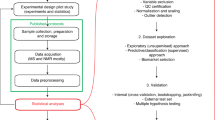
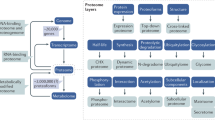
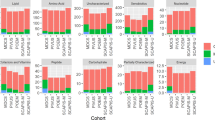
Abstract
Metabolomics—the nontargeted measurement of all metabolites produced by the body—is beginning to show promise in both biomarker discovery and, in the form of pharmacometabolomics, in aiding the choice of therapy for patients with specific diseases. In its two basic forms (pattern recognition and metabolite identification), this developing field has been used to discover potential biomarkers in several renal diseases, including acute kidney injury (attributable to a variety of causes), autosomal dominant polycystic kidney disease and kidney cancer. NMR and gas chromatography or liquid chromatography, together with mass spectrometry, are generally used to separate and identify metabolites. Many hurdles need to be overcome in this field, such as achieving consistency in collection of biofluid samples, controlling for batch effects during the analysis and applying the most appropriate statistical analysis to extract the maximum amount of biological information from the data obtained. Pathway and network analyses have both been applied to metabolomic analysis, which vastly extends its clinical relevance and effects. In addition, pharmacometabolomics analyses, in which a metabolomic signature can be associated with a given therapeutic effect, are beginning to appear in the literature, which will lead to personalized therapies. Thus, metabolomics holds promise for early diagnosis, increased choice of therapy and the identification of new metabolic pathways that could potentially be targeted in kidney disease.
Key Points
-
Metabolomics is the study of all small-molecule metabolites produced by the body and thus provides a functional fingerprint of the physiological and pathophysiological state of the body
-
Two useful clinical methods exist for metabolomics analysis: pattern recognition and metabolite identification
-
The study design (adequate sample size and the statistical analysis strategy) should be carefully planned to detect small differences in metabolic profiles in a population with wide biological variation
-
Metabolomics has resulted in potential biomarkers for several renal diseases, but to become true biomarkers, the clinical merit of potential biomarkers needs to be validated in separate targeted studies
-
Pharmacometabolomics could enable metabolomics to be used in personalized medicine
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
German, J. B., Hammock, B. D. & Watkins, S. M. Metabolomics: building on a century of biochemistry to guide human health. Metabolomics 1, 3–9 (2005).
Kim, K. et al. Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol. Cell. Proteomics 8, 558–570 (2009).
Calvani, R. et al. Gut microbiome-derived metabolites characterize a peculiar obese urinary metabotype. Int. J. Obes. (Lond.) 34, 1095–1098 (2010).
Romick-Rosendale, L. E. et al. NMR-based metabonomics analysis of mouse urine and fecal extracts following oral treatment with the broad-spectrum antibiotic enrofloxacin (Baytril). Magn. Reson. Chem. 47 (Suppl. 1), S36–S46 (2009).
Martin, F. P. et al. Dietary modulation of gut functional ecology studied by fecal metabonomics. J. Proteome Res. 9, 5284–5295 (2010).
Kim, K. B. et al. Toxicometabolomics approach to urinary biomarkers for mercuric chloride (HgCl)-induced nephrotoxicity using proton nuclear magnetic resonance (1H NMR) in rats. Toxicol. Appl. Pharmacol. 249, 114–126 (2010).
Hwang, G.-S., Yang, J.-Y., Ryu, D. H. & Kwon, T.-H. Metabolic profiling of kidney and urine in rats with lithium-induced nephrogenic diabetes insipidus by 1H-NMR-based metabonomics. Am. J. Physiol. Renal Physiol. 298, F461–F470 (2010).
Nicholson, J. K. & Lindon, J. C. Systems biology: metabonomics. Nature 455, 1054–1056 (2008).
Jellum, E., Stokke, O. & Eldjarn, L. Application of gas chromatography, mass spectrometry, and computer methods in clinical biochemistry. Anal. Chem. 46, 1099–1106 (1973).
