Key Points
-
Genome-wide association (GWA) studies have dramatically influenced our ability to identify genetic risk factors for complex immune-mediated diseases.
-
We provide the information necessary for immunologists to better understand the rapid advances in our knowledge of genetic variation and in the technologies to probe this variation, which have enabled GWA studies. In addition, both the analytical challenges and the strengths of these types of studies are discussed.
-
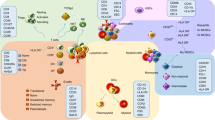
GWA studies of immune-mediated diseases, including Crohn's disease, multiple sclerosis, rheumatoid arthritis and systemic lupus erythematosus, have enabled the discovery of previously unknown factors that influence disease susceptibility and/or pathology.
-
Knowledge gained from these GWA studies can reveal how previously unrecognized biological pathways might contribute to disease pathogenesis. This research points toward the involvement of some pathways that are specific to one immune-mediated disease and others that are common to several different diseases.
-
Despite the success of GWA studies, several challenges lie ahead: how can we complete the mechanistic picture of how genetic variation in multiple different genes leads to disease development? How will these data impact the design of future immunology studies, and how can we apply this knowledge to clinical practice in the future?
Abstract
Given the recent explosion of genetic discoveries, 2007 is becoming known to human geneticists as the year of genome-wide association studies. In fact, more genetic risk factors for common diseases were identified in this one year than had been collectively reported before 2007. In particular, 2007 witnessed the discovery of many genes that influence susceptibility to individual immune-mediated diseases, as well as other genes that are associated with susceptibility to more than one disease. Although much work remains to be done, in this Review we discuss what effect these studies are having on our understanding of disease pathogenesis and their potential impact on future immunology studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Rioux, J. D. & Abbas, A. K. Paths to understanding the genetic basis of autoimmune disease. Nature 435, 584–589 (2005).
Risch, N. & Merikangas, K. The future of genetic studies of complex human diseases. Science 273, 1516–1517 (1996).
Hirschhorn, J. N., Lohmueller, K., Byrne, E. & Hirschhorn, K. A comprehensive review of genetic association studies. Genet. Med. 4, 45–61 (2002).
Kruglyak, L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nature Genet. 22, 139–144 (1999).
Wang, D. G. et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280, 1077–1082 (1998).
Frazer, K. A. et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (2007).
The international HapMap project. Nature 426, 789–796 (2003).
Daly, M. J., Rioux, J. D., Schaffner, S. F., Hudson, T. J. & Lander, E. S. High-resolution haplotype structure in the human genome. Nature Genet. 29, 229–232 (2001).
Redon, R. et al. Global variation in copy number in the human genome. Nature 444, 444–454 (2006).
Hirschhorn, J. N. & Daly, M. J. Genome-wide association studies for common diseases and complex traits. Nature Rev. Genet. 6, 95–108 (2005).
Wang, W. Y., Barratt, B. J., Clayton, D. G. & Todd, J. A. Genome-wide association studies: theoretical and practical concerns. Nature Rev. Genet. 6, 109–118 (2005).
Balding, D. J. A tutorial on statistical methods for population association studies. Nature Rev. Genet. 7, 781–791 (2006).
Chanock, S. J. et al. Replicating genotype-phenotype associations. Nature 447, 655–660 (2007).
Klein, R. J. et al. Complement factor H polymorphism in age-related macular degeneration. Science 308, 385–389 (2005).
Libioulle, C. et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 3, e58 (2007).
Wellcome trust case control consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Nath, S. K. et al. A nonsynonymous functional variant in integrin-αM (encoded by ITGAM) is associated with systemic lupus erythematosus. Nature Genet. 40, 152–154 (2008).
Duerr, R. H. et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006).
Parkes, M. et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nature Genet. 39, 830–832 (2007).
Rioux, J. D. et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nature Genet. 39, 596–604 (2007).
Hafler, D. A. et al. Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 357, 851–862 (2007).
Plenge, R. M. et al. TRAF1–C5 as a risk locus for rheumatoid arthritis — a genomewide study. N. Engl. J. Med. 357, 1199–1209 (2007).
Todd, J. A. et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nature Genet. 39, 857–864 (2007).
van Heel, D. A. et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nature Genet. 39, 827–829 (2007).
