Key Points
-
DNA repair occurs in the context of higher-order chromatin structure.
-
The assembly of the DNA-repair machinery on damaged chromatin is a spatially and temporally highly coordinated event.
-
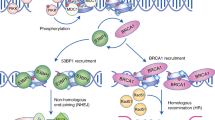
Sites of DNA repair manifest themselves as DNA-repair foci, which form as a consequence of the accumulation of repair factors at sites of damage and serve to amplify the damage signal.
-
Histone modifications, histone exchange and chromatin remodelling are essential steps in DNA repair.
-
In mammalian cells, double-stranded DNA breaks (DSBs) are positionally stable and do not migrate in the nuclear space. In yeast, DSBs can migrate and multiple DSBs coalesce in repair centres.
-
The non-random spatial organization of the genome contributes to the translocation frequency of chromosomes in vivo.
Abstract
DNA repair and maintenance of genome stability are crucial to cellular and organismal function, and defects in these processes have been implicated in cancer and ageing. Detailed molecular, biochemical and genetic analyses have outlined the molecular framework involved in cellular DNA-repair pathways, but recent cell-biological approaches have revealed important roles for the spatial and temporal organization of the DNA-repair machinery during the recognition of DNA lesions and the assembly of repair complexes. It has also become clear that local higher-order chromatin structure, chromatin dynamics and non-random global genome organization are key factors in genome maintenance. These cell-biological features of DNA repair illustrate an emerging role for nuclear architecture in multiple aspects of genome maintenance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Wyman, C. & Kanaar, R. DNA double-strand break repair: all's well that ends well. Annu. Rev. Genet. 40, 363–383 (2006).
Lobrich, M. & Jeggo, P. A. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nature Rev. Cancer 7, 861–869 (2007).
Kanaar, R., Wyman, C. & Rothstein, R. Quality control of DNA break metabolism: in the 'end', it's a good thing. EMBO J. 27, 581–588 (2008).
Lukas, J., Lukas, C. & Bartek, J. Mammalian cell cycle checkpoints: signalling pathways and their organization in space and time. DNA Repair 3, 997–1007 (2004).
Misteli, T. Beyond the sequence: cellular organization of genome function. Cell 128, 787–800 (2007).
Berkovich, E., Monnat, R. J. Jr & Kastan, M. B. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nature Cell Biol. 9, 683–690 (2007).
Paull, T. T. & Lee, J. H. The Mre11/Rad50/Nbs1 complex and its role as a DNA double-strand break sensor for ATM. Cell Cycle 4, 737–740 (2005).
Petrini, J. H. & Stracker, T. H. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 13, 458–462 (2003).
Costanzo, V., Paull, T., Gottesman, M. & Gautier, J. Mre11 assembles linear DNA fragments into DNA damage signaling complexes. PLoS Biol. 2, E110 (2004).
Lee, J. H. & Paull, T. T. ATM activation by DNA double-strand breaks through the Mre11–Rad50–Nbs1 complex. Science 308, 551–554 (2005).
Stucki, M. et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123, 1213–1226 (2005).
Chapman, J. R. & Jackson, S. P. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 9, 795–801 (2008).
Melander, F. et al. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J. Cell Biol. 181, 213–226 (2008).
Spycher, C. et al. Constitutive phosphorylation of MDC1 physically links the MRE11–RAD50–NBS1 complex to damaged chromatin. J. Cell Biol. 181, 227–240 (2008).
Wu, L., Luo, K., Lou, Z. & Chen, J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc. Natl Acad. Sci. USA 105, 11200–11205 (2008).
Huen, M. S. et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914 (2007).
Kolas, N. K. et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318, 1637–1640 (2007).
Mailand, N. et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900 (2007).
Kim, H., Chen, J. & Yu, X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316, 1202–1205 (2007).
Sobhian, B. et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316, 1198–1202 (2007).
Wang, B. et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316, 1194–1198 (2007).
Lisby, M., Barlow, J. H., Burgess, R. C. & Rothstein, R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699–713 (2004). A comprehensive analysis of the temporal sequence of recruitment events in S. cerevisiae.
