Abstract
The field of HIV-1 vaccine research has seen a renaissance with the identification of antibodies that neutralize most circulating HIV-1 strains. An understanding of the structural mode of target recognition that these antibodies use and the immune pathways that lead to their development is emerging. This knowledge has provided fundamental insights into the pathways that elicit broadly neutralizing antibodies and provides a foundation for active and passive immunization strategies to prevent HIV-1 infection.
Similar content being viewed by others
Main
Now this is not the end. It is not even the beginning of the end. But it is, perhaps, the end of the beginning. Winston Churchill, address to Parliament, 10 November 1942
Vaccines represent one of the few medical interventions with the potential to eradicate diseases1. Since HIV-1 was first identified in 1983, efforts to design an effective vaccine have been prominent in the attempts to contain the virus. Despite these efforts, and even after encouraging results from the RV144 vaccine clinical trial (also known as the Thai trial)2, HIV-1 continues to infect almost 1 million individuals each year, and prospects for a highly effective licensed HIV-1 vaccine seem remote3,4,5,6.
To develop an effective vaccine, we must overcome considerable challenges, including a lack of natural immunity to HIV-1 and the formidable mechanisms of immune evasion that are intrinsic to the HIV-1 envelope glycoprotein (Env), which is the target of known HIV-1-directed neutralizing antibodies7,8,9. The viral Env spike is protected by an evolving shield of glycans, variable immunodominant loops and conformational masking of key viral epitopes8,10,11,12. Although immunization with recombinant proteins or gene-based vectors encoding Env proteins can induce high levels of HIV-1-specific antibodies, vaccine-induced antibodies have been unable to neutralize most circulating primary HIV-1 isolates3,4,8,9. Indeed, natural infection predominantly induces non-neutralizing or strain-specific antibodies during the first months of infection11,13,14,15,16. However, approximately 20% of HIV-1-infected individuals develop broadly neutralizing antibodies, which is the type of humoral immune response that we aim to elicit via immunization17,18,19,20,21,22,23.
Efforts to understand the interaction between the virus and the human immune system have led to the identification of human antibodies that effectively neutralize diverse strains of HIV-1 (Refs 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42) (Table 1). These antibodies generally develop in HIV-1-infected individuals who have been infected for at least 2 years and whose sera show broad neutralization of HIV-1. As HIV-1 eventually evades neutralizing antibodies, there may be minimal clinical benefit to the individuals in whom these antibodies eventually develop19,23,43. However, what is clear is that passive delivery of these antibodies in animal models of HIV-1 transmission can completely prevent infection44,45,46,47,48,49.
A key question is, can broadly neutralizing antibodies be used — as templates for vaccine development or directly for passive transfer — to prevent HIV-1 infection in humans? In this Opinion article, we present a brief summary of the currently identified broadly neutralizing antibodies, and we describe what is known about the ontogeny of B cells that give rise to these antibodies. We also discuss the prospects for eliciting similar antibodies through immunization, and approaches for the use of antibodies directly for passive immunization as well as gene-based antibody delivery. The realization that humans can make antibodies that are potentially broadly protective against HIV-1 represents a crucial turning point in the HIV-1 vaccine field: we discuss recent progress, crucial experiments and future prospects for the antibody-based prevention of HIV-1 infection.
Broadly neutralizing HIV-1 antibodies
Human antibodies that effectively neutralize HIV-1 target the HIV-1 viral spike — a trimeric heterodimer composed of gp120 Env, which is involved in recognizing the CD4 receptor, and of gp41 transmembrane glycoprotein, which is responsible for fusing the viral and target cell membrane7,8. Most broadly neutralizing antibodies can be placed into four categories on the basis of the location on the viral spike of the conserved epitopes that they recognize (reviewed in Ref. 50). CD4-binding site-directed antibodies recognize the site of CD4 attachment on gp120; variable region 1 and variable region 2 (V1/V2)-directed antibodies often recognize glycopeptide epitopes that include the N-linked glycan at residue Asn160 on gp120; glycan V3-directed antibodies recognize epitopes that generally include the N-linked glycan at residue Asn332 on gp120; and membrane-proximal external region (MPER)-directed antibodies recognize a site on gp41 that is proximal to the transmembrane-spanning region (Fig. 1). The structural analysis of these broadly reactive neutralizing antibodies suggests that only a small extent of conservation and of exposure (as little as 10–20 amino acid residues or 1–2 N-linked glycans) is sufficient to facilitate specific recognition and, thus, effective antibody binding to the native viral spike.
The HIV-1 envelope spike is the target of known virus-directed neutralizing antibodies. An image of the viral spike obtained by cryo-electron microscopy (light grey)95 is shown fitted with atomic-level ribbon models for three portions of the HIV-1 envelope glycoprotein (Env; red): the membrane-proximal external region (MPER) of gp41 (Ref. 42) is shown towards the top of the image, the structure of the core gp120 with intact amino- and carboxyl termini96 is shown in the middle of the image, and the V1/V2 domain55 is shown towards the bottom of the image. Antibodies that effectively neutralize HIV-1 primarily target four specific regions in Env: the MPER, which is bound by antibody 10E8 (Ref. 42) (cyan), the CD4-binding site on gp120, which is bound by antibody VRC01 (Ref. 34) (fuchsia), and two sites of N-linked glycosylation, one of which is in the V1/ V2 region at residue Asn160 and is bound by antibody PG9 (Refs 55, 57) (green), and the other which is a glycan V3 epitope that generally includes residue Asn332 and is bound by antibody PGT128 (Ref. 60) (blue).