Hom, G. et al. Association of systemic lupus erythematosus with C8orf13–BLK and ITGAM–ITGAX. N. Engl. J. Med. 358, 900–909 (2008).
Zhernakova, A. et al. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am. J. Hum. Genet. 81, 1284–1288 (2007).
Barrett, J. C. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nature Genetics 26 June 2008 (doi: 10.1038/ng.175)
Maas, K. et al. Cutting Edge: molecular portrait of human autoimmune disease. J. Immunol. 169, 5–9 (2002).
Hugot, J. P. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411, 599–603 (2001).
Ogura, Y. et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411, 603–606 (2001).
Rioux, J. D. et al. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nature Genet. 29, 223–228 (2001).
Kanneganti, T. D., Lamkanfi, M. & Nunez, G. Intracellular NOD-like receptors in host defense and disease. Immunity 27, 549–559 (2007).
Kim, Y. G. et al. The cytosolic sensors Nod1 and Nod2 are critical for bacterial recognition and host defense after exposure to Toll-like receptor ligands. Immunity 28, 246–257 (2008).
Wehkamp, J., Schmid, M. & Stange, E. F. Defensins and other antimicrobial peptides in inflammatory bowel disease. Curr. Opin. Gastroenterol. 23, 370–378 (2007).
Levine, B. & Deretic, V. Unveiling the roles of autophagy in innate and adaptive immunity. Nature Rev. Immunol. 7, 767–777 (2007).
Levine, B. & Kroemer, G. Autophagy in the pathogenesis of disease. Cell 132, 27–42 (2008).
Hampe, J. et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nature Genet. 39, 207–211 (2007).
Pua, H. H., Dzhagalov, I., Chuck, M., Mizushima, N. & He, Y. W. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J. Exp. Med. 204, 25–31 (2007).
Birmingham, C. L., Smith, A. C., Bakowski, M. A., Yoshimori, T. & Brumell, J. H. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 281, 11374–11383 (2006).
Taylor, G. A. IRG proteins: key mediators of interferon-regulated host resistance to intracellular pathogens. Cell. Microbiol. 9, 1099–1107 (2007).
Bekpen, C. et al. The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol. 6, R92 (2005).
Singh, S. B., Davis, A. S., Taylor, G. A. & Deretic, V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 313, 1438–1441 (2006).
Ellson, C. D. et al. Neutrophils from p40phox−/− mice exhibit severe defects in NADPH oxidase regulation and oxidant-dependent bacterial killing. J. Exp. Med. 203, 1927–1937 (2006).
Suh, C. I. et al. The phosphoinositide-binding protein p40phox activates the NADPH oxidase during FcγIIA receptor-induced phagocytosis. J. Exp. Med. 203, 1915–1925 (2006).
Rioux, J. D. et al. Genomewide search in Canadian families with inflammatory bowel disease reveals two novel susceptibility loci. Am. J. Hum. Genet. 66, 1863–1870 (2000).
Satsangi, J. et al. Two stage genome-wide search in inflammatory bowel disease provides evidence for susceptibility loci on chromosomes 3, 7 and 12. Nature Genet. 14, 199–202 (1996).
Goyette, P. et al. Gene-centric association mapping of chromosome 3p implicates MST1 in IBD pathogenesis. Mucosal Immunol. 1, 131–138 (2008).
Beckly, J. B. et al. Two-stage candidate gene study of chromosome 3p demonstrates an association between nonsynonymous variants in the MST1R gene and Crohn's disease. Inflamm. Bowel Dis. 14, 500–507 (2008).
Cargill, M. et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am. J. Hum. Genet. 80, 273–390 (2007).
Kastelein, R. A., Hunter, C. A. & Cua, D. J. Discovery and biology of IL-23 and IL-27: related but functionally distinct regulators of inflammation. Annu. Rev. Immunol. 25, 221–242 (2007).
Acosta-Rodriguez, E. V. et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nature Immunol. 8, 639–646 (2007).
Annunziato, F. et al. Phenotypic and functional features of human Th17 cells. J. Exp. Med. 204, 1849–1861 (2007).
Hirota, K. et al. Preferential recruitment of CCR6-expressing Th17 cells to inflamed joints via CCL20 in rheumatoid arthritis and its animal model. J. Exp. Med. 204, 2803–2812 (2007).
