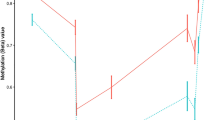
Abstract
Epigenetic changes influence gene expression without altering the DNA sequence. DNA methylation, histone modification and microRNA-associated post-transcriptional gene silencing are three key epigenetic mechanisms. Multiple sclerosis (MS) is a disease of the CNS with both inflammatory and neurodegenerative features. Although studies on epigenetic changes in MS only began in the past decade, a growing body of literature suggests that epigenetic changes may be involved in the development of MS, possibly by mediating the effects of environmental risk factors, such as smoking, vitamin D deficiency and Epstein–Barr virus infection. Such studies are also beginning to deliver important insights into the pathophysiology of MS. For example, inflammation and demyelination in relapsing–remitting MS may be related to the increased differentiation of T cells toward a T-helper 17 phenotype, which is an important epigenetically regulated pathophysiological mechanism. In progressive MS, other epigenetically regulated mechanisms, such as increased histone acetylation and citrullination of myelin basic protein, might exacerbate the disease course. In this Review, we summarize current knowledge on the role of epigenetic changes in the pathophysiology of MS.
Key Points
-
Multiple sclerosis (MS) has both inflammatory and neurodegenerative characteristics, with striking interindividual differences in disease course and severity
-
A low concordance rate for MS in monozygotic twins and enhanced maternal transmission of risk alleles suggest that epigenetic changes influence MS susceptibility
-
The effects of environmental risk factors, such as smoking, vitamin D level and Epstein–Barr virus infection, on the development and course of MS might be mediated by epigenetic mechanisms
-
In patients with relapsing–remitting MS, macrophage activation and differentiation of T-helper 17 cells are important epigenetically regulated proinflammatory mechanisms
-
In patients with progressive MS, citrullination of myelin basic protein and CNS neurosteroid synthesis are important epigenetically regulated neurodegenerative mechanisms
-
Future therapies for MS that target these epigenetic mechanisms might utilize histone deacetylase inhibitors, DNA methyltransferase inhibitors, and oligonucleotides containing modified locked nucleic acids
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Noseworthy, J. H., Lucchinetti, C., Rodriguez, M. & Weinshenker, B. G. Multiple sclerosis. N. Engl. J. Med. 343, 938–952 (2000).
Leray, E. et al. Evidence for a two-stage disability progression in multiple sclerosis. Brain 133, 1900–1913 (2010).
Confavreux, C., Vukusic, S. & Adeleine, P. Early clinical predictors and progression of irreversible disability in multiple sclerosis: an amnesic process. Brain 126, 770–782 (2003).
Koch, M., Kingwell, E., Rieckmann, P. & Tremlett, H. The natural history of primary progressive multiple sclerosis. Neurology 73, 1996–2002 (2009).
Scalfari, A. et al. The natural history of multiple sclerosis: a geographically based study 10: relapses and long-term disability. Brain 133, 1914–1929 (2010).
Confavreux, C., Vukusic, S., Moreau, T. & Adeleine, P. Relapses and progression of disability in multiple sclerosis. N. Engl. J. Med. 343, 1430–1438 (2000).
Carton, H. et al. Risks of multiple sclerosis in relatives of patients in Flanders, Belgium. J. Neurol. Neurosurg. Psychiatry 62, 329–333 (1997).
Sadovnick, A. D., Baird, P. A. & Ward, R. H. Multiple sclerosis: updated risks for relatives. Am. J. Med. Genet. 29, 533–541 (1988).
Robertson, N. P. et al. Age-adjusted recurrence risks for relatives of patients with multiple sclerosis. Brain 119, 449–455 (1996).
Sawcer, S. et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219 (2011).
Jaenisch, R. & Bird, A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33 (Suppl.), 245–254 (2003).
Skinner, M. K., Manikkam, M. & Guerrero-Bosagna, C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol. Metab. 21, 214–222 (2010).
Goll, M. G. & Bestor, T. H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514 (2005).
Okano, M., Bell, D. W., Haber, D. A. & Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999).
Klose, R. J. & Bird, A. P. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31, 89–97 (2006).
Weber, M. & Schübeler, D. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Curr. Opin. Cell Biol. 19, 273–280 (2007).
Dieker, J. & Muller, S. Epigenetic histone code and autoimmunity. Clin. Rev. Allergy Immunol. 39, 78–84 (2010).
