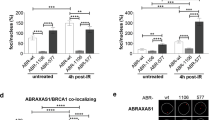
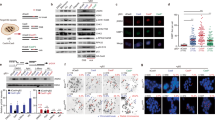
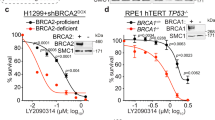
Abstract
Germline missense mutations affecting a single BRCA2 allele predispose humans to cancer. Here we identify a protein-targeting mechanism that is disrupted by the cancer-associated mutation, BRCA2D2723H, and that controls the nuclear localization of BRCA2 and its cargo, the recombination enzyme RAD51. A nuclear export signal (NES) in BRCA2 is masked by its interaction with a partner protein, DSS1, such that point mutations impairing BRCA2-DSS1 binding render BRCA2 cytoplasmic. In turn, cytoplasmic mislocalization of mutant BRCA2 inhibits the nuclear retention of RAD51 by exposing a similar NES in RAD51 that is usually obscured by the BRCA2-RAD51 interaction. Thus, a series of NES-masking interactions localizes BRCA2 and RAD51 in the nucleus. Notably, BRCA2D2723H decreases RAD51 nuclear retention even when wild-type BRCA2 is also present. Our findings suggest a mechanism for the regulation of the nucleocytoplasmic distribution of BRCA2 and RAD51 and its impairment by a heterozygous disease-associated mutation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
The Breast Cancer Linkage Consortium. Cancer risks in BRCA2 mutation carriers. The Breast Cancer Linkage Consortium. J. Natl. Cancer Inst. 91, 1310–1316 (1999).
Collins, N. et al. Consistent loss of the wild type allele in breast cancers from a family linked to the BRCA2 gene on chromosome 13q12–13. Oncogene 10, 1673–1675 (1995).
King, T.A. et al. Heterogenic loss of the wild-type BRCA allele in human breast tumorigenesis. Ann. Surg. Oncol. 14, 2510–2518 (2007).
Knudson, A.G. Jr. Mutation and cancer: statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA 68, 820–823 (1971).
Skoulidis, F. et al. Germline Brca2 heterozygosity promotes KrasG12D-driven carcinogenesis in a murine model of familial pancreatic cancer. Cancer Cell 18, 499–509 (2010).
Xia, F. et al. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc. Natl. Acad. Sci. USA 98, 8644–8649 (2001).
Moynahan, M.E., Pierce, A.J. & Jasin, M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7, 263–272 (2001).
Chen, P.-L. et al. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc. Natl. Acad. Sci. USA 95, 5287–5292 (1998).
Yuan, S.S. et al. BRCA2 is required for ionizing radiation–induced assembly of Rad51 complex in vivo. Cancer Res 59, 3547–3551 (1999).
Yang, H., Li, Q., Fan, J., Holloman, W.K. & Pavletich, N.P. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature 433, 653–657 (2005).
Thorslund, T. et al. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1263–1265 (2010).
Liu, J., Doty, T., Gibson, B. & Heyer, W.D. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1260–1262 (2010).
Jensen, R.B., Carreira, A. & Kowalczykowski, S.C. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467, 678–683 (2010).
Carreira, A. et al. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell 136, 1032–1043 (2009).
Shivji, M.K. et al. The BRC repeats of human BRCA2 differentially regulate RAD51 binding on single- versus double-stranded DNA to stimulate strand exchange. Proc. Natl. Acad. Sci. USA 106, 13254–13259 (2009).
Goldgar, D.E. et al. Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am. J. Hum. Genet. 75, 535–544 (2004).
Karchin, R., Agarwal, M., Sali, A., Couch, F. & Beattie, M.S. Classifying variants of undetermined significance in BRCA2 with protein likelihood ratios. Cancer Inform. 6, 203–216 (2008).
Marston, N.J. et al. Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol. Cell Biol. 19, 4633–4642 (1999).
Yang, H. et al. BRCA2 function in DNA binding and recombination from a BRCA2–DSS1-ssDNA structure. Science 297, 1837–1848 (2002).
