stack, J. et al. Vaccinia virus protein A46R targets multiple Toll-like-interleukin-1 receptor adaptors and contributes to virulence. J. Exp. Med. 201, 1007–1018 (2005).
Kubota, A., Kubota, S., Farrell, H. E., vis-Poynter, N. & Takei, F. Inhibition of NK cells by murine CMV-encoded class I MHC homologue m144. Cell. Immunol. 191, 145–151 (1999).
Lodoen, M. et al. NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J. Exp. Med. 197, 1245–1253 (2003).
Krmpotic, A. et al. NK cell activation through the NKG2D ligand MULT-1 is selectively prevented by the glycoprotein encoded by mouse cytomegalovirus gene m145. J. Exp. Med. 201, 211–220 (2005).
Hasan, M. et al. Selective down-regulation of the NKG2D ligand H60 by mouse cytomegalovirus m155 glycoprotein. J. Virol. 79, 2920–2930 (2005).
Kleijnen, M. F. et al. A mouse cytomegalovirus glycoprotein, gp34, forms a complex with folded class I MHC molecules in the ER which is not retained but is transported to the cell surface. EMBO J. 16, 685–694 (1997). This paper shows a unique and sophisticated evasion mechanism aimed at the CTL response.
Bubeck, A. et al. The glycoprotein gp48 of murine cytomegalovirusL proteasome-dependent cytosolic dislocation and degradation. J. Biol. Chem. 277, 2216–2224 (2002).
Krmpotic, A. et al. The immunoevasive function encoded by the mouse cytomegalovirus gene m152 protects the virus against T cell control in vivo. J. Exp. Med. 190, 1285–1296 (1999).
Yewdell, J. W. & Hill, A. B. Viral interference with antigen presentation. Nature Immunol. 3, 1019–1025 (2002).
Reddehase, M. J. Antigens and immunoevasins: opponents in cytomegalovirus immune surveillance. Nature Rev. Immunol. 2, 831–844 (2002).
Loewendorf, A. et al. Identification of a mouse cytomegalovirus gene selectively targeting CD86 expression on antigen-presenting cells. J. Virol. 78, 13062–13071 (2004).
Menard, C. et al. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J. Virol. 77, 5557–5570 (2003).
Janssen, E. et al. Efficient T cell activation via a Toll–interleukin 1 receptor-independent pathway. Immunity 24, 787–799 (2006).
Valchanova, R. S., Picard-Maureau, M., Budt, M. & Brune, W. Murine cytomegalovirus m142 and m143 are both required to block protein kinase R-mediated shutdown of protein synthesis. J. Virol. 80, 10181–10190 (2006).
Spencer, J. V. et al. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J. Virol. 76, 1285–1292 (2002).
Welsh, R. M., O'Donnell, C. L. & Shultz, L. D. Antiviral activity of NK 1.1+ natural killer cells in C57BL/6 scid mice infected with murine cytomegalovirus. Nature Immunol. 13, 239–245 (1994).
Bukowski, J. F., Woda, B. A., Habu, S., Okumura, K. & Welsh, R. M. Natural killer cell depletion enhances virus synthesis and virus-induced hepatitis in vivo. J. Immunol. 131, 1531–1538 (1983).
Bukowski, J. F., Woda, B. A. & Welsh, R. M. Pathogenesis of murine cytomegalovirus infection in natural killer cell-depleted mice. J. Virol. 52, 119–128 (1984).
Scalzo, A. A., Fitzgerald, N. A., Simmons, A., La Vista, A. B. & Shellam, G. R. Cmv-1, a genetic locus that controls murine cytomegalovirus replication in the spleen. J. Exp. Med. 171, 1469–1483 (1990).
Brown, M. G. et al. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science 292, 934–937 (2001).
Lee, S. H. et al. Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nature Genet. 28, 42–45 (2001). References 78 and 79 describe classical positional-cloning efforts that independently established the importance of Ly49H in sensing MCMV infection.
Smith, H. R. et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl Acad. Sci. USA 99, 8826–8831 (2002).
