Key Points
-
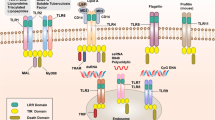
Recognizing the presence of invading pathogens is key to mounting an effective innate immune defence. Mammalian cells express different classes of germline-encoded pattern recognition receptors (PRRs) that monitor the extracellular and the intracellular compartments of host cells for signs of infection and that initiate several conserved signalling pathways.
-
Recent advances have identified several new extracellular and intracellular PRRs and have shed light on the complex interplay of innate immune signalling pathways during pathogen infection.
-
Although the function of most Toll-like receptors (TLRs) has been determined in the past, the functions of orphan mouse TLR11, TLR12 and TLR13, which are not found in humans, have so far eluded researchers. The ligands for these receptors have recently been characterized, which has revealed key differences between the human and mouse innate immune system.
-
Cytoplasmic DNA is a strong activator of immune responses and induces type I interferon (IFN) production through the signalling adaptor stimulator of IFN genes protein (STING). Several candidate proteins have been proposed to bind to double-stranded DNA and to induce type I IFN production. More recently, a previously unrecognized role for cyclic dinucleotides in this signalling pathway has been described.
-
NOD-like receptors are the largest group of intracellular receptors and are known for their ability to induce the assembly of inflammasome complexes. Recent reports have identified new inflammasome complexes and have characterized their function in the recognition of various bacterial pathogens.
-
Canonical inflammasomes have been defined as macromolecular complexes that activate the cysteine protease caspase 1. However, recent findings have highlighted the role of caspase 8 and caspase 11 in inducing pro-inflammatory cytokine maturation and host cell death.
Abstract
Recognizing the presence of invading pathogens is key to mounting an effective innate immune response. Mammalian cells express different classes of germline-encoded pattern recognition receptors that monitor the extracellular and intracellular compartments of host cells for signs of infection and that activate several conserved signalling pathways. An efficient immune response often requires the sequential detection of a pathogen by different receptors in different subcellular compartments, which results in a complex interplay of downstream signalling pathways. In this Review, we discuss the recent identification of previously unknown pattern recognition receptors and how they complement the repertoire of established receptors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Janeway, C. A. Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54, 1–13 (1989).
McGuinness, D. H., Dehal, P. K. & Pleass, R. J. Pattern recognition molecules and innate immunity to parasites. Trends Parasitol. 19, 312–319 (2003).
Martinon, F., Burns, K. & Tschopp, J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 10, 417–426 (2002).
Schroder, K. & Tschopp, J. The inflammasomes. Cell 140, 821–832 (2010).
Miao, E. A. et al. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nature Immunol. 11, 1136–1142 (2010).
Kawai, T. & Akira, S. TLR signaling. Cell Death Differ. 13, 816–825 (2006).
O'Neill, L. A. & Bowie, A. G. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nature Rev. Immunol. 7, 353–364 (2007).
Barbalat, R., Ewald, S. E., Mouchess, M. L. & Barton, G. M. Nucleic acid recognition by the innate immune system. Annu. Rev. Immunol. 29, 185–214 (2011).
Strober, W., Murray, P. J., Kitani, A. & Watanabe, T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nature Rev. Immunol. 6, 9–20 (2006).
Sabbah, A. et al. Activation of innate immune antiviral responses by Nod2. Nature Immunol. 10, 1073–1080 (2009).
Desmet, C. J. & Ishii, K. J. Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination. Nature Rev. Immunol. 12, 479–491 (2012).
Kang, D. C. et al. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl Acad. Sci. USA 99, 637–642 (2002).
Kato, H. et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23, 19–28 (2005).
Yoneyama, M. et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nature Immunol. 5, 730–737 (2004).
Rothenfusser, S. et al. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 175, 5260–5268 (2005).
Satoh, T. et al. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl Acad. Sci. USA 107, 1512–1517 (2010).
Saito, T. et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl Acad. Sci. USA 104, 582–587 (2007).
Upton, J. W., Kaiser, W. J. & Mocarski, E. S. DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe 11, 290–297 (2012).
