Abstract
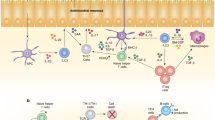
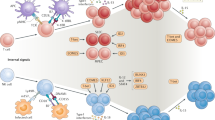
The origins of the adaptive immune system and the basis for its unique association with vertebrate species have been a source of considerable speculation. In light of recent advances in our understanding of the developmental and functional links between the induced regulatory T cell and T helper 17 cell lineages, and their specialized relationship to the gut, we speculate that the co-evolution of these adaptive immune pathways might have given primitive vertebrates a means to benefit from the diversification of their commensal microbiota.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Weaver, C. T., Harrington, L. E., Mangan, P. R., Gavrieli, M. & Murphy, K. M. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity 24, 677–688 (2006).
Bettelli, E. et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006).
Harrington, L. E. et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nature Immunol. 6, 1123–1132 (2005).
Mangan, P. R. et al. Transforming growth factor-β induces development of the TH17 lineage. Nature 441, 231–234 (2006).
Park, H. et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nature Immunol. 6, 1133–1141 (2005).
Josefowicz, S. Z. & Rudensky, A. Control of regulatory T cell lineage commitment and maintenance. Immunity 30, 616–625 (2009).
Veldhoen, M., Hocking, R. J., Atkins, C. J., Locksley, R. M. & Stockinger, B. TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24, 179–189 (2006).
Ziegler, S. F. & Buckner, J. H. FOXP3 and the regulation of Treg/Th17 differentiation. Microbes Infect. 11, 594–598 (2009).
Locksley, R. M. Nine lives: plasticity among T helper cell subsets. J. Exp. Med. 206, 1643–1646 (2009).
Zhou, L., Chong, M. M. & Littman, D. R. Plasticity of CD4+ T cell lineage differentiation. Immunity 30, 646–655 (2009).
Khalturin, K., Panzer, Z., Cooper, M. D. & Bosch, T. C. Recognition strategies in the innate immune system of ancestral chordates. Mol. Immunol. 41, 1077–1087 (2004).
Huang, S. et al. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 18, 1112–1126 (2008).
Liongue, C. & Ward, A. C. Evolution of Class I cytokine receptors. BMC Evol. Biol. 7, 120 (2007).
Terajima, D. et al. Identification and sequence of seventy-nine new transcripts expressed in hemocytes of Ciona intestinalis, three of which may be involved in characteristic cell-cell communication. DNA Res. 10, 203–212 (2003).
Guo, P. et al. Dual nature of the adaptive immune system in lampreys. Nature 459, 796–801 (2009).
Pancer, Z., Mayer, W. E., Klein, J. & Cooper, M. D. Prototypic T cell receptor and CD4-like coreceptor are expressed by lymphocytes in the agnathan sea lamprey. Proc. Natl Acad. Sci. USA 101, 13273–13278 (2004).
Hedrick, S. M. The acquired immune system: a vantage from beneath. Immunity 21, 607–615 (2004).
Pancer, Z. & Cooper, M. D. The evolution of adaptive immunity. Annu. Rev. Immunol. 24, 497–518 (2006).
Hooper, L. V. & Gordon, J. I. Commensal host–bacterial relationships in the gut. Science 292, 1115–1118 (2001).
McFall-Ngai, M. Adaptive immunity: care for the community. Nature 445, 153 (2007).
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R. & Gordon, J. I. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Rev. Microbiol. 6, 776–788 (2008).
Gomez, G. D. & Balcazar, J. L. A review on the interactions between gut microbiota and innate immunity of fish. FEMS Immunol. Med. Microbiol. 52, 145–154 (2008).
Rawls, J. F., Mahowald, M. A., Ley, R. E. & Gordon, J. I. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127, 423–433 (2006).
Colonna, M. Interleukin-22-producing natural killer cells and lymphoid tissue inducer-like cells in mucosal immunity. Immunity 31, 15–23 (2009).
Roark, C. L., Simonian, P. L., Fontenot, A. P., Born, W. K. & O'Brien, R. L. γδ T cells: an important source of IL-17. Curr. Opin. Immunol. 20, 353–357 (2008).
Wostmann, B. S., Larkin, C., Moriarty, A. & Bruckner-Kardoss, E. Dietary intake, energy metabolism, and excretory losses of adult male germfree Wistar rats. Lab. Anim. Sci. 33, 46–50 (1983).
Backhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A. & Gordon, J. I. Host–bacterial mutualism in the human intestine. Science 307, 1915–1920 (2005).