Shoemaker, J. D. & Elliott, W. H. Automated screening of urine samples for carbohydrates, organic and amino acids after treatment with urease. J. Chromatogr. 562, 125–138 (1991).
Kuhara, T. et al. Pilot study of gas chromatographic-mass spectrometric screening of newborn urine for inborn errors of metabolism after treatment with urease. J. Chromatogr. B Biomed. Sci. Appl. 731, 141–147 (1999).
Kuhara, T. Gas chromatographic-mass spectrometric urinary metabolome analysis to study mutations of inborn errors of metabolism. Mass Spectrom. Rev. 24, 814–827 (2005).
Wishart, D. S. et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 35, D521–D526 (2007).
Pauling, L., Robinson, A. B., Teranishi, R. & Cary, P. Quantitative analysis of urine vapor and breath by gas-liquid partition chromatography. Proc. Natl Acad. Sci. USA 68, 2374–2376 (1971).
Horning, E. C. & Horning, M. G. Metabolic profiles: gas-phase methods for analysis of metabolites. Clin. Chem. 17, 802–809 (1971).
Kind, T., Tolstikov, V., Fiehn, O. & Weiss, R. H. A comprehensive urinary metabolomic approach for identifying kidney cancer. Anal. Biochem. 363, 185–195 (2007).
Lindon, J. C. & Nicholson, J. K. Analytical techniques for metabonomics and metabolomics, and multi-omic information recovery. Trends Anal. Chem. 27, 194–204 (2008).
Kind, T. et al. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 81, 10038–10048 (2009).
Dehaven, C. D., Evans, A. M., Dai, H. & Lawton, K. A. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J. Cheminform. 2, 9 (2010).
Saude, E. J. & Sykes, B. D. Urine stability for metabolomic studies: effects of preparation and storage. Metabolomics 3, 19–27 (2007).
Gika, H. G., Theodoridis, G. A. & Wilson, I. D. Liquid chromatography and ultra-performance liquid chromatography-mass spectrometry fingerprinting of human urine: sample stability under different handling and storage conditions for metabonomics studies. J. Chromatogr. A 1189, 314–322 (2008).
Gu, H. et al. Monitoring diet effects via biofluids and their implications for metabolomics studies. Anal. Chem. 79, 89–97 (2007).
Slupsky, C. M. et al. Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Anal. Chem. 79, 6995–7004 (2007).
Gika, H. G., Theodoridis, G. A., Wingate, J. E. & Wilson, I. D. Within-day reproducibility of an HPLC-MS-based method for metabonomic analysis: application to human urine. J. Proteome Res. 6, 3291–3303 (2007).
Katajamaa, M. & Oresic, M. Data processing for mass spectrometry-based metabolomics. J. Chromatogr. A 1158, 318–328 (2007).
Sreekumar, A. et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457, 910–914 (2009).
Kim, K. et al. Urine metabolomic analysis identifies potential biomarkers and pathogenic pathways in kidney cancer. OMICS 15, 293–303 (2011).
Wang, W. et al. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal. Chem. 75, 4818–4826 (2003).
Oresic, M. et al. Phenotype characterisation using integrated gene transcript, protein and metabolite profiling. Appl. Bioinformatics 3, 205–217 (2004).
van den Berg, R. A., Hoefsloot, H. C., Westerhuis, J. A., Smilde, A. K. & van der Werf, M. J. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics 7, 142 (2006).
Westfall, P. H. & Young, S. S. Resampling-based multiple testing: examples and methods for P-value adjustment (John Wiley & Sons, New York, 1993).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. Series B Stat. Methodol. 57, 289–300 (1995).
Taylor, S. L. et al. A metabolomics approach using juvenile cystic mice to identify urinary biomarkers and altered pathways in polycystic kidney disease. Am. J. Physiol. Renal Physiol. 298, F909–F922 (2010).
Rubingh, C. M. et al. Assessing the performance of statistical validation tools for megavariate metabolomics data. Metabolomics 2, 53–61 (2006).
Mahadevan, S., Shah, S. L., Marrie, T. J. & Slupsky, C. M. Analysis of metabolomic data using support vector machines. Anal. Chem. 80, 7562–7570 (2008).