Brooks, W. H., Le Dantec, C., Pers, J.-O., Youinou, P. & Renaudineau, Y. Epigenetics and autoimmunity. J. Autoimmun. 34, J207–J219 (2010).
Bernstein, E. & Allis, C. D. RNA meets chromatin. Genes Dev. 19, 1635–1655 (2005).
Hwang, H.-W. & Mendell, J. T. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer 94, 776–780 (2006).
Sevignani, C., Calin, G. A., Siracusa, L. D. & Croce, C. M. Mammalian microRNAs: a small world for fine-tuning gene expression. Mamm. Genome 17, 189–202 (2006).
Chang, T.-C. & Mendell, J. T. MicroRNAs in vertebrate physiology and human disease. Annu. Rev. Genomics Hum. Genet. 8, 215–239 (2007).
Fabbri, M., Ivan, M., Cimmino, A., Negrini, M. & Calin, G. A. Regulatory mechanisms of microRNAs involvement in cancer. Expert Opin. Biol. Ther. 7, 1009–1019 (2007).
Chao, M. J. et al. Parent-of-origin effects at the major histocompatibility complex in multiple sclerosis. Hum. Mol. Genet. 19, 3679–3689 (2010).
Ebers, G. C. et al. A population-based study of multiple sclerosis in twins. N. Engl. J. Med. 315, 1638–1642 (1986).
Kuusisto, H. et al. Concordance and heritability of multiple sclerosis in Finland: study on a nationwide series of twins. Eur. J. Neurol. 15, 1106–1110 (2008).
Ristori, G. et al. Multiple sclerosis in twins from continental Italy and Sardinia: a nationwide study. Ann. Neurol. 59, 27–34 (2006).
[No authors listed] Multiple sclerosis in 54 twinships: concordance rate is independent of zygosity. French Research Group on Multiple Sclerosis. Ann. Neurol. 32, 724–727 (1992).
Hansen, T. et al. Concordance for multiple sclerosis in Danish twins: an update of a nationwide study. Mult. Scler. 11, 504–510 (2005).
Baranzini, S. E. et al. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature 464, 1351–1356 (2010).
Junker, A. et al. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 132, 3342–3352 (2009).
Haasch, D. et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell. Immunol. 217, 78–86 (2002).
Thai, T.-H. et al. Regulation of the germinal center response by microRNA-155. Science 316, 604–608 (2007).
Teng, G. et al. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity 28, 621–629 (2008).
Teng, G. & Papavasiliou, F. N. Shhh! Silencing by microRNA-155. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 631–637 (2009).
O'Connell, R. M. et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 33, 607–619 (2010).
Steinman, L. A rush to judgment on Th17. J. Exp. Med. 205, 1517–1522 (2008).
Tzartos, J. S. et al. Interleukin-17 production in central nervous system-infiltrating T cells and glial cells is associated with active disease in multiple sclerosis. Am. J. Pathol. 172, 146–155 (2008).
Murugaiyan, G., Beynon, V., Mittal, A., Joller, N. & Weiner, H. L. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J. Immunol. 187, 2213–2221 (2011).
Du, C. et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol. 10, 1252–1259 (2009).
Cox, M. B. et al. MicroRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PLoS ONE 5, e12132 (2010).
Janson, P. C. et al. Profiling of CD4+ T cells with epigenetic immune lineage analysis. J. Immunol. 186, 92–102 (2011).
Liggett, T. et al. Methylation patterns of cell-free plasma DNA in relapsing–remitting multiple sclerosis. J. Neurol. Sci. 290, 16–21 (2010).
Otaegui, D. et al. Differential micro RNA expression in PBMC from multiple sclerosis patients. PLoS ONE 4, e6309 (2009).
Chestnut, B. A. et al. Epigenetic regulation of motor neuron cell death through DNA methylation. J. Neurosci. 31, 16619–16636 (2011).
Moscarello, M. A., Mastronardi, F. G. & Wood, D. D. The role of citrullinated proteins suggests a novel mechanism in the pathogenesis of multiple sclerosis. Neurochem. Res. 32, 251–256 (2007).
Moscarello, M. A., Wood, D. D., Ackerley, C. & Boulias, C. Myelin in multiple sclerosis is developmentally immature. J. Clin. Invest. 94, 146–154 (1994).
Mastronardi, F. G., Noor, A., Wood, D. D., Paton, T. & Moscarello, M. A. Peptidyl argininedeiminase 2 CpG island in multiple sclerosis white matter is hypomethylated. J. Neurosci. Res. 85, 2006–2016 (2007).