Structural features of broadly neutralizing antibodies. Antibodies in each of the four categories can be further classified on the basis of similarities in their structural mode of epitope recognition and on the basis of similarities in B cell ontogeny — that is, the developmental pathway a B cell lineage undergoes to generate the specific features that are required for effective HIV-1 recognition50 (Table 1).
Co-crystal structures comprising both the antibody and the epitope have been resolved for representative antibodies from each of the four categories. Among CD4-binding site-directed antibodies, several antibodies (such as b12 and CH103) use traditional modes of complementarity-determining region (CDR) recognition; however, the most potent and broadly reactive CD4-binding site-directed antibodies (such as the VRC01 class) structurally mimic the CD4 receptor. Such mimicry involves structural and chemical similarity between the heavy chain component of the antibody and the CD4 receptor in their interaction with gp120 (Ref. 34). This type of interaction involves substantial β-strand contacts between the epitope and the second CDR on the heavy chain (CDR H2) of the antibody; this region shows a marked extent of somatic mutation33,37 and is fully encoded within the immunoglobulin heavy-chain variable (VH) gene33,34. So far, only two VH genes (VH1–2 and VH1–46) have been associated with these CD4-binding site-directed antibodies; antibodies with these heavy chains also show an extraordinarily high level of somatic mutations37,39,51. These mutations are not only in the CDR loops but also in the framework region of the antibody, and the CD4 mimicry is probably also related to interactions of the framework region with the CD4-binding site on gp120 (Ref. 52). The restricted VH gene origin of the antibody heavy chain suggests that specific features of the VH1–2 and VH1–46 genes are required to initiate epitope recognition; these VH gene alleles that are associated with the VRC01 class of antibodies are found in most of the human population, which suggests that a general induction of CD4-mimicking antibodies is possible, at least in principle51,53.
Among the V1/V2-directed neutralizing antibodies, epitope recognition involves an uncommonly long third CDR on the heavy chain (CDR H3) of the antibody, which reaches through the glycan shield of gp120 to contact amino acid residues on strand C of V1/V2, while also making contacts with proximal glycans54,55,56,57. CDR H3 regions of the required length (usually >25 amino acid residues) are found in less than 0.2% of antibody heavy chains58. It seems probable that these V1/V2-directed neutralizing antibodies derive from naive B cell precursors that encode the requisite length of CDR H3 and that these antibodies attain the necessary peptide and glycan affinity through additional somatic hypermutation to achieve broad recognition of HIV-1 (Refs 58, 59) (see below).
Both glycan V3-directed antibodies and MPER-directed antibodies have moderately long CDR H3 regions and have undergone substantial levels of affinity maturation. Some antibodies in the glycan V3-directed category (for example, PGT121 to PGT137) seem to require a moderately long CDR H3 region to penetrate the glycan shield, which is similar to the V1/V2-directed antibodies, and they also seem to require extensive somatic hypermutation and crucial deletions or insertions to generate the necessary alterations in antibody framework regions that lead to high-affinity binding38,60,61,62,63. The first MPER-directed antibodies to be described — 2F5 and 4E10 — show substantial polyreactivity and autoreactivity64,65,66, which is perhaps partly due to the requirement for hydrophobic residues in the CDR H3 region to interact with the host cell membrane67,68. However, at least one antibody in this category, 10E8, potently neutralizes the vast majority of HIV-1 strains and does not show autoreactivity in assays of autoimmunity that detect polyreactivity42, which suggests that there might be pathways that could prevent clonal deletion of the B cell precursors and thus increase the likelihood that this antibody can be elicited by vaccination. Antibody 10E8 uses extensive interactions on a moderately long CDR H3 to recognize conserved MPER elements on gp41, and somatic mutation further hones contact within the CDR loops42.
Together, these structural studies have provided insights into the mechanisms by which broadly neutralizing antibodies engage and inactivate HIV-1 Env, and the pathways by which these antibodies arise. Furthermore, these studies facilitate the characterization of the highly conserved structural determinants that are required for antibody recognition. They therefore enable the design of immunogens for vaccination. The idea that antibodies can serve as a guide for immunogen design has been an attractive concept69 that has yet to be realized (see below).
Eliciting effective antibodies
As with other pathogens, HIV-1 vaccine strategies generally seek to use information that has been gleaned from studying the development of effective antibodies in response to natural infection. However, unlike the antibody response generated during many acute viral infections — which matures rapidly to generate high-affinity neutralizing antibodies — broadly neutralizing HIV-1 antibodies take months to years to develop15,23. An unresolved question is: what is the immunological and molecular basis for this delayed response?