Leulier, F. & Lemaitre, B. Toll-like receptors--taking an evolutionary approach. Nature Rev. Genet. 9, 165–178 (2008).
Yarovinsky, F. et al. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science 308, 1626–1629 (2005).
Zhang, D. et al. A Toll-like receptor that prevents infection by uropathogenic bacteria. Science 303, 1522–1526 (2004).
Mathur, R. et al. A mouse model of Salmonella typhi infection. Cell 151, 590–602 (2012). This study reports that TLR11 is a novel flagellin receptor, which functions independently of TLR5.
Hayashi, F. et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103 (2001).
Uematsu, S. et al. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nature Immunol. 7, 868–874 (2006).
Pifer, R., Benson, A., Sturge, C. R. & Yarovinsky, F. UNC93B1 is essential for TLR11 activation and IL-12-dependent host resistance to Toxoplasma gondii. J. Biol. Chem. 286, 3307–3314 (2011).
Kim, Y. M., Brinkmann, M. M., Paquet, M. E. & Ploegh, H. L. UNC93B1 delivers nucleotide-sensing Toll-like receptors to endolysosomes. Nature 452, 234–238 (2008).
Scanga, C. A. et al. Cutting edge: MyD88 is required for resistance to Toxoplasma gondii infection and regulates parasite-induced IL-12 production by dendritic cells. J. Immunol. 168, 5997–6001 (2002).
Debierre-Grockiego, F. et al. Fatty acids from Plasmodium falciparum down-regulate the toxic activity of malaria glycosylphosphatidylinositols. Infect. Immun. 74, 5487–5496 (2006).
Koblansky, A. A. et al. Recognition of profilin by Toll-like receptor 12 is critical for host resistance to Toxoplasma gondii. Immunity 30, 119–130 (2012).
Gratz, N. et al. Group A streptococcus activates type I interferon production and MyD88-dependent signaling without involvement of TLR2, TLR4, and TLR9. J. Biol. Chem. 283, 19879–19887 (2008).
Deshmukh, S. D. et al. Macrophages recognize streptococci through bacterial single-stranded RNA. EMBO Rep. 12, 71–76 (2011).
Oldenburg, M. et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science 337, 1111–1115 (2012). This study describes a role for mouse TLR13 in the recognition of a conserved CGGAAAGACC motif in S. aureus 23S rRNA, which is also targeted by the MLS group of antibiotics. Mutations in this sequence, which are found in certain MLS-resistant strains of S. aureus , not only promote antibiotic resistance but also abolish innate immune recognition.
Li, X. D. & Chen, Z. J. Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. Elife 1, e00102 (2012).
Hidmark, A., von Saint Paul, A. & Dalpke, A. H. Cutting edge: TLR13 is a receptor for bacterial RNA. J. Immunol. 189, 2717–2721 (2012).
Kariko, K., Buckstein, M., Ni, H. & Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005).
Cervantes, J. L., Weinerman, B., Basole, C. & Salazar, J. C. TLR8: the forgotten relative revindicated. Cell. Mol. Immunol. 9, 434–438 (2012).
Takaoka, A. et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505 (2007).
Hornung, V. et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518 (2009).
Fernandes-Alnemri, T., Yu, J. W., Datta, P., Wu, J. & Alnemri, E. S. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509–513 (2009).
Ishii, K. J. et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature 451, 725–729 (2008).
Jones, J. W. et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl Acad. Sci. USA 107, 9771–9776 (2010).
Chiu, Y. H., Macmillan, J. B. & Chen, Z. J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138, 576–591 (2009).
Ablasser, A. et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nature Immunol. 10, 1065–1072 (2009).
Ishikawa, H. & Barber, G. N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008).
Zhong, B. et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 (2008).
Ishikawa, H., Ma, Z. & Barber, G. N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 (2009).
Sauer, J. D. et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 79, 688–694 (2011).
Burdette, D. L. et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 (2011). This report demonstrates the dual function of STING as a sensor for cyclic dinucleotides and as an adaptor for the cytosolic DNA response.
Ouyang, S. et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity 36, 1073–1086 (2012).
