Abstract
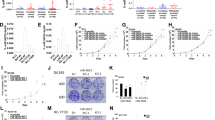
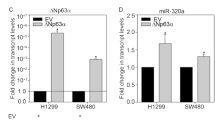
Loss of p53 function is a critical event during tumorigenesis, with half of all cancers harboring mutations within the TP53 gene. Such events frequently result in the expression of a mutated p53 protein with gain-of-function properties that drive invasion and metastasis. Here, we show that the expression of miR-155 was up-regulated by mutant p53 to drive invasion. The miR-155 host gene was directly repressed by p63, providing the molecular basis for mutant p53 to drive miR-155 expression. Significant overlap was observed between miR-155 targets and the molecular profile of mutant p53-expressing breast tumors in vivo. A search for cancer-related target genes of miR-155 revealed ZNF652, a novel zinc-finger transcriptional repressor. ZNF652 directly repressed key drivers of invasion and metastasis, such as TGFB1, TGFB2, TGFBR2, EGFR, SMAD2 and VIM. Furthermore, silencing of ZNF652 in epithelial cancer cell lines promoted invasion into matrigel. Importantly, loss of ZNF652 expression in primary breast tumors was significantly correlated with increased local invasion and defined a population of breast cancer patients with metastatic tumors. Collectively, these findings suggest that miR-155 targeted therapies may provide an attractive approach to treat mutant p53-expressing tumors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Oren M, Rotter V . Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol 2010; 2: a001107.
Brosh R, Rotter V . When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009; 9: 701–713.
Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res 2006; 12: 1157–1167.
Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005; 102: 13550–13555.
Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S et al. Mutant p53 drives invasion by promoting integrin recycling. Cell 2009; 139: 1327–1341.
Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B et al. A mutant-p53/Smad complex opposes p63 to empower TGF²-induced metastasis. Cell 2009; 137: 87–98.
Huang Y, Chuang A, Hao H, Talbot C, Sen T, Trink B et al. Phospho-[Delta]Np63[alpha] is a key regulator of the cisplatin-induced microRNAome in cancer cells. Cell Death Differ 2011; 18: 1220–1230.
Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 2010; 467: 986–990.
Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65: 7065–7070.
Jiang S, Zhang H-W, Lu M-H, He X-H, Li Y, Gu H et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res 2010; 70: 3119–3127.
Blick T, Widodo E, Hugo H, Waltham M, Lenburg ME, Neve RM et al. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin Exp Metastasis 2008; 25: 629–642.
Bergh J, Norberg T, Sjogren S, Lindgren A, Holmberg L . Complete sequencing of the p53 gene provides prognostic information in breast cancer patients, particularly in relation to adjuvant systemic therapy and radiotherapy. Nat Med 1995; 1: 1029–1034.
Davidoff AM, Kerns BJ, Pence JC, Marks JR, Iglehart JD . p53 alterations in all stages of breast cancer. J Surg Oncol 1991; 48: 260–267.
Kouwenhoven EN, van Heeringen SJ, Tena JJ, Oti M, Dutilh BE, Alonso ME et al. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet 2010; 6: e1001065.
Brosh R, Shalgi R, Liran A, Landan G, Korotayev K, Nguyen GH et al. p53-Repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol Syst Biol 2008; 4: 229.
Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD et al. Genes that mediate breast cancer metastasis to lung. Nature 2005; 436: 518–524.
Kumar R, Manning J, Spendlove HE, Kremmidiotis G, McKirdy R, Lee J et al. ZNF652, a novel zinc finger protein, interacts with the putative breast tumor suppressor CBFA2T3 to repress transcription. Mol Cancer Res 2006; 4: 655–665.
Kumar R, Cheney KM, McKirdy R, Neilsen PM, Schulz RB, Lee J et al. CBFA2T3-ZNF652 corepressor complex regulates transcription of the E-box gene HEB. J Biol Chem 2008; 283: 19026–19038.
Yin Q, McBride J, Fewell C, Lacey M, Wang X, Lin Z et al. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J Virol 2008; 82: 5295–5306.
Xu G, Fewell C, Taylor C, Deng N, Hedges D, Wang X et al. Transcriptome and targetome analysis in MIR155 expressing cells using RNA-seq. RNA 2010; 16: 1610–1622.
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006; 10: 515–527.