Melo, J. & Toczyski, D. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14, 237–245 (2002).
Bonilla, C. Y., Melo, J. A. & Toczyski, D. P. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol. Cell 30, 267–276 (2008). Introduces the concept of activation of the DDR in the absence of DNA breaks in S. cerevisiae.
Soutoglou, E. DNA lesions and DNA damage response: Even long lasting relationships need a “break”. Cell Cycle 7, 3653–3658 (2008).
Soutoglou, E. & Misteli, T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science 320, 1507–1510 (2008). Introduces the concept of activation of the DDR in the absence of DNA breaks in mammalian cells.
Lukas, C., Bartek, J. & Lukas, J. Imaging of protein movement induced by chromosomal breakage: tiny 'local' lesions pose great 'global' challenges. Chromosoma 114, 146–154 (2005).
Bekker-Jensen, S. et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 173, 195–206 (2006).
Essers, J. et al. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 21, 2030–2037 (2002).
Lukas, C. et al. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 23, 2674–2683 (2004).
Gorski, S. A., Snyder, S. K., John, S., Grummt, I. & Misteli, T. Modulation of RNA polymerase assembly dynamics in transcriptional regulation. Mol. Cell 30, 486–497 (2008).
Politi, A. et al. Mathematical modeling of nucleotide excision repair reveals efficiency of sequential assembly strategies. Mol. Cell 19, 679–690 (2005). Reports the development of a mathematical model for the assembly process of the DNA-repair machinery in vivo.
Toledo, L. I., Murga, M., Gutierrez-Martinez, P., Soria, R. & Fernandez-Capetillo, O. ATR signaling can drive cells into senescence in the absence of DNA breaks. Genes Dev. 22, 297–302 (2008). Shows that a persistent DDR in the absence of breaks leads to cellular senescence.
Bakkenist, C. J. & Kastan, M. B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421, 499–506 (2003).
Mazumdar, M. et al. Tumor formation via loss of a molecular motor protein. Curr. Biol. 16, 1559–1564 (2006).
Bencokova, Z. et al. ATM activation and signalling under hypoxic conditions. Mol. Cell. Biol. 29, 526–537 (2008).
Downs, J. A., Nussenzweig, M. C. & Nussenzweig, A. Chromatin dynamics and the preservation of genetic information. Nature 447, 951–958 (2007).
Rogakou, E. P., Boon, C., Redon, C. & Bonner, W. M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146, 905–916 (1999).
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 (1998).
Bassing, C. H. et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114, 359–370 (2003).
Celeste, A. et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114, 371–383 (2003). Demonstrates the functional importance of H2AX phosphorylation in genome maintenance.
Celeste, A. et al. Genomic instability in mice lacking histone H2AX. Science 296, 922–927 (2002).
Reina-San-Martin, B. et al. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 197, 1767–1778 (2003).
Xie, A. et al. Control of sister chromatid recombination by histone H2AX. Mol. Cell 16, 1017–1025 (2004).
Zhou, B. B. & Elledge, S. J. The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439 (2000).
Celeste, A. et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nature Cell Biol. 5, 675–679 (2003).
Downs, J. A., Lowndes, N. F. & Jackson, S. P. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408, 1001–1004 (2000).
Fernandez-Capetillo, O. et al. DNA damage-induced G2–M checkpoint activation by histone H2AX and 53BP1. Nature Cell Biol. 4, 993–997 (2002).
Downs, J. A. et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16, 979–990 (2004).
Kusch, T. et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306, 2084–2087 (2004).
Morrison, A. J. et al. INO80 and γ-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119, 767–775 (2004).
Unal, E. et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16, 991–1002 (2004).
van Attikum, H., Fritsch, O., Hohn, B. & Gasser, S. M. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119, 777–788 (2004).
Kruhlak, M. J. et al. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 172, 823–834 (2006).
Ziv, Y. et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nature Cell Biol. 8, 870–876 (2006).
Bao, Y. & Shen, X. Chromatin remodeling in DNA double-strand break repair. Curr. Opin. Genet. Dev. 17, 126–131 (2007).