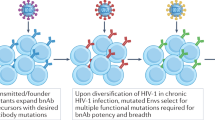
On the one hand, the long time that is required to elicit these antibody responses may partly be a consequence of HIV-1 infection itself, which causes impairment of immune function, including the destruction of CD4+ T cells and damage to lymph node architecture. On the other hand, some aspects of this response may be related to the intrinsic nature of Env as an antigen; for example, it is difficult to elicit HIV-1 neutralizing antibodies in healthy volunteers by immunizing with Env subunit vaccines (such as vaccines containing gp120)3,70. This observation raises the possibility that impaired immune function during natural infection may not be the primary reason that it is difficult to elicit broadly neutralizing antibodies. In addition, the co-evolution of virus and antibody responses may drive the development of effective HIV-1 neutralizing antibodies; for example, broadly neutralizing Asn332-directed antibodies, which bind the glycan V3 epitope, seem to be generated in response to viral escape variants that acquire the appropriate Asn332 glycan as a result of escape from earlier strain-specific antibodies71. Moreover, in a recent study involving longitudinal sampling of a developing broadly neutralizing antibody (CH103) and co-evolving virus, increases in viral divergence were observed just before the development of increased antibody breadth72, which further supports the idea that viral evolution drives the generation of increased breadth in individuals who develop broadly neutralizing HIV-1 antibodies.
Previous attempts to induce broadly neutralizing antibodies by vaccination were fairly unsuccessful. Progress in the past was limited by the unrecognized complexity of the HIV-1 Env and the numerous mechanisms by which HIV-1 evades immunity; for example, early efforts focused on the gp120 monomer as an immunogen and used matched strains to test the neutralization capacity of the resulting antibodies. Although potent immunization could be observed, it became evident that this activity was strain specific, often directed against the V3 loop of gp120. However, when variable regions were removed from the gp120 immunogen it was difficult to elicit any neutralization. This lack of neutralization probably arose from the use of monomeric proteins that presented epitopes that are not exposed on the native viral spike, such as the inner domain. Subsequent attempts to use stabilized soluble trimers also induced responses to immunodominant, strain-specific sites, such as the V1/V2 region or the V3 region. It was evident that even these soluble trimers assumed conformations that were different from the membrane-bound viral spike and therefore they failed to elicit broadly neutralizing antibody responses.
The limitations of these approaches not only led to a failure to induce broadly neutralizing antibodies, but also frustrated efforts to define additional broadly neutralizing antibodies. For more than 15 years, the field relied on four broadly neutralizing antibodies — b12, 2G12, 2F5 and 4E10 — the activity of which was considerably better than strain-specific monoclonal antibodies, but the breadth and the potency of which were nonetheless relatively restricted. Attempts to generate new broadly neutralizing antibodies uniformly failed because screening approaches typically used non-physiological viral glycoproteins. Progress in this field came from a series of different advances. First, it was discovered that viruses passaged in vitro were more easily neutralized than primary isolates from patients. Thus, it became crucial to assemble panels of viral isolates directly from patients, which could be tiered according to their level of neutralization resistance20. Second, antibodies required screening for neutralization against more resistant 'tier 2' or tier 3 viruses to detect broadly neutralizing antibodies31. Third, advances in immunoglobulin gene cloning facilitated high throughput and sensitive screening of individual B cells that enabled the isolation of these rare broadly neutralizing antibodies73. Finally, the description of the viral envelope structure enabled efforts to be focused on functionally constrained determinants that were highly conserved among strains33.
Thus, immunization strategies might need to stimulate the immune system to recognize the functional viral spike through the use of protein mimics and to subsequently produce high-affinity antibodies that recognize conserved epitopes. Alternatively, a vaccination strategy might start with an immunogen that presents a specific conserved epitope and that then boosts the response with the same epitope on a different immunogen to achieve high affinity recognition of the epitope in the context of the native viral spike (Fig. 2a,b). The most effective antibodies target only a few sites of HIV-1 Env vulnerability, and this selective recognition has led to the development of vaccines that use epitope immunogens that stimulate immune responses to these highly conserved functionally important domains.
Over the past 25 years, multiple strategies have been used to elicit effective HIV-1 neutralizing antibodies. These can be placed into three categories. a | Viral spike mimics seek to replicate the functional oligomeric envelope glycoprotein (Env) spike of HIV-1, which is the target of the known broadly neutralizing antibodies. Notable spike mimics include protease purified spikes (or example, gp160) from virions97, gene-delivered spikes (for example, gp160), soluble gp140 protein, a genetically modified form of the trimer that is fully cleaved and stabilized by a disulphide bond between gp120 and gp41 (also known as BG505 664.SOSP; bonds are depicted by lines in the image)57,62, and gp140 spikes lacking a cleavage site stabilized by a domain derived from the T4 phage fibritin protein that facilitates trimerization98 (for example, scaffolded gp140). b | Epitope immunogens focus on either a particular antibody epitope on the Env spike (such as those shown for the 2F5 antibody99) or a site of vulnerability targeted by antibodies that recognize overlapping epitopes (such as antibodies b12, VRC01 and VRC-PG04 shown for the CD4-binding site34,100,101,102; depicted as space-filling models). Epitope scaffold 1 (ES1) to ES5 refer to progressive modifications of the highly conserved epitope that present alternative forms of the same epitope to the immune system by vaccination. A ribbon model of the 2F5 antibody is also depicted. This panel is modified, with permission, from Ref. 99 (2010) © Proceedings of the National Academy of Sciences. c | As specific antibodies with similar B cell ontogenies are induced in multiple HIV-1-infected individuals, strategies to elicit such antibodies can focus on guiding the development of a reproducible B cell lineage, from germline activation, through intermediate maturation, to the expansion of the B cell population that produces mature broadly neutralizing antibodies.