Lamensa, J. W. & Moscarello, M. A. Deimination of human myelin basic protein by a peptidylarginine deiminase from bovine brain. J. Neurochem. 61, 987–996 (1993).
Pedre, X. et al. Changed histone acetylation patterns in normal-appearing white matter and early multiple sclerosis lesions. J. Neurosci. 31, 3435–3445 (2011).
Noorbakhsh, F. et al. Impaired neurosteroid synthesis in multiple sclerosis. Brain 134, 2703–2721 (2011).
Ascherio, A. & Munger, K. L. Environmental risk factors for multiple sclerosis. Part I: the role of infection. Ann. Neurol. 61, 288–299 (2007).
Ascherio, A. & Munger, K. L. Environmental risk factors for multiple sclerosis. Part II: noninfectious factors. Ann. Neurol. 61, 504–513 (2007).
Hernán, M. A., Olek, M. J. & Ascherio, A. Cigarette smoking and incidence of multiple sclerosis. Am. J. Epidemiol. 154, 69–74 (2001).
Healy, B. C. et al. Smoking and disease progression in multiple sclerosis. Arch. Neurol. 66, 858–864 (2009).
Pittas, F. et al. Smoking is associated with progressive disease course and increased progression in clinical disability in a prospective cohort of people with multiple sclerosis. J. Neurol. 256, 577–585 (2009).
Di Pauli, F. et al. Smoking is a risk factor for early conversion to clinically definite multiple sclerosis. Mult. Scler. 14, 1026–1030 (2008).
Hernán, M. A. et al. Cigarette smoking and the progression of multiple sclerosis. Brain 128, 1461–1465 (2005).
Koch, M., van Harten, A., Uyttenboogaart, M. & De Keyser, J. Cigarette smoking and progression in multiple sclerosis. Neurology 69, 1515–1520 (2007).
Wan, E. S. et al. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum. Mol. Genet. 21, 3073–3082 (2012).
Koturbash, I., Beland, F. A. & Pogribny, I. P. Role of epigenetic events in chemical carcinogenesis—a justification for incorporating epigenetic evaluations in cancer risk assessment. Toxicol. Mech. Methods 21, 289–297 (2011).
Ma, Y. T., Collins, S. I., Young, L. S., Murray, P. G. & Woodman, C. B. J. Smoking initiation is followed by the early acquisition of epigenetic change in cervical epithelium: a longitudinal study. Br. J. Cancer 104, 1500–1504 (2011).
Marczylo, E. L., Amoako, A. A., Konje, J. C., Gant, T. W. & Marczylo, T. H. Smoking induces differential miRNA expression in human spermatozoa: A potential transgenerational epigenetic concern? Epigenetics 7, 432–439 (2012).
Ito, K. et al. Cigarette smoking reduces histone deacetylase 2 expression, enhances cytokine expression, and inhibits glucocorticoid actions in alveolar macrophages. FASEB J. 15, 1110–1112 (2001).
Munger, K. L., Levin, L. I., Hollis, B. W., Howard, N. S. & Ascherio, A. Serum 25-hydroxyvitamin D levels and risk of multiple sclerosis. JAMA 296, 2832–2838 (2006).
Smolders, J., Menheere, P., Kessels, A., Damoiseaux, J. & Hupperts, R. Association of vitamin D metabolite levels with relapse rate and disability in multiple sclerosis. Mult. Scler. 14, 1220–1224 (2008).
Simpson, S. et al. Higher 25-hydroxyvitamin D is associated with lower relapse risk in multiple sclerosis. Ann. Neurol. 68, 193–203 (2010).
Simpson, S. Jr, Blizzard, L., Otahal, P., Van der Mei, I. & Taylor, B. Latitude is significantly associated with the prevalence of multiple sclerosis: a meta-analysis. J. Neurol. Neurosurg. Psychiatry http://dx.doi.org/10.1136/jnnp.2011.240432.
Pereira, F. et al. Vitamin D has wide regulatory effects on histone demethylase genes. Cell Cycle 11, 1081–1089 (2012).
Pereira, F. et al. KDM6B/JMJD3 histone demethylase is induced by vitamin D and modulates its effects in colon cancer cells. Hum. Mol. Genet. 20, 4655–4665 (2011).
Joshi, S. et al. 1,25-dihydroxyvitamin D3 ameliorates Th17 autoimmunity via transcriptional modulation of interleukin-17A. Mol. Cell. Biol. 31, 3653–3669 (2011).