Shang, G. et al. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nature Struct. Mol. Biol. 19, 725–727 (2012).
Shu, C., Yi, G., Watts, T., Kao, C. C. & Li, P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nature Struct. Mol. Biol. 19, 722–724 (2012).
Yin, Q. et al. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol. Cell 46, 735–745 (2012).
Huang, Y. H., Liu, X. Y., Du, X. X., Jiang, Z. F. & Su, X. D. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nature Struct. Mol. Biol. 19, 728–730 (2012).
Woodward, J. J., Iavarone, A. T. & Portnoy, D. A. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705 (2010).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2012).
Wu, J. et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2012). References 55 and 56 describe cGAS as a novel DNA sensor that recognizes cytoplasmic DNA and that produces the newly identified endogenous secondary messenger cyclic GMP–AMP, which signals through STING to activate a type I IFN response.
Karaolis, D. K. et al. Bacterial c-di-GMP is an immunostimulatory molecule. J. Immunol. 178, 2171–2181 (2007).
Hu, D. L. et al. c-di-GMP as a vaccine adjuvant enhances protection against systemic methicillin-resistant Staphylococcus aureus (MRSA) infection. Vaccine 27, 4867–4873 (2009).
Zhang, Z. et al. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nature Immunol. 12, 959–965 (2011).
Parvatiyar, K. et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nature Immunol. 13, 1155–1161 (2012).
Unterholzner, L. et al. IFI16 is an innate immune sensor for intracellular DNA. Nature Immunol. 11, 997–1004 (2010).
Orzalli, M. H., DeLuca, N. A. & Knipe, D. M. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc. Natl Acad. Sci. USA 109, e3008–e3017 (2012).
Gariano, G. R. et al. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog. 8, e1002498 (2012).
Yang, P. et al. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a β-catenin-dependent pathway. Nature Immunol. 11, 487–494 (2010).
Kanneganti, T. D. et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature 440, 233–236 (2006).
Sander, L. E. et al. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature 474, 385–389 (2011).
Zhang, X. et al. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J. Immunol. 186, 4541–4545 (2011).
Oshiumi, H., Sakai, K., Matsumoto, M. & Seya, T. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-β-inducing potential. Eur. J. Immunol. 40, 940–948 (2010).
Zhang, Z. et al. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity 34, 866–878 (2011).
Miyashita, M., Oshiumi, H., Matsumoto, M. & Seya, T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol. Cell. Biol. 31, 3802–3819 (2011).
Kim, T. et al. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc. Natl Acad. Sci. USA 107, 15181–15186 (2010).
Fuller-Pace, F. V. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 34, 4206–4215 (2006).
Pichlmair, A. et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nature Immunol. 12, 624–630 (2011).
Abbas, Y. M., Pichlmair, A., Gorna, M. W., Superti-Furga, G. & Nagar, B. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature 494, 60–64 (2013).
Broz, P. et al. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature 490, 288–291 (2012).
Rathinam, V. A. et al. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by Gram-negative bacteria. Cell 150, 606–619 (2012). References 75 and 76 report a link between TRIF-mediated type I IFN production and the activation of the non-canonical caspase 11 inflammasome in response to Gram-negative bacteria.
Gavrilin, M. A. & Wewers, M. D. Francisella recognition by inflammasomes: differences between mice and men. Front. Microbiol. 2, 11 (2011).
Mariathasan, S. et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430, 213–218 (2004).
Amer, A. et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J. Biol. Chem. 281, 35217–35223 (2006).
Franchi, L. et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1β in salmonella-infected macrophages. Nature Immunol. 7, 576–582 (2006).
Miao, E. A. et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin-1β via Ipaf. Nature Immunol. 7, 569–575 (2006).
Miao, E. A. et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl Acad. Sci. USA 107, 3076–3080 (2010).
Lightfield, K. L. et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nature Immunol. 9, 1171–1178 (2008).
Kofoed, E. M. & Vance, R. E. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477, 592–595 (2011).
Zhao, Y. et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 447, 596–600 (2011).