Adding to the complexity of eliciting broadly neutralizing antibodies is the observation that the most effective HIV-1 neutralizing antibodies have unusual structural features (see above), which raises the possibility that the extended time required to induce these antibodies may depend on rare stochastic events or on more unusual or complex IgG gene alterations during development. A detailed understanding of the B cell ontogeny of each broadly neutralizing antibody may thus provide a 'road map' which could be used to elicit similar antibodies through immunization (Fig. 2c).
Guiding somatic mutation. Although the tools of structural biology can be used to create proteins that bind to known neutralizing antibodies, such immunogens must engage B cell receptors (BCRs) on naive B cells, trigger signalling and initiate the process of somatic mutation that is required for affinity maturation. For almost all HIV-1 neutralizing antibodies studied, the reversion of the antibody variable genes to the germline sequences results in markedly decreased affinities to HIV-1 Env34,52,59,74,75,76. Although a germline precursor antibody is expected to have a lower antigenic affinity than the somatically matured antibody that it gives rise to, some variable gene-reverted HIV-1 antibodies — particularly those of the VRC01 class that use their heavy-chain CDR H2 to mimic CD4 binding to gp120 — show no detectable binding to HIV-1 Env34,37,77. This lack of gp120 recognition by the precursor antibodies raises questions about the antigen that engages the BCR on naive B cells. Although BCR triggering might occur if antigens are displayed in a polyvalent manner on the B cell surface, the very poor affinities of these germline precursor antibodies for HIV-1 Env suggest that the initial triggering of the precursor B cells or the early development of B cell responses may be limited.
One potential immunization strategy that may guide the development of broadly neutralizing antibodies is to create a series of Env immunogens that bind to the BCR on naive B cells and subsequently to those on crucial intermediate B cells at early, middle and late stages of the antibody developmental pathway78,79. Such rationally designed immunogens may be best suited to the induction of antibody modalities that are elicited in multiple individuals (Fig. 3), such as those antibodies of the VRC01 class. Immunogens specifically designed to engage naive B cells that express VRC01 class antibodies have recently been designed and were shown to bind to and to trigger the BCR of these cells in in vitro assays53,80,81. Immunization studies carried out in vivo will be required to test the ability of such vaccine antigens to elicit the desired antibody response.
Antibodies are generated by stochastic processes of V(D)J recombination and immunoglobulin heavy and light chain combinatorial recognition, which results in greater than 1012-fold diversity. Standard selection against a typical antigen generates diverse antibodies in different individuals (also known as donors). The antibodies elicited in each individual have different variable (V) gene origins and have undergone different V(D)J recombination events, but all of the antibodies can bind to the same antigen (left branch). However, the recognition of conserved epitopes on the HIV-1 viral spike in some instances results in remarkably similar antibodies even though they develop in different individuals; these antibodies share the same V genes and B cell ontogenies (right branch)37,39,51. Similarities in epitope recognition may relate to constraints in epitope recognition and/or they may relate to the use of select V gene elements, as has been suggested for the VRC01 class of antibodies34,103 and for CD4-binding site-directed antibodies104. The donor codes indicate which antibodies were isolated from different subjects and from independent cohort series that have been previously described.
In addition to structure-based design efforts, recent advances in next-generation sequencing of antibody gene transcripts enable the genetic record of the developing B cell response to be determined. This provides a way to elucidate the development pathway of an antibody from an unmutated antibody progenitor to a high-affinity neutralizing antibody39,51,82. Similar genetic analysis of the antigen-specific B cell repertoire may then allow us to follow antibody development in the context of HIV-1 immunization to determine if we can direct naive B cells towards the most relevant pathway of affinity maturation and differentiation. Once such pathway-dependent B cell maturation is adequately defined, it may then be possible to target specific VH genes that optimize the chances of success in a vaccine (such as VH1–2 and VH1–46 for the VRC01 class of antibodies). In addition, the development of potential vaccines may require testing in animals that have immune repertoires sufficiently close to a 'typical' human; for example, mice with knock-in of the human IgG locus or non-human primates with an IgG locus similar to that in humans83,84.
Engaging the naive B cell repertoire. In some instances, rare recombination events create the crucial genetic elements needed for effective HIV-1 neutralization. Long CDR H3 loops seem to preferentially result from specific V(D)J gene recombinations or, in some cases, from unusual DD gene fusions58,85,86. Thus, the time it takes to elicit uncommonly long CDR H3 of the V1/V2-directed category of broadly neutralizing antibodies could be due to the low frequency of the appropriate primary recombination event, together with the additional requisite somatic mutations. An additional feature of these broadly neutralizing antibodies is the high frequency of small insertions and deletions that probably arise as a by-product of repair during somatic mutation, which is similar to what occurs in other highly mutated IgGs. Some of these deletions are repeatedly seen in broadly neutralizing antibodies that are derived from different individuals, which suggests that additional specific changes are required to generate them.
During natural infection, the continual generation of viral escape mutants provides fresh antigenic templates to engage naive BCRs, including rarely generated recombinants. Such escape mutants would engage the naive B cell rather than stimulate the memory B cell responses needed to generate broadly neutralizing antibodies. By contrast, immunization generally involves the sequential use of a single antigen that stimulates the generation of high-affinity antigen-specific antibodies. The resulting antibody could bind antigens and thus prevent the vaccine antigens from binding to and stimulating other rare naive B cells. Therefore, homologous boosting for HIV-1 may differ from that for immunization against other pathogens that have more restricted antigenic diversity, in which immunodominance may have a less prominent role. It is not entirely clear how to surmount this difficulty, but one means might be through the continual use of HIV-1 Env variants, each sufficiently different to avoid binding by non-neutralizing antibodies. Strategies that seek to vaccinate with trimers that resemble the viral spike or those that use highly conserved epitope structures may benefit from such longitudinal variation, which could be used as a general strategy for boosting subdominant immune responses (Fig. 2).
