Key Points
-
The gut microbiota can positively and negatively affect host health, and the specific composition of the microbiota may be important for host health and fitness.
-
Environment factors (diet, host environment, maternal effect, stochasticity, and so on) interact to shape the gut microbiota, but how they interact with host genetics to influence the composition of the microbiota is not well understood.
-
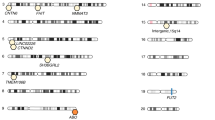
A recent quantitative trait loci (QTL) study highlighted host genetic factors that control the composition of the gut microbial community.
-
Candidate gene approaches (that monitor genetic diversity at a given locus in humans and the effects of gene insertions or deletions in mice) implicate host genomic loci in shaping gut microbial diversity.
-
Host genes that have been identified as important for determining microbial diversity in the gut have roles primarily in the innate and adaptive immune system, and in metabolism.
-
Genome-wide association studies can consider the gut microbiota as a host phenotypic trait, but can also incorporate the microbiota into studies of host gene–environment interactions in the context of chronic inflammatory disease.
Abstract
To what extent do host genetics control the composition of the gut microbiome? Studies comparing the gut microbiota in human twins and across inbred mouse lines have yielded inconsistent answers to this question. However, candidate gene approaches, in which one gene is deleted or added to a model host organism, show that a single host gene can have a tremendous effect on the diversity and population structure of the gut microbiota. Now, quantitative genetics is emerging as a highly promising approach that can be used to better understand the overall architecture of host genetic influence on the microbiota, and to discover additional host genes controlling microbial diversity in the gut. In this Review, we describe how host genetics and the environment shape the microbiota, and how these three factors may interact in the context of chronic disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Koenig, J. E. et al. Microbes and Health Sackler Colloquium: succession of microbial consortia in the developing infant gut microbiome. Proc. Natl Acad. Sci. USA 28 Jul 2010 (doi:10.1073/pnas.1000081107). A pilot study describing the colonization process of the infant gut microbiome from birth to 2.5 years of age; this study reveals that seemingly chaotic shifts in the microbiome are explained by events in the infant's life, such as diet change, antibiotic treatment and others.
Palmer, C., Bik, E. M., DiGiulio, D. B., Relman, D. A. & Brown, P. O. Development of the human infant intestinal microbiota. PLoS Biol. 5, e177 (2007).
Dethlefsen, L., Huse, S., Sogin, M. L. & Relman, D. A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6, e280 (2008).
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
Ley, R. E., Turnbaugh, P. J., Klein, S. & Gordon, J. I. Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023 (2006).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010). A large metagenomics-based characterization of the microbiome in multiple subjects finds a set of widely shared bacterial species.
Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009). A key paper using human twin data to measure the effect of host genotype on the microbiome and the link between dysbiosis and obesity.
Moodley, Y. et al. The peopling of the Pacific from a bacterial perspective. Science 323, 527–530 (2009).
Schwarz, S. et al. Horizontal versus familial transmission of Helicobacter pylori. PLoS Pathog. 4, e1000180 (2008).
Vaishampayan, P. A. et al. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol. Evol. 6, 53–66 (2010).
Dethlefsen, L., Eckburg, P. B., Bik, E. M. & Relman, D. A. Assembly of the human intestinal microbiota. Trends. Ecol. Evol. 21, 517–523 (2006).
Ley, R. E., Peterson, D. A. & Gordon, J. I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848 (2006).
Tims, S., Zoetendal, E. G., de Vos, W. M. & Kleerebezem, M. in: Metagenomics of the Human Body (ed. Nelson, K. E.) 15–41 (Springer, Berlin, 2011).
Frank, D. N. et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA 104, 13780–13785 (2007).
Sartor, R. B. Microbial influences in inflammatory bowel diseases. Gastroenterology 134, 577–594 (2008).
Larsen, N. et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 5, e9085 (2010).
Peterson, D. A., Frank, D. N., Pace, N. R. & Gordon, J. I. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 3, 417–427 (2008).
Wang, Y. et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 3, 944–954 (2009).
Turnbaugh, P. J., Backhed, F., Fulton, L. & Gordon, J. I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223 (2008).