Arase, H., Mocarski, E. S., Campbell, A. E., Hill, A. B. & Lanier, L. L. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 296, 1323–1326 (2002). Showed that Ly49i is important as an inhibitory receptor that increases susceptibility to MCMV in 129-strain mice.
Tomasello, E. & Vivier, E. KARAP/DAP12/TYROBP: three names and a multiplicity of biological functions. Eur. J. Immunol. 35, 1670–1677 (2005).
Lanier, L. L., Corliss, B. C., Wu, J., Leong, C. & Phillips, J. H. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature 391, 703–707 (1998).
McVicar, D. W. et al. DAP12-mediated signal transduction in natural killer cells. A dominant role for the Syk protein-tyrosine kinase. J. Biol. Chem. 273, 32934–32942 (1998).
Crozat, K. et al. Analysis of the MCMV resistome by ENU mutagenesis. Mamm. Genome 17, 398–406 (2006).
Hoebe, K. et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature 424, 743–748 (2003).
Allman, D. et al. Ikaros is required for plasmacytoid dendritic cell differentiation. Blood 108, 4025–4034 (2006).
Krug, A. et al. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity 21, 107–119 (2004).
Andrews, D. M., Scalzo, A. A., Yokoyama, W. M., Smyth, M. J. & Degli-Esposti, M. A. Functional interactions between dendritic cells and NK cells during viral infection. Nature Immunol. 4, 175–181 (2003).
van Dommelen, S. L. et al. Perforin and granzymes have distinct roles in defensive immunity and immunopathology. Immunity 25, 835–848 (2006).
Stinchcombe, J., Bossi, G. & Griffiths, G. M. Linking albinism and immunity: the secrets of secretory lysosomes. Science 305, 55–59 (2004).
Ward, D. M. et al. Use of expression constructs to dissect the functional domains of the CHS/beige protein: identification of multiple phenotypes. Traffic 4, 403–415 (2003).
Ward, D. M., Griffiths, G. M., Stinchcombe, J. C. & Kaplan, J. Analysis of the lysosomal storage disease Chediak-Higashi syndrome. Traffic 1, 816–822 (2000).
Tchernev, V. T. et al. The Chediak–Higashi protein interacts with SNARE complex and signal transduction proteins. Mol. Med. 8, 56–64 (2002).
Clark, R. & Griffiths, G. M. Lytic granules, secretory lysosomes and disease. Curr. Opin. Immunol. 15, 516–521 (2003).
Menasche, G. et al. Mutations in RAB27A cause Griscelli syndrome associated with haemophagocytic syndrome. Nature Genet. 25, 173–176 (2000).
Feldmann, J. et al. Munc13–4 is essential for cytolytic granules fusion and is mutated in a form of familial hemophagocytic lymphohistiocytosis (FHL3). Cell 115, 461–473 (2003).
Crozat, K. et al. Jinx, an MCMV susceptibility phenotype caused by disruption of Unc13d: a mouse model of type 3 familial hemophagocytic lymphohistiocytosis. J. Exp. Med. 204, 853–863 (2007). An example of ENU mutagenesis used to create a novel MCMV immunodeficiency phenotype, with subsequent identification of the mutation in an important effector gene that had not previously been targeted.
Stow, J. L., Manderson, A. P. & Murray, R. Z. SNAREing immunity: the role of SNAREs in the immune system. Nature Rev. Immunol. 6, 919–929 (2006).
Hong, W. Cytotoxic T lymphocyte exocytosis: bring on the SNAREs! Trends Cell Biol. 15, 644–650 (2005).
Croker, B. A. et al. ATP-sensitive potassium channels mediate survival during infection in mammals and insects. Nature Genet. (in the press).
Dighe, A. et al. Requisite H2k role in NK cell-mediated resistance in acute murine cytomegalovirus-infected MA/My mice. J. Immunol. 175, 6820–6828 (2005).
Desrosiers, M. P. et al. Epistasis between mouse Klra and major histocompatibility complex class I loci is associated with a new mechanism of natural killer cell-mediated innate resistance to cytomegalovirus infection. Nature Genet. 37, 593–599 (2005).