Although we have made remarkable progress in elucidating the structure of vulnerable regions on the HIV-1 Env spike and have begun to translate this knowledge into the generation of novel immunogens, key scientific obstacles must be overcome to induce broadly reactive neutralizing antibodies. Immunization with various forms of gene-based vectors encoding env genes, or with soluble Env proteins, generates high levels of Env-specific antibodies, but the neutralizing antibody responses induced are not nearly as potent or broadly reactive as most of those that are mediated by neutralizing monoclonal antibodies. The factors that influence this limited neutralizing antibody response are still being elucidated but may include the low-affinity binding of germline antibodies noted above, the immunodominance of Env variable regions and the potential poor immunoreactivity of specific neutralization regions of Env.
Antibody-mediated prevention
The challenges of active immunization have led to a search for alternative methods to harness the potential benefits of neutralizing antibodies. The passive transfer of IgG offers an alternative way to use the broadly neutralizing HIV-1 antibodies that have recently been identified. The issues related to this strategy of direct antibody prophylaxis are quite different from those related to active immunization. In the case of passive immunization, no understanding of the B cell ontogeny is required; rather, this approach relies on the selection of highly effective neutralizing antibodies, the clinical grade production of IgG and the ultimate delivery and maintenance of a suitable concentration of transferred antibody (Box 1).
Passive antibody administration. Although there are over 30 licensed therapeutic monoclonal antibodies87, only one antibody has been approved for the prevention of an infectious pathogen. Palivizumab (Synagis; MedImmune) is a humanized mouse monoclonal antibody directed against the fusion glycoprotein of respiratory syncytial virus (RSV). When it is delivered monthly by intramuscular injection, Synagis effectively protects against clinical RSV disease in premature infants88. Similarly, it may be possible to prevent HIV-1 infection of humans via passive antibody transfer. In support of this idea is the demonstration that neutralizing antibodies prevent infection in the chimeric simian–human immunodeficiency virus (SHIV) macaque model or in the HIV-1 humanized mouse model of infection46,47. In studies using these animal models, the levels of antibody that was required for protection varied depending on the source of the antibody and the strain of challenge virus. However, recent studies using repeated viral challenges through a mucosal route have shown that an antibody plasma concentration in the range of 25 to 50 μg per ml, which results in serum neutralizing antibody titres in the range of ∼1:200 (that is, serum at a 1 in 200 dilution produced 90% neutralization in the assay), could prevent mucosal SHIV infection in the macaque model48.
Until the discovery of the currently used generation of highly potent HIV-1 monoclonal antibodies, the concept of antibody-based prevention of HIV-1 infection seemed untenable, as antibodies showed modest potency and limited breadth of neutralization. However, currently used antibodies, such as those in the VRC01 class, the recently described PG and PGT glycan-reactive antibodies of the glycan V3 category, and the MPER-directed 10E8 antibody, show potencies at concentrations below 1 μg per ml against most HIV-1 strains in vitro31,33,38,42,89. At least one broadly neutralizing antibody, PG121, shows protection at low concentrations following passive transfer into macaques90. Recent advances in antibody engineering suggest the potential to increase both antibody potency and the antibody half-life87,89,91. In addition, the combined use of HIV-1 antibodies directed to distinct epitopes has the potential to potently neutralize the vast majority of globally circulating HIV-1 strains37,38,92.
However, there is currently no proof-of-concept that an HIV-1 monoclonal antibody can prevent human HIV-1 infection, although Phase I clinical trials using the VRC01 antibody are being planned. The ability of an antibody to protect against infection in humans would not only provide a potential tool to reduce HIV-1 transmission, but would also validate the antibody target for further development as a vaccine antigen.
Genetic delivery of antibody molecules. Should clinical trials show that the passive transfer of monoclonal antibodies can successfully prevent HIV-1 infection in humans, there would remain considerable challenges regarding the cost and scaling up the antibody product. Hence, an alternative approach is to use vector-based delivery of genes encoding antibody molecules as a means to produce sufficient levels of antibodies over a sustained period of time. Indeed, high levels of sustained antibody expression in mice and monkeys have been achieved in two studies; both studies used an adeno-associated virus vector to deliver the genes encoding the immunoglobulin heavy and light chains and showed protection against SIV infection in non-human primates93,94 and against HIV-1 infection in humanized mice39. These studies provide a proof-of-concept that the delivery of antibody genes can confer long-term protection against lentiviral infection.
For such a method to be widely adopted, several technical issues and regulatory concerns would need to be addressed; for example, immune responses to either the vector or to the antibody transgene might limit the magnitude and/or the duration of antibody gene expression. The modulation of this response and the ability to promote specific tolerance to these gene products might therefore be required to optimize efficacy. For widespread use of this approach, it would also be desirable to enable the regulation of gene expression to eliminate the encoded antibody in the event of an unanticipated adverse effect.