Halff, E. F. et al. Formation and structure of a NAIP5–NLRC4 inflammasome induced by direct interactions with conserved N- and C-terminal regions of flagellin. J. Biol. Chem. 287, 38460–38472 (2012).
Qu, Y. et al. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature 490, 539–542 (2012). This report shows that phosphorylation of NLRC4 is crucial for the response to flagellin and identifies PKCδ as the relevant kinase.
Normand, S. et al. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc. Natl Acad. Sci. USA 108, 9601–9606 (2011).
Chen, G. Y., Liu, M., Wang, F., Bertin, J. & Nunez, G. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J. Immunol. 186, 7187–7194 (2011).
Elinav, E. et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757 (2011). This study provides the first evidence for the existence of an NLRP6 inflammasome in gut epithelial cells and describes its essential role in maintaining gut homeostasis.
Dupaul-Chicoine, J. et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity 32, 367–378 (2010).
Strowig, T., Henao-Mejia, J., Elinav, E. & Flavell, R. Inflammasomes in health and disease. Nature 481, 278–286 (2012).
Zaki, M. H. et al. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 32, 379–391 (2010).
Khare, S. et al. An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity 36, 464–476 (2012).
Lich, J. D. & Ting, J. P. Monarch-1/PYPAF7 and other CATERPILLER (CLR, NOD, NLR) proteins with negative regulatory functions. Microbes Infect. 9, 672–676 (2007).
Allen, I. C. et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-κB signaling. Immunity 36, 742–754 (2012).
Vladimer, G. I. et al. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity 37, 96–107 (2012).
Brodsky, I. E. et al. A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell Host Microbe 7, 376–387 (2010).
Zheng, Y., Lilo, S., Mena, P. & Bliska, J. B. YopJ-induced caspase-1 activation in Yersinia-infected macrophages: independent of apoptosis, linked to necrosis, dispensable for innate host defense. PLoS ONE 7, e36019 (2012).
Henry, T., Brotcke, A., Weiss, D. S., Thompson, L. J. & Monack, D. M. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J. Exp. Med. 204, 987–994 (2007).
Holler, N. et al. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nature Immunol. 1, 489–495 (2000).
Mocarski, E. S., Upton, J. W. & Kaiser, W. J. Viral infection and the evolution of caspase 8-regulated apoptotic and necrotic death pathways. Nature Rev. Immunol. 12, 79–88 (2012).
Sun, L. et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148, 213–227 (2012).
Wang, Z., Jiang, H., Chen, S., Du, F. & Wang, X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 148, 228–243 (2012).
Feoktistova, M. et al. cIAPs block ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol. Cell 43, 449–463 (2011).
Tenev, T. et al. The ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol. Cell 43, 432–448 (2011).
Upton, J. W., Kaiser, W. J. & Mocarski, E. S. Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe 7, 302–313 (2010).
Robinson, N. et al. Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nature Immunol. 13, 954–962 (2012).
Kayagaki, N. et al. Non-canonical inflammasome activation targets caspase-11. Nature 479, 117–121 (2011). This study describes a novel non-canonical caspase 11 inflammasome and shows there to be an essential role for caspase 11 in LPS-induced lethality in a mouse model of septic shock.
Case, C. L. et al. Caspase-11 stimulates rapid flagellin-independent pyroptosis in response to Legionella pneumophila. Proc. Natl Acad. Sci. USA 110, 1851–1856 (2013).
Gringhuis, S. I. et al. Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1β via a noncanonical caspase-8 inflammasome. Nature Immunol. 13, 246–254 (2012).
Vince, J. E. et al. Inhibitor of apoptosis proteins limit RIP3 kinase-dependent interleukin-1 activation. Immunity 36, 215–227 (2012).
Pierini, R. et al. AIM2/ASC triggers caspase-8-dependent apoptosis in Francisella-infected caspase-1-deficient macrophages. Cell Death Differ. 19, 1709–1721 (2012).