Kuznetsov, S.G., Liu, P. & Sharan, S.K. Mouse embryonic stem cell-based functional assay to evaluate mutations in BRCA2. Nat. Med. 14, 875–881 (2008).
Zhou, Q. et al. Dss1 interaction with Brh2 as a regulatory mechanism for recombinational repair. Mol. Cell Biol. 27, 2512–2526 (2007).
Jares-Erijman, E.A. & Jovin, T.M. FRET imaging. Nat. Biotechnol. 21, 1387–1395 (2003).
Goedhart, J., Vermeer, J.E.M., Adjobo-Hermans, M.J.W., van Weeren, L. & Gadella, T.W.J. Jr. Sensitive detection of p65 homodimers using red-shifted and fluorescent protein-based FRET couples. PLoS ONE 2, e1011 (2007).
Spain, B.H., Larson, C.J., Shihabuddin, L.S., Gage, F.H. & Verma, I.M. Truncated BRCA2 is cytoplasmic: implications for cancer-linked mutations. Proc. Natl. Acad. Sci. USA 96, 13920–13925 (1999).
Wu, K. et al. Functional evaluation and cancer risk assessment of BRCA2 unclassified variants. Cancer Res. 65, 417–426 (2005).
la Cour, T. et al. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 17, 527–536 (2004).
Sonoda, E. et al. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17, 598–608 (1998).
Pellegrini, L. et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature 420, 287–293 (2002).
Rajendra, E. & Venkitaraman, A.R. Two modules in the BRC repeats of BRCA2 mediate structural and functional interactions with the RAD51 recombinase. Nucleic Acids Res. 38, 82–96 (2010).
Yu, D.S. et al. Dynamic control of Rad51 recombinase by self-association and interaction with BRCA2. Mol. Cell 12, 1029–1041 (2003).
Rittinger, K. et al. Structural analysis of 14–3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14–3-3 in ligand binding. Mol. Cell 4, 153–166 (1999).
Hantschel, O. et al. Structural basis for the cytoskeletal association of Bcr-Abl/c-Abl. Mol. Cell 19, 461–473 (2005).
Güttler, T. et al. NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat. Struct. Mol. Biol. 17, 1367–1376 (2010).
Davies, A.A. et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell 7, 273–282 (2001).
Xu, D., Farmer, A., Collett, G., Grishin, N.V. & Chook, Y.M. Sequence and structural analyses of nuclear export signals in the NESdb database. Mol. Biol. Cell 23, 3677–3693 (2012).
Dong, X. et al. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature 458, 1136–1141 (2009).
Mladenov, E., Anachkova, B. & Tsaneva, I. Sub-nuclear localization of Rad51 in response to DNA damage. Genes Cells 11, 513–524 (2006).
Gildemeister, O.S., Sage, J.M. & Knight, K.L. Cellular redistribution of Rad51 in response to DNA damage: a novel role for Rad51C. J. Biol. Chem. 284, 31945–31952 (2009).
Ciccia, A. & Elledge, S.J. The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 (2010).
San Filippo, J., Sung, P. & Klein, H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 (2008).
Dingwall, C. & Laskey, R.A. Protein import into the cell nucleus. Annu. Rev. Cell Biol. 2, 367–390 (1986).
Krogan, N.J. et al. Proteasome involvement in the repair of DNA double-strand breaks. Mol. Cell 16, 1027–1034 (2004).
Mannen, T., Andoh, T. & Tani, T. Dss1 associating with the proteasome functions in selective nuclear mRNA export in yeast. Biochem. Biophys. Res. Commun. 365, 664–671 (2008).
Gudmundsdottir, K., Lord, C.J., Witt, E., Tutt, A.N. & Ashworth, A. DSS1 is required for RAD51 focus formation and genomic stability in mammalian cells. EMBO Rep. 5, 989–993 (2004).
Saeki, H. et al. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc. Natl. Acad. Sci. USA 103, 8768–8773 (2006).
Edwards, S.L. et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451, 1111–1115 (2008).
Sakai, W. et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 451, 1116–1120 (2008).