Despite these technical challenges, various gene delivery vectors are now under consideration that may facilitate long-term antibody production in vivo. Although the adeno-associated virus vector has shown promise as a replication-defective vector that can mediate long-term gene expression, other gene delivery modalities, including DNA and RNA expression vectors, offer alternative strategies to confer long-term protection against HIV-1 infection by genetic immunization.
The end of the beginning
The extraordinary ability of HIV-1 to evade the humoral immune response has long been recognized. This resistance has led to some pessimism about the potential of a vaccine to induce neutralizing antibodies that could recognize diverse strains of HIV-1, and thus prevent infection.
Renewed, yet cautious, optimism has nonetheless arisen from the understanding that the immune system can generate potent and broadly neutralizing antibodies. The isolation and the characterization of such monoclonal antibodies has helped to elucidate key vulnerable regions on the HIV-1 viral spike. Co-crystal structures of antibodies bound to their epitopes on gp120 or gp41 have provided atomic level detail of these conserved regions and this facilitates the design of novel immunogens to present these viral epitopes to the immune system.
In addition to structural information, a better understanding of the immune pathways that lead to the generation of HIV-1 neutralizing antibodies will enable the development of protective immunity. The most potent of the known neutralizing antibodies have unusual characteristics that may hinder their induction. Depending on the antibody, these features include genetic restriction to specific VH genes, long CDR H3 loops and high levels of affinity maturation. Next-generation sequencing of B cell antibody genes is providing a mechanistic understanding of how such neutralizing antibodies acquire these characteristics and, thus, can provide insights into how to elicit similar antibodies via immunization. An understanding of antibody genetic pathways enables us to track the immune response during immunization as a means to guide further immunogen design and immunization strategies.
Finally, highly potent and broadly reactive neutralizing antibodies provide the possibility of direct antibody passive transfer and new monoclonal antibody gene delivery strategies, as a means to prevent HIV-1 infection. The proof-of-concept of such antibody-mediated protection in humans would validate specific viral epitopes for vaccine design and could provide immune correlates that could be measured in future vaccine efficacy trials, all of which would accelerate the development of an effective HIV-1 vaccine.
However, a number of challenges remain in the effort to develop a highly effective HIV-1 vaccine, either for active or passive immunization. For active immunization, it will be crucial not only to understand antigenicity — for example, the structural basis of interactions with broadly neutralizing antibodies — but also to understand how to stimulate antibody responses (that is, immunogenicity) of the highly conserved target sites. Although the idea of antigen-guided development of B cells that produce these broadly neutralizing antibodies is attractive, questions remain about how and when to vaccinate to achieve these effects and how to select the optimal combination of antigens. These parameters must first be analysed in experimental animals whose immune systems are genetically different from the human immune system, which may result in both the generation of false-positive and false-negative leads. Therefore, human studies will ultimately be required. For these clinical studies, there is a need for a greater production capacity at good manufacturing practice standards to generate vaccine candidates, as well as finding the financial means to conduct large, lengthy and costly efficacy trials.
For passive immunization, several unknown factors must also be addressed. As the antibody doses required for in vivo protection are unknown, it is not clear whether passive immunization with monoclonal antibodies is practical or feasible. Can this approach be implemented on a large scale and in the developing world? For antibody gene delivery, the optimal vectors and antibody genes must be defined, as well as the antibody levels that are protective and that can be maintained in the long-term in humans using this method. Safety concerns related to long-term antibody gene expression will also need to be addressed in future clinical trials; whether gene-delivered antibodies will elicit destructive host immune responses — to human tissues, to the vector or to the antibodies themselves — will need to be determined. For these reasons, the recent successes do not represent the end or the beginning of the end, but rather the end of the beginning, of the quest for an HIV-1 vaccine.
References
Nabel, G. J. Designing tomorrow's vaccines. N. Engl. J. Med. 368, 551–560 (2013).
Rerks-Ngarm, S. et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361, 2209–2220 (2009).
Mascola, J. R. & Montefiori, D. C. The role of antibodies in HIV vaccines. Annu. Rev. Immunol. 28, 413–444 (2010).
McElrath, M. J. & Haynes, B. F. Induction of immunity to human immunodeficiency virus type-1 by vaccination. Immunity 33, 542–554 (2010).
McMichael, A. J., Borrow, P., Tomaras, G. D., Goonetilleke, N. & Haynes, B. F. The immune response during acute HIV-1 infection: clues for vaccine development. Nature Rev. Immunol. 10, 11–23 (2010).
Kong, L. & Sattentau, Q. J. Antigenicity and immunogenicity in HIV-1 antibody-based vaccine design. J. AIDS Clin. Res. S8, 3 (2012).
Wyatt, R. & Sodroski, J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280, 1884–1888 (1998).
Pantophlet, R. & Burton, D. R. GP120: target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 24, 739–769 (2006).
Overbaugh, J. & Morris, L. The antibody response against HIV-1. Cold Spring Harb. Perspect. Med. 2, a007039 (2012).
Kwong, P. D. et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682 (2002).
Wei, X. et al. Antibody neutralization and escape by HIV-1. Nature 422, 307–312 (2003).
Burton, D. R., Stanfield, R. L. & Wilson, I. A. Antibody versus HIV in a clash of evolutionary titans. Proc. Natl Acad. Sci. USA 102, 14943–14948 (2005).
Richman, D. D., Wrin, T., Little, S. J. & Petropoulos, C. J. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl Acad. Sci. USA 100, 4144–4149 (2003).