Chai, B., Huang, J., Cairns, B. R. & Laurent, B. C. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 19, 1656–1661 (2005).
Tsukuda, T., Fleming, A. B., Nickoloff, J. A. & Osley, M. A. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438, 379–383 (2005).
Kobor, M. S. et al. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2, E131 (2004).
Mizuguchi, G. et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348 (2004).
Papamichos-Chronakis, M., Krebs, J. E. & Peterson, C. L. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 20, 2437–2449 (2006).
van Attikum, H., Fritsch, O. & Gasser, S. M. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 26, 4113–4125 (2007).
Park, J. H. et al. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting γ-H2AX induction. EMBO J. 25, 3986–3997 (2006).
Haldar, D. & Kamakaka, R. T. Schizosaccharomyces pombe Hst4 functions in DNA damage response by regulating histone H3 K56 acetylation. Eukaryot. Cell 7, 800–813 (2008).
Jazayeri, A., McAinsh, A. D. & Jackson, S. P. Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc. Natl Acad. Sci. USA 101, 1644–1649 (2004).
Tamburini, B. A. & Tyler, J. K. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol. Cell. Biol. 25, 4903–4913 (2005).
Keogh, M. C. et al. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature 439, 497–501 (2006).
Chowdhury, D. et al. γ-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell 20, 801–809 (2005).
Chowdhury, D. et al. A PP4-phosphatase complex dephosphorylates γ-H2AX generated during DNA replication. Mol. Cell 31, 33–46 (2008).
Krogan, N. J. et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl Acad. Sci. USA 101, 13513–13518 (2004).
Ikura, T. et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell. Biol. 27, 7028–7040 (2007).
Jha, S., Shibata, E. & Dutta, A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol. Cell. Biol. 28, 2690–2700 (2008).
Heo, K. et al. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol. Cell 30, 86–97 (2008).
Polo, S. E., Roche, D. & Almouzni, G. New histone incorporation marks sites of UV repair in human cells. Cell 127, 481–493 (2006).
Chen, C. C. et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134, 231–243 (2008).
Mousson, F., Ochsenbein, F. & Mann, C. The histone chaperone Asf1 at the crossroads of chromatin and DNA checkpoint pathways. Chromosoma 116, 79–93 (2007).
Tyler, J. K. et al. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402, 555–560 (1999).
Mello, J. A. et al. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 3, 329–334 (2002).
O'Hagan, H. M., Mohammad, H. P. & Baylin, S. B. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 4, e1000155 (2008).
Oberdoerffer, P. et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 135, 907–918 (2008).
Goodarzi, A. A. et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell 31, 167–177 (2008). Provides evidence for a role of higher-order chromatin structure in DDR activation.
Ayoub, N., Jeyasekharan, A. D., Bernal, J. A. & Venkitaraman, A. R. HP1-β mobilization promotes chromatin changes that initiate the DNA damage response. Nature 453, 682–686 (2008).
Kim, Y. C. et al. The activation of ATM depends on chromatin interactions occurring prior to DNA damage induction. Nature Cell Biol. 11, 92–96 (2008).
Murga, M. et al. Global chromatin compaction limits the strength of the DNA damage response. J. Cell Biol. 178, 1101–1108 (2007). Provides evidence for an inhibitory effect of chromatin compaction on DNA-damage signalling.
Kim, J. A., Kruhlak, M., Dotiwala, F., Nussenzweig, A. & Haber, J. E. Heterochromatin is refractory to γ-H2AX modification in yeast and mammals. J. Cell Biol. 178, 209–218 (2007).
Efroni, S. et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2, 437–447 (2008).
Lisby, M., Antunez de Mayolo, A., Mortensen, U. H. & Rothstein, R. Cell cycle-regulated centers of DNA double-strand break repair. Cell Cycle 2, 479–483 (2003).
Lisby, M., Mortensen, U. H. & Rothstein, R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nature Cell Biol. 5, 572–577 (2003).