Hooper, L. V. Do symbiotic bacteria subvert host immunity? Nature Rev. Microbiol. 7, 367–374 (2009).
Weaver, C. T., Hatton, R. D., Mangan, P. R. & Harrington, L. E. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu. Rev. Immunol. 25, 821–852 (2007).
Mucida, D. et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science 317, 256–260 (2007).
Sun, C. M. et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 204, 1775–1785 (2007).
Benson, M. J., Pino-Lagos, K., Rosemblatt, M. & Noelle, R. J. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J. Exp. Med. 204, 1765–1774 (2007).
Coombes, J. L. et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid-dependent mechanism. J. Exp. Med. 204, 1757–1764 (2007).
Ouyang, W., Kolls, J. K. & Zheng, Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity 28, 454–467 (2008).
Raftery, L. A. & Sutherland, D. J. TGF-β family signal transduction in Drosophila development: from Mad to Smads. Dev. Biol. 210, 251–268 (1999).
Kaiser, P., Rothwell, L., Avery, S. & Balu, S. Evolution of the interleukins. Dev. Comp. Immunol. 28, 375–394 (2004).
Malagoli, D., Conklin, D., Sacchi, S., Mandrioli, M. & Ottaviani, E. A putative helical cytokine functioning in innate immune signalling in Drosophila melanogaster. Biochim. Biophys. Acta 1770, 974–978 (2007).
Schulenburg, H., Hoeppner, M. P., Weiner, J. & Bornberg-Bauer, E. Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology 213, 237–250 (2008).
Wahl, S. M. Transforming growth factor-β: innately bipolar. Curr. Opin. Immunol. 19, 55–62 (2007).
Franchini, A., Malagoli, D. & Ottaviani, E. Cytokines and invertebrates: TGF-β and PDGF. Curr. Pharm. Des. 12, 3025–3031 (2006).
Allen, J. B. et al. Rapid onset synovial inflammation and hyperplasia induced by transforming growth factor β. J. Exp. Med. 171, 231–247 (1990).
Brandes, M. E., Allen, J. B., Ogawa, Y. & Wahl, S. M. Transforming growth factor β1 suppresses acute and chronic arthritis in experimental animals. J. Clin. Invest. 87, 1108–1113 (1991).
Haddad, G., Hanington, P. C., Wilson, E. C., Grayfer, L. & Belosevic, M. Molecular and functional characterization of goldfish (Carassius auratus L.) transforming growth factor β. Dev. Comp. Immunol. 32, 654–663 (2008).
Yang, M. & Zhou, H. Grass carp transforming growth factor-β1 (TGF-β1): molecular cloning, tissue distribution and immunobiological activity in teleost peripheral blood lymphocytes. Mol. Immunol. 45, 1792–1798 (2008).
Brustle, A. et al. The development of inflammatory TH-17 cells requires interferon-regulatory factor 4. Nature Immunol. 8, 958–966 (2007).
Quintana, F. J. et al. Control of Treg and TH17 cell differentiation by the aryl hydrocarbon receptor. Nature 453, 65–71 (2008).
Schraml, B. U. et al. The AP-1 transcription factor Batf controls TH17 differentiation. Nature 460, 405–409 (2009).
Veldhoen, M. et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 453, 106–109 (2008).
Zhang, F., Meng, G. & Strober, W. Interactions among the transcription factors Runx1, RORγt and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nature Immunol. 9, 1297–1306 (2008).
Ivanov, I. I. et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006).
Yang, X. O. et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors RORα and RORγ. Immunity 28, 29–39 (2008).
Zheng, Y. & Rudensky, A. Y. Foxp3 in control of the regulatory T cell lineage. Nature Immunol. 8, 457–462 (2007).
Yang, X. O. et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity 29, 44–56 (2008).
Zhou, L. et al. IL-6 programs TH-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nature Immunol. 8, 967–974 (2007).
Zhou, L. et al. TGF-β-induced Foxp3 inhibits TH17 cell differentiation by antagonizing RORγt function. Nature 453, 236–240 (2008).
Du, J., Huang, C., Zhou, B. & Ziegler, S. Isoform-specific inhibition of RORα-mediated transcriptional activation by human FOXP3. J. Immunol. 180, 4785–4792 (2008).
Ichiyama, K. et al. Foxp3 inhibits RORγt-mediated IL-17A mRNA transcription through direct interaction with RORγt. J. Biol. Chem. 283, 17003–17008 (2008).
Deknuydt, F., Bioley, G., Valmori, D. & Ayyoub, M. IL-1β and IL-2 convert human Treg into TH17 cells. Clin. Immunol. 131, 298–307 (2009).