Vijay-Kumar, M. et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231 (2010). This article shows that TLR5 deficiency in mice results in an altered microbiota that is associated with metabolic syndrome. Transplanting this altered microbiome confers the disease state to germ-free recipient wild-type mice.
Garrett, W. S. et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131, 33–45 (2007).
Wen, L. et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455, 1109–1113 (2008).
Borody, T. J. et al. Bacteriotherapy using fecal flora: toying with human motions. J. Clin. Gastroenterol. 38, 475–483 (2004).
You, D. M., Franzos, M. A. & Holman, R. P. Successful treatment of fulminant Clostridium difficile infection with fecal bacteriotherapy. Ann. Intern. Med. 148, 632–633 (2008).
Grehan, M. J. et al. Durable alteration of the colonic microbiota by the administration of donor fecal flora. J. Clin. Gastroenterol. 44, 551–561 (2010).
Benson, A. K. et al. Individuality in gut microbiota composition is a complex, polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl Acad. Sci. USA 107, 18933–18938 (2010). The first study taking a QTL-based approach to relate genomic variation in the host to microbial diversity in the gut. Testing of the CMM abundances for co-segregation with 530 SNPs identifies 18 host-associated QTLs that show a linkage with the abundances of specific microbial taxa.
Ley, R. E. et al. Obesity alters gut microbial ecology. Proc. Natl Acad. Sci. USA 102, 11070–11075 (2005).
Whittaker, R. H. Evolution and measurement of species diversity. Taxon 21, 213–251 (1972).
Orcutt, R. P., Gianni, F. J. & Judge, R. J. Development of an “Altered Schaedler Flora” for NCI gnotobiotic rodents. Microecol. Ther. 17, 59 (1987).
Deloris Alexander, A. et al. Quantitative PCR assays for mouse enteric flora reveal strain-dependent differences in composition that are influenced by the microenvironment. Mamm. Genome 17, 1093–1104 (2006).
Dethlefsen, L., McFall-Ngai, M. & Relman, D. A. An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature 449, 811–818 (2007).
Louis, P., Scott, K. P., Duncan, S. H. & Flint, H. J. Understanding the effects of diet on bacterial metabolism in the large intestine. J. Appl. Microbiol. 102, 1197–1208 (2007).
Tilg, H. Obesity, metabolic syndrome, and microbiota: multiple interactions. J. Clin. Gastroenterol. 44, S16–S18 (2010).
Turnbaugh, P. J. et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1, 6ra14 (2009).
Zhang, C. et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 4, 232–241 (2010).
Duncan, S. H. et al. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 73, 1073–1078 (2007).
Sonnenburg, E. D. et al. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141, 1241–1252 (2010).
Stewart, J. A., Chadwick, V. S. & Murray, A. Investigations into the influence of host genetics on the predominant eubacteria in the faecal microflora of children. J. Med. Microbiol. 54, 1239–1242 (2005).
Zoetendal, E. G., Akkermans, A. D., Akkermans-van Vliet, W. M., de Visser, J. A. & de Vos, W. M. The host genotype affects the bacterial community in the human gastronintestinal tract. Microb. Ecol. Health Dis. 13, 129–134 (2001).
Turnbaugh, P. et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc. Natl Acad. Sci. USA 107, 7503–7508 (2010).
Teucher, B. et al. Dietary patterns and heritability of food choice in a UK female twin cohort. Twin Res. Hum. Genet. 10, 734–748 (2007).
Vinkhuyzen, A. A., van der Sluis, S., de Geus, E. J., Boomsma, D. I. & Posthuma, D. Genetic influences on 'environmental' factors. Genes Brain Behav. 9, 276–287 (2010).
Matsumoto, M. et al. Voluntary running exercise alters microbiota composition and increases n-butyrate concentration in the rat cecum. Biosci. Biotechnol. Biochem. 72, 572–576 (2008).
Santacruz, A. et al. Interplay between weight loss and gut microbiota composition in overweight adolescents. Obesity 17, 1906–1915 (2009).
Friswell, M. K. et al. Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS ONE 5, e8584 (2010).