Yeo, T. W. et al. A second major histocompatibility complex susceptibility locus for multiple sclerosis. Ann. Neurol. 61, 228–236 (2007).
Gregory, S. G. et al. Interleukin 7 receptor α-chain (IL7R) shows allelic and functional association with multiple sclerosis. Nature Genet. 39, 1083–1091 (2007).
Lundmark, F. et al. Variation in interleukin 7 receptor α-chain (IL7R) influences risk of multiple sclerosis. Nature Genet. 39, 1108–1113 (2007).
Vella, A. et al. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am. J. Hum. Genet. 76, 773–779 (2005).
Plenge, R. M. et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nature Genet. 39, 1477–1482 (2007).
Tsitsikov, E. N. et al. TRAF1 is a negative regulator of TNF signaling: enhanced TNF signaling in TRAF1-deficient mice. Immunity 15, 647–657 (2001).
Chung, J. Y., Park, Y. C., Ye, H. & Wu, H. All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell Sci. 115, 679–688 (2002).
Elliott, M. J. et al. Randomised double-blind comparison of chimeric monoclonal antibody to tumour necrosis factor α (cA2) versus placebo in rheumatoid arthritis. Lancet 344, 1105–1110 (1994).
Weinblatt, M. E. et al. A trial of etanercept, a recombinant tumor necrosis factor receptor:Fc fusion protein, in patients with rheumatoid arthritis receiving methotrexate. N. Engl. J. Med. 340, 253–259 (1999).
Remmers, E. F. et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N. Engl. J. Med. 357, 977–986 (2007).
Vang, T. et al. Protein tyrosine phosphatases in autoimmunity. Annu. Rev. Immunol. 26, 29–55 (2008).
Criswell, L. A. et al. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am. J. Hum. Genet. 76, 561–571 (2005).
Kozyrev, S. V. et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nature Genet. 40, 211–216 (2008).
Harley, J. B. et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nature Genet. 40, 204–210 (2008).
Graham, D. S. et al. Polymorphism at the TNF superfamily gene TNFSF4 confers susceptibility to systemic lupus erythematosus. Nature Genet. 40, 83–89 (2008).
Fernando, M. M. et al. Identification of two independent risk factors for lupus within the MHC in United Kingdom families. PLoS Genet. 3, e192 (2007).
Baechler, E. C. et al. Gene expression profiling in human autoimmunity. Immunol. Rev. 210, 120–137 (2006).
Bauer, J. W. et al. Elevated serum levels of interferon-regulated chemokines are biomarkers for active human systemic lupus erythematosus. PLoS Med. 3, e491 (2006).
Graham, R. R. et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc. Natl Acad. Sci. USA 104, 6758–6763 (2007).
Lim, J. et al. An essential role for talin during αMβ2-mediated phagocytosis. Mol. Biol. Cell 18, 976–985 (2007).
Schymeinsky, J., Mocsai, A. & Walzog, B. Neutrophil activation via β2 integrins (CD11/CD18): molecular mechanisms and clinical implications. Thromb. Haemost. 98, 262–273 (2007).
Yokoyama, K. et al. BANK regulates BCR-induced calcium mobilization by promoting tyrosine phosphorylation of IP3 receptor. EMBO J. 21, 83–92 (2002).
Kaplan, M. H. STAT4: a critical regulator of inflammation in vivo. Immunol. Res. 31, 231–242 (2005).
Kathiresan, S. et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nature Genet. 40, 189–197 (2008).
Lettre, G. et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nature Genet. 40, 584–591 (2008).
Zeggini, E. et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nature Genet. 40, 638–645 (2008).
Hollox, E. J. et al. Psoriasis is associated with increased β-defensin genomic copy number. Nature Genet. 40, 23–25 (2008).
Pujana, M. A. et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nature Genet. 39, 1338–1349 (2007).
Brass, A. L. et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319, 921–926 (2008).
Chen, Y. et al. Variations in DNA elucidate molecular networks that cause disease. Nature 452, 429–435 (2008).
Emilsson, V. et al. Genetics of gene expression and its effect on disease. Nature 452, 423–428 (2008).
Keller, M. P. et al. A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Res. 18, 706–716 (2008).