Hoffmann, J. A. The immune response of Drosophila. Nature 426, 33–38 (2003).
Cherry, S. & Perrimon, N. Entry is a rate-limiting step for viral infection in a Drosophila melanogaster model of pathogenesis. Nature Immunol. 5, 81–87 (2004).
Cherry, S. et al. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 19, 445–452 (2005).
Sabatier, L. et al. Pherokine-2 and -3: two Drosophila molecules related to pheromone/odor-binding proteins induced by viral and bacterial infections. Eur. J. Biochem. 270, 3398–3407 (2003).
Dostert, C. et al. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nature Immunol. 6, 946–953 (2005).
Agaisse, H. & Perrimon, N. The roles of JAK/STAT signaling in Drosophila immune responses. Immunol. Rev. 198, 72–82 (2004).
Lin, C. C. et al. Characterization of two mosquito STATs, AaSTAT and CtSTAT. Differential regulation of tyrosine phosphorylation and DNA binding activity by lipopolysaccharide treatment and by Japanese encephalitis virus infection. J. Biol. Chem. 279, 3308–3317 (2004).
Liu, W. J., Chang, Y. S., Wang, A. H., Kou, G. H. & Lo, C. F. White spot syndrome virus annexes a shrimp STAT to enhance expression of the immediate-early gene ie1. J. Virol. 81, 1461–1471 (2007).
Thoetkiattikul, H., Beck, M. H. & Strand, M. R. Inhibitor κB-like proteins from a polydnavirus inhibit NF-κB activation and suppress the insect immune response. Proc. Natl Acad. Sci. USA 102, 11426–11431 (2005).
Zambon, R. A., Nandakumar, M., Vakharia, V. N. & Wu, L. P. The Toll pathway is important for an antiviral response in Drosophila. Proc. Natl Acad. Sci. USA 102, 7257–7262 (2005).
Lee, Y. S. et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117, 69–81 (2004).
Galiana-Arnoux, D., Dostert, C., Schneemann, A., Hoffmann, J. A. & Imler, J. L. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nature Immunol. 7, 590–597 (2006).
Wang, X. H. et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science 312, 452–454 (2006).
van Rij, R. P. et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 20, 2985–2995 (2006). References 112–117 establish that RNAi has a crucial role in the control of RNA virus infection in flies, and that the RNAi machinery provides sequence-specific immunity, specifically degrading viral RNAs.
Zambon, R. A., Vakharia, V. N. & Wu, L. P. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell. Microbiol. 8, 880–889 (2006).
Obbard, D. J., Jiggins, F. M., Halligan, D. L. & Little, T. J. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 16, 580–585 (2006).
Li, H., Li, W. X. & Ding, S. W. Induction and suppression of RNA silencing by an animal virus. Science 296, 1319–1321 (2002).
Chao, J. A. et al. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nature Struct. Mol. Biol. 12, 952–957 (2005). The authors provide a detailed structural analysis of the first suppressor of RNAi identified in an animal virus.
Bucheton, A. The relationship between the flamenco gene and gypsy in Drosophila: how to tame a retrovirus. Trends Genet. 11, 349–353 (1995).
Brennecke, J. et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103 (2007).
Pelisson, A., Sarot, E., Payen-Groschene, G. & Bucheton, A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J. Virol. 81, 1951–1960 (2007).
Carmell, M. A. et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12, 503–514 (2007).
Houwing, S. et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129, 69–82 (2007).
Aravin, A. A., Sachidanandam, R., Girard, A., Fejes-Toth, K. & Hannon, G. J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744–747 (2007).
Deleris, A. et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313, 68–71 (2006).
Dunoyer, P., Himber, C. & Voinnet, O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nature Genet. 37, 1356–1360 (2005).
Roignant, J. Y. et al. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA 9, 299–308 (2003).
Saleh, M. C. et al. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nature Cell Biol. 8, 793–802 (2006).
Lecellier, C. H. et al. A cellular microRNA mediates antiviral defense in human cells. Science 308, 557–560 (2005).