Lisby, M. & Rothstein, R. DNA damage checkpoint and repair centers. Curr. Opin. Cell Biol. 16, 328–334 (2004).
Nagai, S. et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322, 597–602 (2008).
Soutoglou, E. et al. Positional stability of single double-strand breaks in mammalian cells. Nature Cell Biol. 9, 675–682 (2007). Reports the development and use of an experimental system to visualize DSBs in living mammalian cells and demonstrates that mammalian DSBs are immobile in the cell nucleus.
Aten, J. A. et al. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science 303, 92–95 (2004).
Nelms, B. E., Maser, R. S., MacKay, J. F., Lagally, M. G. & Petrini, J. H. In situ visualization of DNA double-strand break repair in human fibroblasts. Science 280, 590–592 (1998).
Dimitrova, N., Chen, Y. C., Spector, D. L. & de Lange, T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456, 524–528 (2008). Shows that 53BP1 promotes chromatin mobility and end joining of deprotected telomeres.
Difilippantonio, S. et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature 456, 529–533 (2008).
Soutoglou, E. & Misteli, T. Mobility and immobility of chromatin in transcription and genome stability. Curr. Opin. Genet. Dev. 17, 435–442 (2007).
Lanctot, C., Cheutin, T., Cremer, M., Cavalli, G. & Cremer, T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nature Rev. Genet. 8, 104–115 (2007).
Meaburn, K. J. & Misteli, T. Cell biology: chromosome territories. Nature 445, 379–781 (2007).
Meaburn, K. J., Misteli, T. & Soutoglou, E. Spatial genome organization in the formation of chromosomal translocations. Semin. Cancer Biol. 17, 80–90 (2007).
Liyanage, M. et al. Abnormal rearrangement within the α/δ T-cell receptor locus in lymphomas from Atm-deficient mice. Blood 96, 1940–1946 (2000).
Parada, L. A., McQueen, P. G., Munson, P. J. & Misteli, T. Conservation of relative chromosome positioning in normal and cancer cells. Curr. Biol. 12, 1692–1697 (2002).
Parada, L. A., McQueen, P. G. & Misteli, T. Tissue-specific spatial organization of genomes. Genome Biol. 5, R44 (2004).
Neves, H., Ramos, C., da Silva, M. G., Parreira, A. & Parreira, L. The nuclear topography of ABL, BCR, PML, and RARα genes: evidence for gene proximity in specific phases of the cell cycle and stages of hematopoietic differentiation. Blood 93, 1197–1207 (1999).
Roix, J. J., McQueen, P. G., Munson, P. J., Parada, L. A. & Misteli, T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nature Genet. 34, 287–291 (2003). Demonstrates a correlation between the spatial proximity of genome regions and their likelihood of translocating, pointing to a significant role of non-random genome organization in determining translocation outcome.
Branco, M. R. & Pombo, A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 4, e138 (2006). Demonstrates physical intermingling of the chromatin fibres of neighbouring chromosomes.
Mitelman, F., Johansson, B. & Mertens, F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nature Genet. 36, 331–334 (2004).
Bird, A. W. et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419, 411–415 (2002).
Megee, P. C., Morgan, B. A. & Smith, M. M. Histone H4 and the maintenance of genome integrity. Genes Dev. 9, 1716–1727 (1995).
Qin, S. & Parthun, M. R. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell. Biol. 22, 8353–8365 (2002).
Qin, S. & Parthun, M. R. Recruitment of the type B histone acetyltransferase Hat1p to chromatin is linked to DNA double-strand breaks. Mol. Cell. Biol. 26, 3649–3658 (2006).
Murr, R. et al. Histone acetylation by Trrap–Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nature Cell Biol. 8, 91–99 (2006).
Murr, R., Vaissiere, T., Sawan, C., Shukla, V. & Herceg, Z. Orchestration of chromatin-based processes: mind the TRRAP. Oncogene 26, 5358–5372 (2007).
Gupta, A. et al. Involvement of human MOF in ATM function. Mol. Cell. Biol. 25, 5292–5305 (2005).