Acknowledgements
This work was supported by the following grants: PP00P3_139120/1 from the Swiss National Science Foundation to P.B., and AI095396 and AI08972 to D.M.M. from the US National Institute of Allergy and Infectious Diseases. We apologize to investigators whose contributions were not cited more extensively because of space limitations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Cyclic dinucleotides
-
Small bacterial or host-derived nucleic acids — such as cyclic diguanylate monophosphate (c-di-GMP), cyclic diadenylate monophosphate (c-di-AMP) or cyclic GMP–AMP — that function as secondary messengers and that can induce an innate immune response when present in the cytosol.
- Leaderless cytokines
-
Cytokines that lack a classical amino-terminal secretion signal sequence (also referred to as leader peptide or leader sequence) and that are thought to be secreted by an endoplasmic reticulum- and Golgi-independent mechanism.
- Pyroptosis
-
A lytic pro-inflammatory form of programmed cell death that is initiated by the activation of inflammatory caspases.
- Plasmacytoid dendritic cells
-
(pDCs). A dendritic cell subset that morphologically resembles a plasmablast. pDCs produce large amounts of type I interferons in response to viral infection.
- Necroptosis
-
A form of programmed necrosis that is initiated by the kinases receptor-interacting protein 1 (RIP1) and RIP3 in response to external signals, in conditions in which caspase 8 activity is compromised.
- Ribonuclease A
-
(RNase A). An endoribonuclease that specifically cleaves single-stranded RNA and that is often used to remove RNA from samples.
- Macrolide, lincosamide and streptogramin B
-
(MLS). A group of antibiotics that function as translational inhibitors by targeting the 50S ribosomal subunit, which contains 23S ribosomal RNA.
- DExD/H box helicase
-
An enzyme that can unwind double-stranded RNA using energy derived from ATP hydrolysis. The DExD/H box is a characteristic amino acid signature motif of many RNA-binding proteins.
- Small interfering RNA
-
(siRNA). Short double-stranded RNAs of 19 to 23 nucleotides that induce RNA interference, which is a post-transcriptional process that leads to gene silencing in a sequence-specific manner.
- Short hairpin RNA
-
(shRNA). A sequence of RNA that makes a tight hairpin turn, which can be used to silence target gene expression via RNA interference.
- Leucine-rich repeat
-
(LRR). A protein structural motif composed of repeating stretches of 20 to 30 amino acids that are unusually rich in the hydrophobic amino acid leucine and that form an α/β-horseshoe fold. LRRs are found in many pattern recognition receptors, such as Toll-like receptors and NOD-like receptors, but also in many functionally unrelated proteins.
- β-catenin
-
This protein functions both as a transcriptional activator and as a membrane–cytoskeleton linker protein by binding to E-cadherin. Following detachment from E-cadherin, β-catenin can relocate to the nucleus.
- Type III secretion system
-
(T3SS). A virulence-associated specialized molecular machine present in some bacteria that facilitates the translocation of bacterial proteins into host cells.
- Colitis
-
An inflammatory disease of the colon. In humans, colitis is most commonly classified as ulcerative colitis or as Crohn's disease, which are two inflammatory bowel diseases that have unknown aetiologies. Various hereditary and induced mouse models of human colitis have been developed.
Rights and permissions
About this article
Cite this article
Broz, P., Monack, D. Newly described pattern recognition receptors team up against intracellular pathogens. Nat Rev Immunol 13, 551–565 (2013). https://doi.org/10.1038/nri3479
Published:
Issue Date:
DOI: https://doi.org/10.1038/nri3479
This article is cited by
-
RBP–RNA interactions in the control of autoimmunity and autoinflammation
Cell Research (2023)
-
Microbiota in a long survival discourse with the human host
Archives of Microbiology (2023)
-
Association of rare variants in genes of immune regulation with pediatric autoimmune CNS diseases
Journal of Neurology (2022)
-
Comparative Transcriptome Analysis of Head Kidney of Aeromonas hydrophila-infected Hypoxia-tolerant and Normal Large Yellow Croaker
Marine Biotechnology (2022)
-
Interaction of Salmonella Gallinarum and Salmonella Enteritidis with peripheral leucocytes of hens with different laying performance
Veterinary Research (2021)