Bottini, N., Vang, T., Cucca, F. & Mustelin, T. Role of PTPN22 in type 1 diabetes and other autoimmune diseases. Semin. Immunol. 18, 207–213 (2006).
Rieck, M. et al. Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. J. Immunol. 179, 4704–4710 (2007).
Palsson-McDermott, E. M. & O'Neill, L. A. Building an immune system from nine domains. Biochem. Soc. Trans. 35, 1437–1444 (2007).
De Jager, P. L. et al. The role of inflammatory bowel disease susceptibility loci in multiple sclerosis and systemic lupus erythematosus. Genes Immun. 7, 327–334 (2006).
Ueda, H. et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 423, 506–511 (2003).
Brand, O. J. et al. Association of the interleukin-2 receptor α (IL-2Rα)/CD25 gene region with Graves' disease using a multilocus test and tag SNPs. Clin. Endocrinol. (Oxf.) 66, 508–512 (2007).
Graham, R. R. et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nature Genet. 38, 550–555 (2006).
Velaga, M. R. et al. The codon 620 tryptophan allele of the lymphoid tyrosine phosphatase (LYP) gene is a major determinant of Graves' disease. J. Clin. Endocrinol. Metab. 89, 5862–5865 (2004).
Hakonarson, H. et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature 448, 591–594 (2007).
Smyth, D. J. et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nature Genet. 38, 617–619 (2006).
Sutherland, A. et al. Genomic polymorphism at the interferon-induced helicase (IFIH1) locus contributes to Graves' disease susceptibility. J. Clin. Endocrinol. Metab. 92, 3338–3341 (2007).
Goyette P et al. Gene-centric association mapping of chromosome 3p implicates MST1 in IBD pathogenesis. Mucosal. Immunol. 1, 131–138 (2008).
Oda, K. & Kitano, H. A comprehensive map of the toll-like receptor signaling network. Mol. Syst. Biol. 2, 2006.0015 (2006).
Acknowledgements
The authors would like to thank C. Lefebvre, A. Huett, A. Ng and C. Labbé for their help in the preparation of this manuscript. R.J.X. is supported by grants from the National Institutes of Allergy and Immunology (AI062773), National Institutes of Diabetes, Digestive and Kidney Disease (DK43,351) and CCIB development funds. J.D.R. is funded by grants from the National Institutes of Allergy and Infectious Diseases (AI065687; AI067152), from the National Institute of Diabetes and Digestive and Kidney Diseases (DK064869; DK062432) and from the Crohn's and Colitis Foundation of America (SRA512).
Author information
Authors and Affiliations
Corresponding author
Related links
Glossary
- Causal gene
-
The specific gene that is responsible for disease risk conferred by a genomic region that has been identified by an association.
- Linkage studies
-
Studies aimed at establishing linkage between a genetic marker and a disease locus. Linkage is based on the tendency of genes and genetic markers to be inherited together because of their proximity on the same chromosome.
- Tag SNPs
-
Representative SNPs in a region of the genome with high linkage disequilibrium. Tag SNPs allow a reduction in the number of SNPs that must be genotyped while allowing the generation of the same amount of information. The HapMap Project catalogued tag SNPs in the entire genome.
- Hardy–Weinberg equilibrium
-
A fundamental principle in genetics stating that the genotypic frequencies of a large, randomly mating population remain constant in the absence of mutation, migration, natural selection or random drift.
- Penetrance
-
The probability, under given environmental conditions, with which a specific phenotype is expressed by individuals with a specific genotype. For example, if 50% of the people with a gene 'X' that is known to cause a disease do in fact develop the disease, then the penetrance of that gene is 0.5.
- Genome-wide linkage scan
-
A linkage study carried out with markers located across the entire genome. Linkage studies were traditionally carried out with ∼300 markers of simple sequence-length repeats and more recently with ∼5,000 single-nucleotide polymorphisms.
- Genetic markers
-
Genomic variants used as positional tools to find associations between specific DNA fragments and a certain phenotype or disease.
- Single-nucleotide polymorphisms
-
(SNPs). Genomic variants in which a single base in the DNA differs from the usual base at that position. SNPs are the most common type of variation in the human genome.
- Linkage disequilibrium
-
A situation in which alleles in a chromosomal region occur together more often than can be accounted for by chance, indicating that the alleles are in close proximity on the DNA strand and are most likely to be passed on together within a population.