Driscoll, R., Hudson, A. & Jackson, S. P. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315, 649–652 (2007).
Masumoto, H., Hawke, D., Kobayashi, R. & Verreault, A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436, 294–298 (2005).
Huyen, Y. et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432, 406–411 (2004).
Wysocki, R. et al. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell. Biol. 25, 8430–8443 (2005).
Botuyan, M. V. et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373 (2006).
Houston, S. I. et al. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J. Biol. Chem. 283, 19478–19488 (2008).
Jorgensen, S. et al. The histone methyltransferase SET8 is required for S-phase progression. J. Cell Biol. 179, 1337–1345 (2007).
Du, L. L., Nakamura, T. M. & Russell, P. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev. 20, 1583–1596 (2006).
Sanders, S. L. et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119, 603–614 (2004).
Fernandez-Capetillo, O., Allis, C. D. & Nussenzweig, A. Phosphorylation of histone H2B at DNA double-strand breaks. J. Exp. Med. 199, 1671–1677 (2004).
Ahn, S. H., Henderson, K. A., Keeney, S. & Allis, C. D. H2B (Ser10) phosphorylation is induced during apoptosis and meiosis in S. cerevisiae. Cell Cycle 4, 780–783 (2005).
Acknowledgements
We thank V. Roukos for critically reading the manuscript. Misteli's laboratory is supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute and Center for Cancer Research. Soutoglou's laboratory is supported by the National Centre for Scientific Research and Institute of Genetics and Molecular and Cellular Biology.
Author information
Authors and Affiliations
Related links
Glossary
- D-loop
-
A DNA structure in which the two strands of a double-stranded DNA are separated by a third strand. It is created after strand invasion during homologous recombination.
- Holliday junction
-
A mobile junction between four strands of DNA, with homology between pairs of neighbouring DNA molecules.
- Histone variant
-
A core histone other than the classical histones (H2A, H2B, H3 or H4).
- Chromatin-remodelling complex
-
A protein complex that alters the higher-order structure of the chromatin fibre by changing its compaction or repositioning nucleosomes, often using ATP hydrolysis.
- Clamp loader
-
A protein that loads the clamp complex onto the DNA.
- Clamp complex
-
A protein complex that wraps around DNA and facilitates the processivity of DNA replication.
- Nucleotide-excision repair
-
The repair mechanism that removes damaged single nucleotides, mostly in response to ultraviolet damage.
- Histone deacetylase
-
An enzymatic activity that removes an acetyl group from a histone tail.
- Histone acetyl transferase
-
An enzymatic activity that adds an acetyl group to a histone tail.
- Heterochromatin
-
Compact, closed chromatin that generally contains silenced genome regions.
- Euchromatin
-
Decondensed, open chromatin that generally contains transcriptionally active genome regions.
- Histone methyltransferase
-
An enzymatic activity that adds a methyl group to a histone tail.
- V(D)J recombination
-
A recombination reaction that assembles genes encoding diverse T-cell receptor and immunoglobulin molecules from variable (V), diversity (D) and joining (J) gene segments. V(D)J recombination is necessary for the recognition of diverse foreign antigens.
Rights and permissions
About this article
Cite this article
Misteli, T., Soutoglou, E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 10, 243–254 (2009). https://doi.org/10.1038/nrm2651
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm2651
This article is cited by
-
Transmembrane nuclease NUMEN/ENDOD1 regulates DNA repair pathway choice at the nuclear periphery
Nature Cell Biology (2023)
-
Role of H4K16 acetylation in 53BP1 recruitment to double-strand break sites in in vitro aged cells
Biogerontology (2022)
-
Der Rolle der DNA-Schadensantwort bei granulomatösen Erkrankungen
Zeitschrift für Rheumatologie (2022)
-
Chromatin tracing and multiplexed imaging of nucleome architectures (MINA) and RNAs in single mammalian cells and tissue
Nature Protocols (2021)
-
Chromosome aberrations induced by the non-mutagenic carcinogen acetamide involve in rat hepatocarcinogenesis through micronucleus formation in hepatocytes
Archives of Toxicology (2021)