Abstract
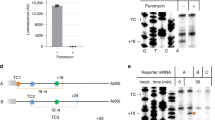
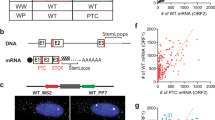
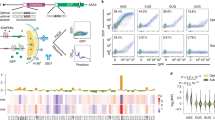
The accuracy of eukaryotic gene expression is monitored at multiple levels. Surveillance pathways have been identified that degrade messenger RNAs containing nonsense mutations, harboring stalled ribosomes or lacking termination codons. Here we report a previously uncharacterized surveillance pathway triggered by ribosome extension into the 3′ untranslated region. This ribosome extension–mediated decay, REMD, accounts for marked repression of protein synthesis from a human α-globin gene containing a prevalent antitermination mutation. REMD can be mechanistically distinguished from other surveillance pathways by its functional linkage to accelerated deadenylation, by its independence from the NMD factor Upf1 and by cell-type restriction. This unusual pathway of mRNA surveillance is likely to act as a modifier of additional genetic defects and may reflect post-transcriptional controls particular to erythroid and other differentiated cell lineages.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Nagy, E. & Maquat, L.E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 23, 198–199 (1998).
Inacio, A. et al. Nonsense mutations in close proximity to the initiation codon fail to trigger full nonsense-mediated mRNA decay. J. Biol. Chem. 279, 32170–32180 (2004).
Zhang, J. & Maquat, L.E. Evidence that translation reinitiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO J. 16, 826–833 (1997).
Stockklausner, C. et al. The uORF-containing thrombopoietin mRNA escapes nonsense-mediated decay (NMD). Nucleic Acids Res. 34, 2355–2363 (2006).
Silva, A.L. et al. The canonical UPF1-dependent nonsense-mediated mRNA decay is inhibited in transcripts carrying a short open reading frame independent of sequence context. RNA 12, 2160–2170 (2006).
Conti, E. & Izaurralde, E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 17, 316–325 (2005).
Frischmeyer, P.A. et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295, 2258–2261 (2002).
van Hoof, A., Frischmeyer, P.A., Dietz, H.C. & Parker, R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295, 2262–2264 (2002).
Doma, M.K. & Parker, R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440, 561–564 (2006).
Cao, D. & Parker, R. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell 113, 533–545 (2003).
Chen, C.Y. & Shyu, A.B. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol. Cell. Biol. 23, 4805–4813 (2003).
Yamashita, A. et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 12, 1054–1063 (2005).
Clegg, J.B., Weatherall, D.J. & Milner, P.F. Haemoglobin Constant Spring—a chain termination mutant? Nature 234, 337–340 (1971).
Liebhaber, S.A. & Kan, Y.W. Differentiation of the mRNA transcripts originating from the alpha 1- and alpha 2-globin loci in normals and alpha-thalassemics. J. Clin. Invest. 68, 439–446 (1981).
Kong, J., Ji, X. & Liebhaber, S.A. The KH-domain protein alpha CP has a direct role in mRNA stabilization independent of its cognate binding site. Mol. Cell. Biol. 23, 1125–1134 (2003).
Wang, X., Kiledjian, M., Weiss, I.M. & Liebhaber, S.A. Detection and characterization of a 3′ untranslated region ribonucleoprotein complex associated with human alpha-globin mRNA stability. Mol. Cell. Biol. 15, 1769–1777 (1995).
Ji, X., Kong, J. & Liebhaber, S.A. In vivo association of the stability control protein alphaCP with actively translating mRNAs. Mol. Cell. Biol. 23, 899–907 (2003).
Holcik, M. & Liebhaber, S.A. Four highly stable eukaryotic mRNAs assemble 3′ untranslated region RNA-protein complexes sharing cis and trans components. Proc. Natl. Acad. Sci. USA 94, 2410–2414 (1997).
Decker, C.J. & Parker, R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 7, 1632–1643 (1993).
Morales, J., Russell, J.E. & Liebhaber, S.A. Destabilization of human alpha-globin mRNA by translation anti-termination is controlled during erythroid differentiation and is paralleled by phased shortening of the poly(A) tail. J. Biol. Chem. 272, 6607–6613 (1997).
Chang, T.C. et al. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev. 18, 2010–2023 (2004).
Tucker, M. et al. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104, 377–386 (2001).
Boeck, R. et al. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J. Biol. Chem. 271, 432–438 (1996).
Brown, C.E., Tarun, S.Z., Jr., Boeck, R. & Sachs, A.B. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 5744–5753 (1996).
Korner, C.G. & Wahle, E. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J. Biol. Chem. 272, 10448–10456 (1997).
He, F. & Jacobson, A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 9, 437–454 (1995).
Unterholzner, L. & Izaurralde, E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell 16, 587–596 (2004).
Ishigaki, Y., Li, X., Serin, G. & Maquat, L.E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106, 607–617 (2001).
Weiss, I.M. & Liebhaber, S.A. Erythroid cell-specific determinants of alpha-globin mRNA stability. Mol. Cell. Biol. 14, 8123–8132 (1994).
Bilger, A., Fox, C.A., Wahle, E. & Wickens, M. Nuclear polyadenylation factors recognize cytoplasmic polyadenylation elements. Genes Dev. 8, 1106–1116 (1994).
McGrew, L.L., Dworkin-Rastl, E., Dworkin, M.B. & Richter, J.D. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev. 3, 803–815 (1989).
Waggoner, S.A. & Liebhaber, S.A. Regulation of alpha-globin mRNA stability. Exp. Biol. Med. (Maywood) 228, 387–395 (2003).
Yu, J. & Russell, J.E. Structural and functional analysis of an mRNP complex that mediates the high stability of human beta-globin mRNA. Mol. Cell. Biol. 21, 5879–5888 (2001).
Paillard, L., Maniey, D., Lachaume, P., Legagneux, V. & Osborne, H.B. Identification of a C-rich element as a novel cytoplasmic polyadenylation element in Xenopus embryos. Mech. Dev. 93, 117–125 (2000).
Acknowledgements
We thank A.-B. Shyu of The University of Texas Medical School at Houston for the kind gift of plasmids encoding the dominant-negative mutants of deadenylases Ccr4, Pan2 and Parn, and N. Cooke of The University of Pennsylvania for her critical comments. This work was supported by US National Institutes of Health grants R37-HL65449 and PO1 CA72765 (S.A.L.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Extension of the antiterminated ribosome into the 3′ untranslated region triggers two independent destabilization events, an α-complex–dependent action and an α-complex–independent ribosome-extension effect. (PDF 267 kb)
Rights and permissions
About this article
Cite this article
Kong, J., Liebhaber, S. A cell type–restricted mRNA surveillance pathway triggered by ribosome extension into the 3′ untranslated region. Nat Struct Mol Biol 14, 670–676 (2007). https://doi.org/10.1038/nsmb1256
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1256
This article is cited by
-
Novel and recurrent AID mutations underlie prevalent autosomal recessive form of HIGM in consanguineous patients
Immunogenetics (2016)
-
Structural determinants of human ζ-globin mRNA stability
Journal of Hematology & Oncology (2014)
-
An RNA-protein complex links enhanced nuclear 3′ processing with cytoplasmic mRNA stabilization
The EMBO Journal (2011)