Brougham, D. F. et al. Artificial neural networks for classification in metabolomic studies of whole cells using 1H nuclear magnetic resonance. J. Biomed. Biotechnol. 2011, 158094 (2011).
Ein-Dor, L., Zuk, O. & Domany, E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc. Natl Acad. Sci. USA 103, 5923–5928 (2006).
Niemann, C. U. & Serkova, N. J. Biochemical mechanisms of nephrotoxicity: application for metabolomics. Expert Opin. Drug Metab. Toxicol. 3, 527–544 (2007).
Wishart, D. S. Metabolomics: the principles and potential applications to transplantation. Am. J. Transplant. 5, 2814–2820 (2005).
Niwa, T. Update of uremic toxin research by mass spectrometry. Mass Spectrom. Rev. 30, 510–521 (2011).
Kikuchi, K. et al. Metabolomic analysis of uremic toxins by liquid chromatography/electrospray ionization-tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 878, 1662–1668 (2010).
Niwa, T., Maeda, K., Ohki, T., Saito, A. & Kobayashi, K. A gas chromatographic-mass spectrometric analysis for phenols in uremic serum. Clin. Chim. Acta 110, 51–57 (1981).
Toyohara, T. et al. Metabolomic profiling of uremic solutes in CKD patients. Hypertens. Res. 33, 944–952 (2010).
Boudonck, K. J. et al. Discovery of metabolomics biomarkers for early detection of nephrotoxicity. Toxicol. Pathol. 37, 280–292 (2009).
Star, R. A. Treatment of acute renal failure. Kidney Int. 54, 1817–1831 (1998).
Xu, E. Y. et al. Integrated pathway analysis of rat urine metabolic profiles and kidney transcriptomic profiles to elucidate the systems toxicology of model nephrotoxicants. Chem. Res. Toxicol. 21, 1548–1561 (2008).
Perroud, B. et al. Pathway analysis of kidney cancer using proteomics and metabolic profiling. Mol. Cancer 5, 64 (2006).
Melnick, J. Z., Baum, M. & Thompson, J. R. Aminoglycoside-induced Fanconi's syndrome. Am. J. Kidney Dis. 23, 118–122 (1994).
Pratt, C. B. et al. Ifosfamide, Fanconi's syndrome, and rickets. J. Clin. Oncol. 9, 1495–1499 (1991).
Portilla, D. et al. Metabolomic study of cisplatin-induced nephrotoxicity. Kidney Int. 69, 2194–2204 (2006).
Perroud, B., Ishimaru, T., Borowsky, A. D. & Weiss, R. H. Grade-dependent proteomics characterization of kidney cancer. Mol. Cell. Proteomics 8, 971–985 (2009).
Park, J. C. et al. A metabonomic study on the biochemical effects of doxorubicin in rats using (1)H-NMR spectroscopy. J. Toxicol. Environ. Health A 72, 374–384 (2009).
Xie, G. et al. Metabonomic evaluation of melamine-induced acute renal toxicity in rats. J. Proteome Res. 9, 125–133 (2010).
Wei, L. et al. Metabolic profiling studies on the toxicological effects of realgar in rats by 1H NMR spectroscopy. Toxicol. Appl. Pharmacol. 234, 314–325 (2009).
Broumand, B. Diabetes: changing the fate of diabetics in the dialysis unit. Blood Purif. 25, 39–47 (2007).
Lu, D. et al. 13C NMR isotopomer analysis reveals a connection between pyruvate cycling and glucose-stimulated insulin secretion (GSIS). Proc. Natl Acad. Sci. USA 99, 2708–2713 (2002).
Jensen, M. V. et al. Metabolic cycling in control of glucose-stimulated insulin secretion. Am. J. Physiol. Endocrinol. Metab. 295, E1287–E1297 (2008).
Cline, G. W., Lepine, R. L., Papas, K. K., Kibbey, R. G. & Shulman, G. I. 13C NMR isotopomer analysis of anaplerotic pathways in INS-1 cells. J. Biol. Chem. 279, 44370–44375 (2004).