Loh, G., Brodziak, F. & Blaut, M. The Toll-like receptors TLR2 and TLR4 do not affect the intestinal microbiota composition in mice. Environ. Microbiol. 10, 709–715 (2008).
Kovacs, A. et al. Genotype is a stronger determinant than sex of the mouse gut microbiota. Microb. Ecol. 6, 423–428 (2011).
Darvasi, A. & Soller, M. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics 141, 1199–1207 (1995).
Wang, X. et al. Using advanced intercross lines for high-resolution mapping of HDL cholesterol quantitative trait loci. Genome Res. 13, 1654–1664 (2003).
Kelly, S. A. et al. Genetic architecture of voluntary exercise in an advanced intercross line of mice. Physiol. Genomics 42, 190–200 (2010).
Mohamadzadeh, M. et al. Lactobacilli activate human dendritic cells that skew T cells toward T helper 1 polarization. Proc. Natl Acad. Sci. USA 102, 2880–2885 (2005).
Presley, L. L., Wei, B., Braun, J. & Borneman, J. Bacteria associated with immunoregulatory cells in mice. Appl. Environ. Microbiol. 76, 936–941 (2010).
Devlin, B., Daniels, M. & Roeder, K. The heritability of IQ. Genetics 137, 597–606 (1994).
Visscher, P. M. et al. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet. 2, e41 (2006).
Khachatryan, Z. A. et al. Predominant role of host genetics in controlling the composition of gut microbiota. PLoS ONE 3, e3064 (2008). Genetic variation of the MEFV gene is associated with shifts in the bacterial populations within the phyla Bacteroidetes, Firmicutes and Proteobacteria.
Ting, J. P., Kastner, D. L. & Hoffman, H. M. CATERPILLERs, pyrin and hereditary immunological disorders. Nature Rev. Immunol. 6, 183–195 (2006).
Kelesidis, T., Kelesidis, I., Chou, S. & Mantzoros, C. S. Narrative review: the role of leptin in human physiology: emerging clinical applications. Ann. Intern. Med. 152, 93–100 (2010).
Fernandez-Riejos, P. et al. Role of leptin in the activation of immune cells. Mediators Inflamm. 2010, 568343 (2010).
Jeon, J. P. et al. Copy number variation at leptin receptor gene locus associated with metabolic traits and the risk of type 2 diabetes mellitus. BMC Genomics 11, 426 (2010).
Park, K. S. et al. Polymorphisms in the leptin receptor (LEPR)-putative association with obesity and T2DM. J. Hum. Genet. 51, 85–91 (2006).
Sun, Q. et al. Genome-wide association study identifies polymorphisms in LEPR as determinants of plasma soluble leptin receptor levels. Hum. Mol. Genet. 19, 1846–1855 (2010).
Waldram, A. et al. Top-down systems biology modeling of host metabotype-microbiome associations in obese rodents. J. Proteome Res. 8, 2361–2375 (2009).
Turnbaugh, P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006).
El Homsi, M. et al. Leptin modulates the expression of secreted and membrane-associated mucins in colonic epithelial cells by targeting PKC, PI3K, and MAPK pathways. Am. J. Physiol. Gastrointest. Liver Physiol. 293, G365–G373 (2007).
Plaisancie, P. et al. Luminal leptin activates mucin-secreting goblet cells in the large bowel. Am. J. Physiol. Gastrointest. Liver Physiol. 290, G805–G812 (2006).
Crawford, P. A. et al. Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc. Natl Acad. Sci. USA 106, 11276–11281 (2009).
Barbosa, T. & Rescigno, M. Host-bacteria interactions in the intestine: homeostasis to chronic inflammation. Wiley Interdiscip. Rev. Syst. Biol. Med. 2, 80–97 (2010).
Barrett, J. C. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nature Genet. 40, 955–962 (2008). A GWA study that reveals many gene variants associated with Crohn's disease.
Imielinski, M. et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nature Genet. 41, 1335–1340 (2009).
Potter, C. et al. Association between anti-tumour necrosis factor treatment response and genetic variants within the TLR and NFκB signalling pathways. Ann. Rheum. Dis. 69, 1315–1320 (2010).