Siaud, N. et al. Plasticity of BRCA2 function in homologous recombination: genetic interactions of the PALB2 and DNA binding domains. PLoS Genet. 7, e1002409 (2011).
Kojic, M., Zhou, Q., Lisby, M. & Holloman, W.K. Brh2-Dss1 interplay enables properly controlled recombination in Ustilago maydis. Mol. Cell Biol. 25, 2547–2557 (2005).
Han, X., Saito, H., Miki, Y. & Nakanishi, A.A. CRM1-mediated nuclear export signal governs cytoplasmic localization of BRCA2 and is essential for centrosomal localization of BRCA2. Oncogene 27, 2969–2977 (2008).
Honrado, E. et al. Immunohistochemical expression of DNA repair proteins in familial breast cancer differentiate BRCA2-associated tumors. J. Clin. Oncol. 23, 7503–7511 (2005).
Mitra, A. et al. Overexpression of RAD51 occurs in aggressive prostatic cancer. Histopathology 55, 696–704 (2009).
Hattori, H., Skoulidis, F., Russell, P. & Venkitaraman, A.R. Context dependence of checkpoint kinase 1 as a therapeutic target for pancreatic cancers deficient in the BRCA2 tumor suppressor. Mol. Cancer Ther. 10, 670–678 (2011).
Jeyasekharan, A.D. et al. DNA damage regulates the mobility of Brca2 within the nucleoplasm of living cells. Proc. Natl. Acad. Sci. USA 107, 21937–21942 (2010).
Frey, S., Richter, R.P. & Gorlich, D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314, 815–817 (2006).
Ayoub, N., Jeyasekharan, A.D., Bernal, J.A. & Venkitaraman, A.R. HP1-β mobilization promotes chromatin changes that initiate the DNA damage response. Nature 453, 682–686 (2008).
Acknowledgements
We acknowledge the generous gifts of WT and BRCA2D2723H ES cells from S. Sharan (US National Institutes of Health) and sYFP cDNA from T. Gadella (University of Amsterdam). We thank D. Görlich (Max Planck Institute) for generously providing constructs encoding RanQ69L and CRM1 and for information concerning assay conditions. We thank members of the Venkitaraman laboratory for technical assistance and constructive discussions and M. Goode for help with the feeder-free culture for ES cell cultures. A.D.J. was supported by a career development fellowship from the UK MRC Cancer Unit. Work in the laboratory of C.F.K. was funded by the UK Biosciences and Biotechnology Research Council, and work in the laboratory of A.R.V. was funded by the UK MRC.
Author information
Authors and Affiliations
Contributions
A.D.J. and A.R.V. conceived the project. Experiments were performed by A.D.J., A.B.J., J.S. and M.L. (microscopy); Y.L. and M.L. (biochemistry); and A.D.J., H.H., V.P., A.B.J., E.S., Y.R., K.S. and N.A. (cell biology). Structural analysis was performed by E.R., and FRET-FLIM analysis was performed by S.S. and C.F.K. A.D.J. and A.R.V. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 (PDF 9547 kb)
Rights and permissions
About this article
Cite this article
Jeyasekharan, A., Liu, Y., Hattori, H. et al. A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization. Nat Struct Mol Biol 20, 1191–1198 (2013). https://doi.org/10.1038/nsmb.2666
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2666
This article is cited by
-
Characterization of BRCA2 R3052Q variant in mice supports its functional impact as a low-risk variant
Cell Death & Disease (2023)
-
Dynamic interaction of BRCA2 with telomeric G-quadruplexes underlies telomere replication homeostasis
Nature Communications (2022)
-
CRIP1 cooperates with BRCA2 to drive the nuclear enrichment of RAD51 and to facilitate homologous repair upon DNA damage induced by chemotherapy
Oncogene (2021)
-
Tracking the expression of therapeutic protein targets in rare cells by antibody-mediated nanoparticle labelling and magnetic sorting
Nature Biomedical Engineering (2020)
-
Depletion of nuclear import protein karyopherin alpha 7 (KPNA7) induces mitotic defects and deformation of nuclei in cancer cells
BMC Cancer (2018)