Savage, D. B., Petersen, K. F. & Shulman, G. I. Disordered lipid metabolism and the pathogenesis of insulin resistance. Physiol. Rev. 87, 507–520 (2007).
Holland, W. L. & Summers, S. A. Sphingolipids, insulin resistance, and metabolic disease: new insights from in vivo manipulation of sphingolipid metabolism. Endocr. Rev. 29, 381–402 (2008).
Lucio, M. et al. Insulin sensitivity is reflected by characteristic metabolic fingerprints—a Fourier transform mass spectrometric non-targeted metabolomics approach. PLoS ONE 5, e13317 (2010).
Suhre, K. et al. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS ONE 5, e13953 (2010).
Zhao, X. et al. Metabonomic fingerprints of fasting plasma and spot urine reveal human pre-diabetic metabolic traits. Metabolomics 6, 362–374 (2010).
Connor, S. C., Hansen, M. K., Corner, A., Smith, R. F. & Ryan, T. E. Integration of metabolomics and transcriptomics data to aid biomarker discovery in type 2 diabetes. Mol. Biosyst. 6, 909–921 (2010).
Ganti, S. et al. Urinary acylcarnitines are altered in kidney cancer. Int. J. Cancer http://dx.doi.org/10.1002/ijc.26274.
Marchesini, G. et al. Elimination of infused branched-chain amino-acids from plasma of patients with non-obese type 2 diabetes mellitus. Clin. Nutr. 10, 105–113 (1991).
Alam, A. & Perrone, R. D. Management of ESRD in patients with autosomal dominant polycystic kidney disease. Adv. Chronic Kidney Dis. 17, 164–172 (2010).
Saude, E. J., Adamko, D., Rowe, B. H., Marrie, T. & Sykes, B. D. Variation of metabolites in normal human urine. Metabolomics 3, 439–451 (2007).
van der Greef, J., Hankemeier, T. & McBurney, R. N. Metabolomics-based systems biology and personalized medicine: moving towards n = 1 clinical trials? Pharmacogenomics 7, 1087–1094 (2006).
Bayet-Robert, M., Morvan, D., Chollet, P. & Barthomeuf, C. Pharmacometabolomics of docetaxel-treated human MCF7 breast cancer cells provides evidence of varying cellular responses at high and low doses. Breast Cancer Res. Treat. 120, 613–626 (2010).
Ji, Y. et al. Glycine and a glycine dehydrogenase (GLDC) SNP as citalopram/escitalopram response biomarkers in depression: pharmacometabolomics-informed pharmacogenomics. Clin. Pharmacol. Ther. 89, 97–104 (2011).
Acknowledgements
R. H. Weiss would like to acknowledge the support of NIH grants 5UO1CA86402 (Early Detection Research Network), 1R01CA135401-01A1 and 1R01DK082690-01A1, and the Medical Service of the US Department of Veterans' Affairs.
Author information
Authors and Affiliations
Contributions
Both authors contributed to all aspects of this article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Weiss, R., Kim, K. Metabolomics in the study of kidney diseases. Nat Rev Nephrol 8, 22–33 (2012). https://doi.org/10.1038/nrneph.2011.152
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrneph.2011.152
This article is cited by
-
Metabolomic profiling of overnight peritoneal dialysis effluents predicts the peritoneal equilibration test type
Scientific Reports (2023)
-
Characterization of oral and gut microbiome and plasma metabolomics in COVID-19 patients after 1-year follow-up
Military Medical Research (2022)
-
In-depth characterisation of the urine metabolome in cats with and without urinary tract diseases
Metabolomics (2022)
-
Rare genetic variants affecting urine metabolite levels link population variation to inborn errors of metabolism
Nature Communications (2021)
-
Pressurized planar electrochromatography of DNS amino acids derivatives in silica gel and silanized silica gel systems with formic acid addition to the water mobile phase
JPC – Journal of Planar Chromatography – Modern TLC (2021)