Teran-Ventura, E. et al. Characterization of housing-related spontaneous variations of gut microbiota and expression of toll-like receptors 2 and 4 in rats. Microb. Ecol. 60, 691–702 (2010).
Albert, E. J., Sommerfeld, K., Gophna, S., Marshall, J. S. & Gophna, U. The gut microbiota of toll-like receptor 2-deficient mice exhibits lineage-specific modifications. Environ. Microbiol. Rep. 1, 65–70 (2009).
Thompson, C. L., Hofer, M. J., Campbell, I. L. & Holmes, A. J. Community dynamics in the mouse gut microbiota: a possible role for IRF9-regulated genes in community homeostasis. PLoS ONE 5, e10335 (2010).
Gonsky, R. et al. Distinct IFNG methylation in a subset of ulcerative colitis patients based on reactivity to microbial antigens. Inflamm. Bowel Dis. 17, 171–178 (2011).
Gootenberg, D. B. & Turnbaugh, P. J. Humanized animal models of the microbiome. J. Anim. Sci. 10 Sep 2010 (doi:10.2527/jas.2010-3371).
Waterston, R. H. et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002).
Ley, R. E. et al. Evolution of mammals and their gut microbes. Science 320, 1647–1651 (2008).
Dominguez-Bello, M. G. et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA 107, 11971–11975 (2010).
Matsumiya, Y., Kato, N., Watanabe, K. & Kato, H. Molecular epidemiological study of vertical transmission of vaginal Lactobacillus species from mothers to newborn infants in Japanese, by arbitrarily primed polymerase chain reaction. J. Infect. Chemother. 8, 43–49 (2002).
Tannock, G., Fuller, R., Smith, S. & Hall, M. Plasmid profiling of members of the family Enterobacteriaceae, lactobacilli, and bifidobacteria to study the transmission of bacteria from mother to infant. J. Clin. Microbiol. 28, 1225–1228 (1990).
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R. & Gordon, J. I. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Rev. Microbiol. 6, 776–788 (2008).
Jones, B. V., Begley, M., Hill, C., Gahan, C. G. & Marchesi, J. R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl Acad. Sci. USA 105, 13580–13585 (2008).
Lozupone, C. A. et al. The convergence of carbohydrate active gene repertoires in human gut microbes. Proc. Natl Acad. Sci. USA 105, 15076–15081 (2008).
Samuel, B. S. et al. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc. Natl Acad. Sci. USA 104, 10643–10648 (2007).
Clarke, T. B. et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nature Med. 16, 228–231 (2010).
Hooper, L. V. Do symbiotic bacteria subvert host immunity. Nature Rev. Microbiol. 7, 367–374 (2009).
Comstock, L. E. & Kasper, D. L. Bacterial glycans: key mediators of diverse host immune responses. Cell 126, 847–850 (2006).
Peterson, D. A., McNulty, N. P., Guruge, J. L. & Gordon, J. I. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2, 328–339 (2007).
Walter, J., Britton, R. A. & Roos, S. Microbes and Health Sackler Colloquium: Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc. Natl Acad. Sci. USA 25 Jun 2010 (doi: 10.1073/pnas.1000099107). A study describing the evolutionary processes that shape the symbiotic interactions between microorganisms and the host gut, focusing on Lactobacillus reuteri.
Crowell-Davis, S. & Caudle, A. Coprophagy by foals: recognition of maternal feces. Appl. Anim. Behav. Sci. 24, 267–272 (1989).
Osawa, R., Blanshard, W. H. & Ocallaghan, P. G. Microbiological studies of the intestinal microflora of the koala, Phascolarctos cinereus. II. Pap, a special maternal feces consumed by juvenile koalas. Aust. J. Zool. 41, 611–620 (1993).
Lombardo, M. Access to mutualistic endosymbiotic microbes: an underappreciated benefit of group living. Behav. Ecol. Sociobiol. 62, 479–497 (2008).
Bik, E. M. et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 4, 962–974 (2010).
Costello, E. K. et al. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009).