Otsuka, M et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer-1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 27, 123–134 (2007).
Taganov, K. D., Boldin, M. P. & Baltimore, D. MicroRNAs and immunity: tiny players in a big field. Immunity 26, 133–137 (2007).
Cullen, B. R. Is RNA interference involved in intrinsic antiviral immunity in mammals? Nature Immunol. 7, 563–567 (2006).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Acknowledgements
We thank M. Hashimoto and B. Layton for their assistance in completing this manuscript, and C. Hetru for discussions and preparation of Fig. 5. This work was supported in part by grants from the Special Coordination Funds of the Japanese Ministry of Education, Culture, Sports, Science and Technology, and from the 21st Century Center of Excellence Program of Japan, and by the National Institutes of Health, USA (P01 AI070,167).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Cross-presentation
-
The ability of certain antigen-presenting cells to load peptides that are derived from exogenous antigens onto MHC class I molecules. This property is atypical, because most cells exclusively present peptides from their endogenous proteins on MHC class I molecules. Cross-presentation is essential for the initiation of immune responses to viruses that do not infect antigen-presenting cells.
- Plasmacytoid dendritic cells
-
(pDCs). A subset of DCs that was named 'plasmacytoid' because their appearance under the microscope is similar to that of plasmablasts. In humans, these DCs can be derived from lineage (Lin)− haematopoietic stem cells from the peripheral blood. These DCs are the main producers of type I interferons in response to viral infections.
- ENU-induced mutation
-
A point mutation that is induced by the alkylating agent N-ethyl-N-nitrosourea (ENU).
- Ubiquitylation
-
The attachment of the small protein ubiquitin to lysine residues that are present in other proteins; this often tags these proteins for rapid cellular degradation.
- Myeloid dendritic cells
-
A subset of CD8α− dendritic cells that might be important for initiating vigorous immune responses.
- SCID mice
-
(Severe combined immunodeficient mice). A naturally occurring mouse mutant with SCID disease owing to an inability to rearrange antigen-receptor-chain genes.
- Quantitative trait locus
-
A locus that specifies a phenotypic difference between two different strains of mice, which may be distinguished by millions of genetic differences in all.
- Hypomorphic
-
A type of mutation in which either the altered gene product has a decreased level of activity or the wild-type gene product is expressed at a decreased level.
- Ikaros family
-
A family of zinc-finger-containing transcription factors. These factors are pleiotropic regulators of haematopoiesis and are required for the generation of lymphocyte and dendritic-cell lineages, as well as lymph nodes and Peyer's patches.
- Resistome
-
The set of all genes with non-redundant function in the resistance to a particular microbial agent.
- SNARE proteins
-
(Soluble-N-ethylmaleimide-sensitive-factor accessory-protein receptor proteins). A class of proteins that is required for membrane fusion events that occur in the course of vesicle trafficking and secretion.
- Orthologous genes
-
Genes that are present in a different species but are derived from a common ancestral gene.
- Haemolymph
-
The insect equivalent of blood.
Rights and permissions
About this article
Cite this article
Beutler, B., Eidenschenk, C., Crozat, K. et al. Genetic analysis of resistance to viral infection. Nat Rev Immunol 7, 753–766 (2007). https://doi.org/10.1038/nri2174
Issue Date:
DOI: https://doi.org/10.1038/nri2174
This article is cited by
-
Secreted LRPAP1 binds and triggers IFNAR1 degradation to facilitate virus evasion from cellular innate immunity
Signal Transduction and Targeted Therapy (2023)
-
Host genetic susceptibility to viral infections: the role of type I interferon induction
Genes & Immunity (2020)
-
Transcription modulation by CDK9 regulates inflammatory genes and RIPK3-MLKL-mediated necroptosis in periodontitis progression
Scientific Reports (2019)
-
Dengue virus causes changes of MicroRNA-genes regulatory network revealing potential targets for antiviral drugs
BMC Systems Biology (2018)
-
IFITM3 inhibits virus-triggered induction of type I interferon by mediating autophagosome-dependent degradation of IRF3
Cellular & Molecular Immunology (2018)