Beriou, G. et al. IL-17 producing human peripheral regulatory T cells retain suppressive function. Blood 113, 4240–4249 (2009).
Koenen, H. J. et al. Human CD25highFoxp3pos regulatory T cells differentiate into IL-17-producing cells. Blood 112, 2340–2352 (2008).
Osorio, F. et al. DC activated via dectin-1 convert Treg into IL-17 producers. Eur. J. Immunol. 38, 3274–3281 (2008).
Sharma, M. D. et al. Indoleamine 2, 3-dioxygenase controls conversion of Foxp3+ Tregs to TH17-like cells in tumor-draining lymph nodes. Blood 113, 6102–6111 (2009).
Voo, K. S. et al. Identification of IL-17-producing FOXP3+ regulatory T cells in humans. Proc. Natl Acad. Sci. USA 106, 4793–4798 (2009).
Lochner, M. et al. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORγt+ T cells. J. Exp. Med. 205, 1381–1393 (2008).
Chung, Y. et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity 30, 576–587 (2009).
Ono, M. et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature 446, 685–689 (2007).
Zheng, Y. et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control TH2 responses. Nature 458, 351–356 (2009).
Maynard, C. L. & Weaver, C. T. Diversity in the contribution of interleukin-10 to T-cell-mediated immune regulation. Immunol. Rev. 226, 219–233 (2008).
Berger, S. L. The complex language of chromatin regulation during transcription. Nature 447, 407–412 (2007).
Wei, G. et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167 (2009).
Wilson, C. B., Rowell, E. & Sekimata, M. Epigenetic control of T-helper-cell differentiation. Nature Rev. Immunol. 9, 91–105 (2009).
Lexberg, M. H. et al. Th memory for interleukin-17 expression is stable in vivo. Eur. J. Immunol. 38, 2654–2664 (2008).
Lee, Y. K. et al. Late developmental plasticity in the T helper 17 lineage. Immunity 30, 92–107 (2009).
Li, B. et al. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc. Natl Acad. Sci. USA 104, 4571–4576 (2007).
Samanta, A. et al. TGF-β and IL-6 signals modulate chromatin binding and promoter occupancy by acetylated FOXP3. Proc. Natl Acad. Sci. USA 105, 14023–14027 (2008).
Macpherson, A. J., McCoy, K. D., Johansen, F. E. & Brandtzaeg, P. The immune geography of IgA induction and function. Mucosal Immunol. 1, 11–22 (2008).
Tsuji, M. et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer's patches. Science 323, 1488–1492 (2009).
Du Pasquier, L. & Bernard, C. C. Active suppression of the allogeneic histocompatibility reactions during the metamorphosis of the clawed toad Xenopus. Differentiation 16, 1–7 (1980).
Tsutsui, S., Nakamura, O. & Watanabe, T. Lamprey (Lethenteron japonicum) IL-17 upregulated by LPS-stimulation in the skin cells. Immunogenetics 59, 873–882 (2007).
Rolff, J. Why did the acquired immune system of vertebrates evolve? Dev. Comp. Immunol. 31, 476–482 (2007).
Acknowledgements
This work was supported by grants AI35783, AI57956 and DK71176 from the United States National Institutes of Health and by the University of Alabama at Birmingham Mucosal HIV and Immunobiology Center. We thank members of our laboratory for helpful comments and offer our apologies to colleagues whose work could not be adequately discussed or cited owing to space limitations.
Author information
Authors and Affiliations
Related links
Rights and permissions
About this article
Cite this article
Weaver, C., Hatton, R. Interplay between the TH17 and TReg cell lineages: a (co-)evolutionary perspective. Nat Rev Immunol 9, 883–889 (2009). https://doi.org/10.1038/nri2660
Issue Date:
DOI: https://doi.org/10.1038/nri2660
This article is cited by
-
Protective Effect of Akkermansia muciniphila on the Preeclampsia-Like Mouse Model
Reproductive Sciences (2023)
-
Maternal immune activation during pregnancy is associated with more difficulties in socio-adaptive behaviors in autism spectrum disorder
Scientific Reports (2023)
-
Heat shock protein-70 is elevated in childhood primary immune thrombocytopenia
European Journal of Medical Research (2022)
-
The role of Th17 cells in the pathogenesis and treatment of breast cancer
Cancer Cell International (2022)
-
A defective interleukin-17 receptor A1 causes weight loss and intestinal metabolism-related gene downregulation in Japanese medaka, Oryzias latipes
Scientific Reports (2021)