Kim, T. K. et al. Heterogeneity of vaginal microbial communities within individuals. J. Clin. Microbiol. 47, 1181–1189 (2009).
Pei, Z. et al. Bacterial biota in the human distal esophagus. Proc. Natl Acad. Sci. USA 101, 4250–4255 (2004).
Price, L. B. et al. The effects of circumcision on the penis microbiome. PLoS ONE 5, e8422 (2010).
Chen, E. S. et al. APOA1/A5 variants and haplotypes as a risk factor for obesity and better lipid profiles in a Brazilian elderly cohort. Lipids 45, 511–517 (2010).
Liu, Z. K., Hu, M., Baum, L., Thomas, G. N. & Tomlinson, B. Associations of polymorphisms in the apolipoprotein A1/C3/A4/A5 gene cluster with familial combined hyperlipidaemia in Hong Kong Chinese. Atherosclerosis 208, 427–432 (2010).
Asquith, M. J., Boulard, O., Powrie, F. & Maloy, K. J. Pathogenic and protective roles of MyD88 in leukocytes and epithelial cells in mouse models of inflammatory bowel disease. Gastroenterology 139, 519–529.e2 (2010).
Hampe, J. et al. Association of NOD2 (CARD 15) genotype with clinical course of Crohn's disease: a cohort study. Lancet 359, 1661–1665 (2002).
Frank, D. N. et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 17, 179–184 (2011).
Petnicki-Ocwieja, T. et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc. Natl Acad. Sci. USA 106, 15813–15818 (2009).
Fellermann, K. et al. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am. J. Hum. Genet. 79, 439–448 (2006).
Wehkamp, J. et al. Reduced Paneth cell α-defensins in ileal Crohn's disease. Proc. Natl Acad. Sci. USA 102, 18129–18134 (2005).
Wehkamp, J. et al. The Paneth cell α-defensin deficiency of ileal Crohn's disease is linked to Wnt/Tcf-4. J. Immunol. 179, 3109–3118 (2007).
Baker, P. I., Love, D. R. & Ferguson, L. R. Role of gut microbiota in Crohn's disease. Expert Rev. Gastroenterol. Hepatol. 3, 535–546 (2009).
Salzman, N. H. et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nature Immunol. 11, 76–83 (2010). This article demonstrates that mice which are genetically engineered to express a human defensin in the intestine have an altered gut community composition, indicating that host-derived antimicrobial peptides can control microbial diversity in the gut.
Ivanov, I. I. et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349 (2008).
Ivanov, I. I & Littman, D. R. Segmented filamentous bacteria take the stage. Mucosal Immunol. 3, 209–212 (2010).
Artis, D. et al. RELMβ/FIZZ2 is a goblet cell-specific immune-effector molecule in the gastrointestinal tract. Proc. Natl Acad. Sci. USA 101, 13596–13600 (2004).
Hogan, S. P. et al. Resistin-like molecule β regulates innate colonic function: barrier integrity and inflammation susceptibility. J. Allergy Clin. Immunol. 118, 257–268 (2006).
Cash, H. L., Whitham, C. V., Behrendt, C. L. & Hooper, L. V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313, 1126–1130 (2006).
Hildebrandt, M. A. et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 137, 1716–1724.e2 (2009).
Yel, L. Selective IgA deficiency. J. Clin. Immunol. 30, 10–16 (2010).
Suzuki, K. et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl Acad. Sci. USA 101, 1981–1986 (2004).
Wijburg, O. L. et al. Innate secretory antibodies protect against natural Salmonella typhimurium infection. J. Exp. Med. 203, 21–26 (2006).
Nadal, I. et al. Shifts in clostridia, bacteroides and immunoglobulin-coating fecal bacteria associated with weight loss in obese adolescents. Int. J. Obes. 33, 758–767 (2009).
Toivanen, P., Vaahtovuo, J. & Eerola, E. Influence of major histocompatibility complex on bacterial composition of fecal flora. Infect. Immun. 69, 2372–2377 (2001).
Vaahtovuo, J., Toivanen, P. & Eerola, E. Bacterial composition of murine fecal microflora is indigenous and genetically guided. FEMS Microbiol. Ecol. 44, 131–136 (2003).