Pankova K, Rosel D, Novotny M, Brabek J . The molecular mechanisms of transition between mesenchymal and amoeboid invasiveness in tumor cells. Cell Mol Life Sci 2010; 67: 63–71.
Machesky LM . Lamellipodia and filopodia in metastasis and invasion. FEBS Lett 2008; 582: 2102–2111.
Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D et al. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem 2010; 285: 17869–17879.
Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol 2008; 28: 6773–6784.
Xiang X, Zhuang X, Ju S, Zhang S, Jiang H, Mu J et al. miR-155 promotes macroscopic tumor formation yet inhibits tumor dissemination from mammary fat pads to the lung by preventing EMT. Oncogene 2011; 30: 3440–3453.
Nieves-Alicea R, Colburn NH, Simeone AM, Tari AM . Programmed cell death 4 inhibits breast cancer cell invasion by increasing tissue inhibitor of metalloproteinases-2 expression. Breast Cancer Res Treat 2009; 114: 203–209.
Santhanam AN, Baker AR, Hegamyer G, Kirschmann DA, Colburn NH . Pdcd4 repression of lysyl oxidase inhibits hypoxia-induced breast cancer cell invasion. Oncogene 2010; 29: 3921–3932.
Wen YH, Shi X, Chiriboga L, Matsahashi S, Yee H, Afonja O . Alterations in the expression of PDCD4 in ductal carcinoma of the breast. Oncol Rep 2007; 18: 1387–1393.
Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell 2004; 5: 607–616.
Do TV, Kubba LA, Du H, Sturgis CD, Woodruff TK . Transforming growth factor-beta1, transforming growth factor-beta2, and transforming growth factor-beta3 enhance ovarian cancer metastatic potential by inducing a Smad3-dependent epithelial-to-mesenchymal transition. Mol Cancer Res 2008; 6: 695–705.
Vuoriluoto K, Haugen H, Kiviluoto S, Mpindi JP, Nevo J, Gjerdrum C et al. Vimentin regulates EMT induction by Slug and oncogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene 2011; 30: 1436–1448.
Yu Q, Stamenkovic I . Transforming growth factor-beta facilitates breast carcinoma metastasis by promoting tumor cell survival. Clin Exp Metastasis 2004; 21: 235–242.
Akhurst RJ, Derynck R . TGF-beta signaling in cancer – a double-edged sword. Trends Cell Biol 2001; 11: S44–S51.
Noll JE, Jeffery J, Al-Ejeh F, Kumar R, Khanna KK, Callen DF et al. Mutant p53 drives multinucleation and invasion through a process that is suppressed by ANKRD11. Oncogene 2011; 31: 2836–2848.
Utku Y, Dehan E, Ouerfelli O, Piano F, Zuckermann RN, Pagano M et al. A peptidomimetic siRNA transfection reagent for highly effective gene silencing. Mol Biosyst 2006; 2: 312–317.
Pishas KI, Al-Ejeh F, Zinonos I, Brown MP, Evdokiou A, Callen DF et al. Nutlin-3a is a potential therapeutic for Ewing Sarcoma. Clin Cancer Res 2011; 17: 494–504.
Park SM, Gaur AB, Lengyel E, Peter ME . The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008; 22: 894–907.
Acknowledgements
We thank Dr Erik Flemington for his kind gift of the pMSCV-Puro-GFP-miR-155 plasmid. We also thank Dr Emily Paterson and Bee Suan Tay for technical assistance. This study was supported by the National Health and Medical Research Council (NHMRC) of Australia (project #1009447).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Neilsen, P., Noll, J., Mattiske, S. et al. Mutant p53 drives invasion in breast tumors through up-regulation of miR-155. Oncogene 32, 2992–3000 (2013). https://doi.org/10.1038/onc.2012.305
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2012.305
Keywords
This article is cited by
-
microRNA-205 represses breast cancer metastasis by perturbing the rab coupling protein [RCP]-mediated integrin β1 recycling on the membrane
Apoptosis (2024)
-
First case of endometrial cancer after yolk sac tumor in a patient with Li-Fraumeni syndrome
BMC Women's Health (2023)
-
Systemic Listeria monocytogenes infection in aged mice induces long-term neuroinflammation: the role of miR-155
Immunity & Ageing (2022)
-
Mutual exclusivity of ESR1 and TP53 mutations in endocrine resistant metastatic breast cancer
npj Breast Cancer (2022)
-
p53 signaling in cancer progression and therapy
Cancer Cell International (2021)