- Haplotype
-
A combination or pattern of alleles at multiple linked loci that are inherited together.
- HapMap
-
A genetic resource created by the International HapMap Project (see the International HapMap website). The HapMap is a catalogue of common genetic variants that occur in humans. It describes what these variants are, where they occur in the genome and how they are distributed among people within populations and among populations in different parts of the world.
- Genotyping
-
A test designed to identify the genetic constitution of an individual — that is, the alleles present at one or more specific loci.
- Alleles
-
Alleles are alternative forms of genes that are located at a specific position on a specific chromosome. In association studies, the term allele is most commonly used to refer to one variant of a marker.
- Causal variant
-
The specific DNA sequence that functionally gives rise to the increased risk conferred by the causal gene or genomic region identified by an association.
- Copy-number variant
-
(CNV). A genomic variant characterized by differences in the number of copies of specific repeated DNA fragments that range from 1 kb to several Mb long. CNVs can contain entire genes and can be used as markers in association studies.
- Association testing
-
A statistical approach that tests for association between marker or candidate gene alleles and diseases. If a correlation is observed between genotype and phenotype, there is said to be an association between the marker or allele and the disease or trait.
- Replication studies
-
Studies designed to test (or replicate) a prior and explicit genetic hypothesis. For example, replication studies are carried out to validate the findings of a genome-wide association study, thereby discriminating false positives from true positives identified in the study.
- Autophagy
-
A cellular process in which cytoplasmic organelles and macromolecular complexes are entrapped in double-membrane-bound vesicles for delivery to lysosomes and subsequent degradation. This process is involved in constitutive turnover of proteins and organelles and is central to cellular activities that maintain a balance between synthesis and breakdown of various proteins.
- Gene desert
-
A large genomic segment devoid of genes. Gene deserts tend to harbour limited sequence conservation. The rare conserved region can contain transcriptional regulatory elements.
- Odds ratios
-
Measures of relative risk, usually estimated from studies that compare patients with control subjects. In a genetic disease context, the odds ratio is defined as the ratio of the odds of having an allele or a genotype while being affected by the disease to the odds of having the same allele or a genotype while being a healthy control.
- B-1 cells
-
B cells that express the cell-surface molecule CD5, which can bind to another B-cell-surface protein, CD72. CD5–CD72 interactions are thought to mediate B-cell–B-cell interactions. B-1 cells are mainly found in the peritoneum and have a role in T-cell-independent antibody production.
- Locus
-
The specific position of a gene or a marker on a chromosome.
- TH17 cells
-
(T helper 17 cells). A subset of CD4+ T helper cells that produce interleukin-17 (IL-17) and that are thought to be important in inflammatory and autoimmune diseases. Their generation involves transforming growth factor-β (TGFβ), IL-6, IL-23 or IL-21, IL-1β and the transcription factors RORγt and STAT3.
- Immunoreceptor tyrosine-based activation motifs
-
(ITAMs). ITAMs are protein motifs that exhibit unique abilities to bind and activate Lyn and Syk tyrosine kinases. This motif may be dually phosphorylated on tyrosines that link antigen receptors to downstream signalling molecules.
- Knock-in and knockout models
-
Animal models generated through molecular biology experiments that aim to delete the expression of endogenous genes (knock-out) or to introduce a specific allele of a given gene (knock-in).
Rights and permissions
About this article
Cite this article
Xavier, R., Rioux, J. Genome-wide association studies: a new window into immune-mediated diseases. Nat Rev Immunol 8, 631–643 (2008). https://doi.org/10.1038/nri2361
Issue Date:
DOI: https://doi.org/10.1038/nri2361
This article is cited by
-
Personalized Medicine for IBD Patients
BioNanoScience (2023)
-
In silico SNP analysis of the breast cancer antigen NY-BR-1
BMC Cancer (2016)
-
Unifying immunology with informatics and multiscale biology
Nature Immunology (2014)
-
Janus Kinases: An Ideal Target for the Treatment of Autoimmune Diseases
Journal of Investigative Dermatology Symposium Proceedings (2013)
-
The Arrival of JAK Inhibitors: Advancing the Treatment of Immune and Hematologic Disorders
BioDrugs (2013)