Gray, E. S. et al. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 81, 6187–6196 (2007).
Mikell, I. et al. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog. 7, e1001251 (2011).
Tomaras, G. D. et al. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J. Virol. 85, 11502–11519 (2011).
Binley, J. M. et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78, 13232–13252 (2004).
Li, Y. et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nature Med. 13, 1032–1034 (2007).
Sather, D. N. et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83, 757–769 (2009).
Stamatatos, L., Morris, L., Burton, D. R. & Mascola, J. R. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nature Med. 15, 866–870 (2009).
Doria-Rose, N. A. et al. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J. Virol. 84, 1631–1636 (2010).
Walker, L. M. et al. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 6, e1001028 (2010).
Gray, E. S. et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J. Virol. 85, 4828–4840 (2011).
Muster, T. et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67, 6642–6647 (1993).
Burton, D. R. et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266, 1024–1027 (1994).
Trkola, A. et al. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 69, 6609–6617 (1995).
Zwick, M. B. et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75, 10892–10905 (2001).
Calarese, D. A. et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300, 2065–2071 (2003).
Ofek, G. et al. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 78, 10724–10737 (2004).
Cardoso, R. M. et al. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22, 163–173 (2005).
Walker, L. M. et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289 (2009).
Corti, D. et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS ONE 5, e8805 (2010).
Wu, X. et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 (2010).
Zhou, T. et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817 (2010).
Bonsignori, M. et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 85, 9998–10009 (2011).
Morris, L. et al. Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PLoS ONE 6, e23532 (2011).
Scheid, J. F. et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637 (2011).
Walker, L. M. et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 (2011).
Wu, X. et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602 (2011).
Zhu, Z. et al. Cross-reactive HIV-1-neutralizing human monoclonal antibodies identified from a patient with 2F5-like antibodies. J. Virol. 85, 11401–11408 (2011).
Bonsignori, M. et al. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: Implications for vaccine design. J. Virol. 86, 4688–4692 (2012).
Huang, J. et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412 (2012).
McMichael, A. & Hanke, T. The quest for an AIDS vaccine: is the CD8+ T-cell approach feasible? Nature Rev. Immunol. 2, 283–291 (2002).
Shibata, R. et al. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nature Med. 5, 204–210 (1999).
Mascola, J. R. et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nature Med. 6, 207–210 (2000).
Mascola, J. R. Defining the protective antibody response for HIV-1. Curr. Mol. Med. 3, 209–216 (2003).
Burke, B. & Barnett, S. W. Broadening our view of protective antibody responses against HIV. Curr. HIV Res. 5, 625–641 (2007).
Hessell, A. J. et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nature Med. 15, 951–954 (2009).
Hessell, A. J. et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 5, e1000433 (2009).
Kwong, P. D. & Mascola, J. R. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 37, 412–425 (2012).
Zhou, T. et al. Multi-donor analysis reveals structural elements, genetic determinants and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity http://dx.doi.org/10.1016/j.immuni.2013.04.012 (2013).
Klein, F. et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153, 126–138 (2013).
Jardine, J. et al. Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716 (2013).
Pejchal, R. et al. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc. Natl Acad. Sci. USA 107, 11483–11488 (2010).
McLellan, J. S. et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480, 336–343 (2011).
Pancera, M. et al. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat. Struct. Mol. Biol. 20, 804–813 (2013).
Julien, J. P. et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc. Natl Acad. Sci. USA 110, 4351–4356 (2013).
Briney, B. S. et al. Frequency and genetic characterization of V(DD)J recombinants in the human peripheral blood antibody repertoire. Immunology 137, 56–64 (2012).
Pancera, M. et al. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J. Virol. 84, 8098–8110 (2010).
Pejchal, R. et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334, 1097–1103 (2011).
Mouquet, H., Warncke, M., Scheid, J. F., Seaman, M. S. & Nussenzweig, M. C. Enhanced HIV-1 neutralization by antibody heteroligation. Proc. Natl Acad. Sci. USA 109, 875–880 (2012).
Julien, J. P. et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 9, e1003342 (2013).
Kong, L. et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat. Struct. Mol. Biol. 20, 796–803 (2013).
Haynes, B. F. et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308, 1906–1908 (2005).
Alam, S. M. et al. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 178, 4424–4435 (2007).
Matyas, G. R. et al. Neutralizing antibodies induced by liposomal HIV-1 glycoprotein 41 peptide simultaneously bind to both the 2F5 or 4E10 epitope and lipid epitopes. AIDS 23, 2069–2077 (2009).
Sun, Z. Y. et al. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity 28, 52–63 (2008).
Ofek, G. et al. Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J. Virol. 84, 2955–2962 (2010).
Burton, D. R. Antibodies, viruses and vaccines. Nature Rev. Immunol. 2, 706–713 (2002).
McCoy, L. E. & Weiss, R. A. Neutralizing antibodies to HIV-1 induced by immunization. J. Exp. Med. 210, 209–223 (2013).
Moore, P. L. et al. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nature Med. 18, 1688–1692 (2012).
Liao, H. X. et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476 (2013).
Scheid, J. F. et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458, 636–640 (2009).
Xiao, X. et al. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem. Biophys. Res. Commun. 390, 404–409 (2009).
Alam, S. M. et al. Differential reactivity of germ line allelic variants of a broadly neutralizing HIV-1 antibody to a gp41 fusion intermediate conformation. J. Virol. 85, 11725–11731 (2011).