De Palma, G. et al. Interplay between human leukocyte antigen genes and the microbial colonization process of the newborn intestine. Curr. Issues Mol. Biol. 12, 1–10 (2010).
Acknowledgements
We thank A. Benson, D. Pomp, L. Angenent and the three reviewers for helpful comments on the manuscript, and J. Koenig for early discussions. We are grateful for a Beckman Young Investigator Award, and to The Hartwell Foundation, the David and Lucile Packard Foundation, the US National Science Foundation (IOS-0958,184), and the US National Institutes of Health Human Microbiome Project Data Analysis and Coordination Center (U01 HG004866) for support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Related links
Glossary
- Microbiota
-
A microbial community or assemblage.
- Dysbiosis
-
A shift in the relative abundances of the microbial taxa compared with the adundances that are observed in healthy animals.
- Microbiome
-
The complete set of genes within a microbiota.
- Germ-free
-
Pertaining to an animal: lacking a microbiome; born and raised under sterile conditions for research purposes.
- Adiposity
-
The property of containing fat.
- β-diversity
-
A measure of diversity that describes the differences between any two ecosystems (for example, the UniFrac distance metric). Related to α-diversity and γ-diversity, which are measures of the diversity in a single ecosystem and across a group of ecosystems, respectively.
- UniFrac
-
A β-diversity measure that is phylogeny based. Microbial communities are more similar if they are composed of members that are more closely related, phylogenetically, as this implies a shared evolutionary past. UniFrac units range from 0 (identical communities) to 1 (totally different communities).
- Altered Schaedler flora
-
A standard enteric flora containing eight species that are known to exhibit tissue tropism, occupying different niches in the mouse gastrointestinal tract.
- Quantitative trait locus
-
A genomic region for which variation is associated with the quantitative variation in a phenotypic trait.
- Heritability
-
The proportion of phenotypic variation in a population that is attributable to genetic variation among individuals.
- Fingerprinting-based comparison
-
A molecular technique for the study of nucleic acids using either denaturing-gradient gel electrophoresis (DGGE) or temporal temperature gradient gel electrophoresis (TTGE). Another method is terminal restriction fragment length polymorphism (T-RFLP), in which the DNA is amplified using fluorescence-labelled primers, digested using restriction enzymes and then detected. In microbial ecology, these techniques are used to compare microbial communities.
- Dam
-
A female animal parent.
- Advanced intercross line
-
An experimental mouse population used for the study of quantitative trait loci (QTL). This population is created by randomly intercrossing strains and then intercrossing the mice in subsequent generations until the desired number of generations is achieved. This results in mice with many recombinations in the genome, which is required for QTL studies.
- Genome-wide association study
-
A genetic approach aiming to relate genome-wide variation in genetic markers to variation in any given phenotype. The microbiota, or its components, can be considered as a host phenotype.
- ob/ob mouse
-
A mouse carrying two copies of a non-functional leptin gene (Ob; also known as Lep). These mice are leptin deficient and obese.
- Zucker rat
-
A rat that is deficient in the leptin receptor (FA; also known as LEPR and OBR) and is obese.
- Hyperphagia
-
Over-consumption of food.
Rights and permissions
About this article
Cite this article
Spor, A., Koren, O. & Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9, 279–290 (2011). https://doi.org/10.1038/nrmicro2540
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2540
This article is cited by
-
Integrated omics analysis reveals the alteration of gut microbiota and fecal metabolites in Cervus elaphus kansuensis
Applied Microbiology and Biotechnology (2024)
-
Geography and elevation as drivers of cloacal microbiome assemblages of a passerine bird distributed across Sulawesi, Indonesia
Animal Microbiome (2023)
-
Influence of scat ageing on the gut microbiome: how old is too old?
BMC Genomics (2023)
-
Gut microbiome diversity and composition is associated with exploratory behavior in a wild-caught songbird
Animal Microbiome (2023)
-
No evidence for associations between brood size, gut microbiome diversity and survival in great tit (Parus major) nestlings
Animal Microbiome (2023)