Ascherio, A. et al. Epstein–Barr virus antibodies and risk of multiple sclerosis: a prospective study. JAMA 286, 3083–3088 (2001).
Handel, A. E. et al. An updated meta-analysis of risk of multiple sclerosis following infectious mononucleosis. PLoS ONE 5, (2010).
Niller, H. H., Wolf, H. & Minarovits, J. Epigenetic dysregulation of the host cell genome in Epstein–Barr virus-associated neoplasia. Semin. Cancer Biol. 19, 158–164 (2009).
Tsai, C.-N., Tsai, C.-L., Tse, K.-P., Chang, H.-Y. & Chang, Y.-S. The Epstein–Barr virus oncogene product, latent membrane protein 1, induces the downregulation of E-cadherin gene expression via activation of DNA methyltransferases. Proc. Natl Acad. Sci. USA 99, 10084–10089 (2002).
Tsai, C.-L. et al. Activation of DNA methyltransferase 1 by EBV LMP1 Involves c-Jun NH2-terminal kinase signaling. Cancer Res. 66, 11668–11676 (2006).
Kwong, J. et al. Promoter hypermethylation of multiple genes in nasopharyngeal carcinoma. Clin. Cancer Res. 8, 131–137 (2002).
Reddy, P. et al. Histone deacetylase inhibition modulates indoleamine 2,3-dioxygenase-dependent DC functions and regulates experimental graft-versus-host disease in mice. J. Clin. Invest. 118, 2562–2573 (2008).
Nencioni, A. et al. Histone deacetylase inhibitors affect dendritic cell differentiation and immunogenicity. Clin. Cancer Res. 13, 3933–3941 (2007).
Shen, S., Li, J. & Casaccia-Bonnefil, P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J. Cell Biol. 169, 577–589 (2005).
Dasgupta, S., Zhou, Y., Jana, M., Banik, N. L. & Pahan, K. Sodium phenylacetate inhibits adoptive transfer of experimental allergic encephalomyelitis in SJL/J. mice at multiple steps. J. Immunol. 170, 3874–3882 (2003).
Camelo, S. et al. Transcriptional therapy with the histone deacetylase inhibitor trichostatin A ameliorates experimental autoimmune encephalomyelitis. J. Neuroimmunol. 164, 10–21 (2005).
Gray, S. G. & Dangond, F. Rationale for the use of histone deacetylase inhibitors as a dual therapeutic modality in multiple sclerosis. Epigenetics 1, 67–75 (2006).
Faraco, G., Cavone, L. & Chiarugi, A. The therapeutic potential of HDAC inhibitors in the treatment of multiple sclerosis. Mol. Med. 17, 442–447 (2011).
Penas, C. et al. Valproate reduces CHOP levels and preserves oligodendrocytes and axons after spinal cord injury. Neuroscience 178, 33–44 (2011).
Candelaria, M. et al. Hydralazine and magnesium valproate as epigenetic treatment for myelodysplastic syndrome. Preliminary results of a phase-II trial. Ann. Hematol. 90, 379–387 (2011).
Elmén, J. et al. LNA-mediated microRNA silencing in non-human primates. Nature 452, 896–899 (2008).
Author information
Authors and Affiliations
Contributions
M. W. Koch and O. Kovalchuk researched data for the article. M. W. Koch, L. M. Metz and O. Kovalchuk provided substantial contributions to discussion of the article, writing and review and/or editing before submission of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Koch, M., Metz, L. & Kovalchuk, O. Epigenetic changes in patients with multiple sclerosis. Nat Rev Neurol 9, 35–43 (2013). https://doi.org/10.1038/nrneurol.2012.226
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrneurol.2012.226
This article is cited by
-
Vitamins A and D Enhance the Expression of Ror-γ-Targeting miRNAs in a Mouse Model of Multiple Sclerosis
Molecular Neurobiology (2023)
-
Analysis of the entire mitochondrial genome reveals Leber’s hereditary optic neuropathy mitochondrial DNA mutations in an Arab cohort with multiple sclerosis
Scientific Reports (2022)
-
Citrullination in the pathology of inflammatory and autoimmune disorders: recent advances and future perspectives
Cellular and Molecular Life Sciences (2022)
-
Histone citrullination: a new target for tumors
Molecular Cancer (2021)
-
Angiogenic responses in a 3D micro-engineered environment of primary endothelial cells and pericytes
Angiogenesis (2021)