Liao, H. X. et al. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J. Exp. Med. 208, 2237–2249 (2011).
Hoot, S. et al. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS Pathog. 9, e1003106 (2013).
Moir, S., Malaspina, A. & Fauci, A. S. Prospects for an HIV vaccine: leading B cells down the right path. Nature Struct. Mol. Biol. 18, 1317–1321 (2011).
Haynes, B. F., Kelsoe, G., Harrison, S. C. & Kepler, T. B. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nature Biotech. 30, 423–433 (2012).
McGuire, A. T. et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J. Exp. Med. 210, 655–663 (2013).
Scharf, L. et al. Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody. Proc. Natl Acad. Sci. USA 110, 6049–6054 (2013).
Zhu, J. et al. Somatic populations of PGT135-137 HIV-1-neutralizing antibodies identified by 454 pyrosequencing and bioinformatics. Front. Microbiol. 3, 315 (2012).
Devoy, A., Bunton-Stasyshyn, R. K., Tybulewicz, V. L., Smith, A. J. & Fisher, E. M. Genomically humanized mice: technologies and promises. Nature Rev. Genet. 13, 14–20 (2012).
Sundling, C. et al. High-resolution definition of vaccine-elicited B cell responses against the HIV primary receptor binding site. Sci. Transl. Med. 4, 142ra196 (2012).
Briney, B. S., Willis, J. R. & Crowe, J. E. Jr. Human peripheral blood antibodies with long HCDR3s are established primarily at original recombination using a limited subset of germline genes. PLoS ONE 7, e36750 (2012).
Larimore, K., McCormick, M. W., Robins, H. S. & Greenberg, P. D. Shaping of human germline IgH repertoires revealed by deep sequencing. J. Immunol. 189, 3221–3230 (2012).
Chan, A. C. & Carter, P. J. Therapeutic antibodies for autoimmunity and inflammation. Nature Rev. Immunol. 10, 301–316 (2010).
Shadman, K. A. & Wald, E. R. A review of palivizumab and emerging therapies for respiratory syncytial virus. Expert Opin. Biol. Ther. 11, 1455–1467 (2011).
Diskin, R. et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 334, 1289–1293 (2011).
Moldt, B. et al. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc. Natl Acad. Sci. USA 109, 18921–18925 (2012).
Zalevsky, J. et al. Enhanced antibody half-life improves in vivo activity. Nature Biotech. 28, 157–159 (2010).
Doria-Rose, N. A. et al. HIV-1 neutralization coverage is improved by combining monoclonal antibodies that target independent epitopes. J. Virol. 86, 3393–3397 (2012).
Balazs, A. B. et al. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature 481, 81–84 (2011).
Johnson, P. R. et al. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nature Med. 15, 901–906 (2009).
Mao, Y. et al. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nature Struct. Mol. Biol. 19, 893–899 (2012).
Pancera, M. et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl Acad. Sci. USA 107, 1166–1171 (2010).
Tong, T., Crooks, E. T., Osawa, K. & Binley, J. M. HIV-1 virus-like particles bearing pure env trimers expose neutralizing epitopes but occlude nonneutralizing epitopes. J. Virol. 86, 3574–3587 (2012).
Yang, X. et al. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 76, 4634–4642 (2002).
Ofek, G. et al. Elicitation of structure-specific antibodies by epitope scaffolds. Proc. Natl Acad. Sci. USA 107, 17880–17887 (2010).
Zhou, T. et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445, 732–737 (2007).
Azoitei, M. L. et al. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science 334, 373–376 (2011).
Joyce, M. G. et al. Outer domain of HIV-1 gp120: Antigenic optimization, structural malleability, and crystal structure with antibody VRC-PG04. J. Virol. 87, 2294–2306 (2013).
West, A. P. Jr., Diskin, R., Nussenzweig, M. C. & Bjorkman, P. J. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc. Natl Acad. Sci. USA 109, E2083–E2090 (2012).
Huang, C. C. et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc. Natl Acad. Sci. USA 101, 2706–2711 (2004).
Acknowledgements
We thank B. Hartman and J. Stuckey for assistance with display items, J. Gorman for modelling for Figure 1, A. Tislerics for manuscript preparation, N. Michael for helpful discussions, and members of the Virology Laboratory, the Structural Biology Section, the Structural Bioinformatics Core, and the Humoral Immunology Section, Vaccine Research Center, for discussions and comments on the manuscript. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, US National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Kwong, P., Mascola, J. & Nabel, G. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat Rev Immunol 13, 693–701 (2013). https://doi.org/10.1038/nri3516
Published:
Issue Date:
DOI: https://doi.org/10.1038/nri3516
This article is cited by
-
Conformational antigenic heterogeneity as a cause of the persistent fraction in HIV-1 neutralization
Retrovirology (2023)
-
Structures and immune recognition of Env trimers from two Asia prevalent HIV-1 CRFs
Nature Communications (2023)
-
Leveraging lessons learned from the COVID-19 pandemic for HIV
Communications Medicine (2022)
-
Monoclonal antibodies from humans with Mycobacterium tuberculosis exposure or latent infection recognize distinct arabinomannan epitopes
Communications Biology (2021)
-
On the irrationality of rational design of an HIV vaccine in light of protein intrinsic disorder
Archives of Virology (2021)