Abstract
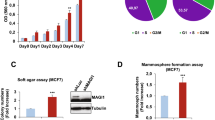
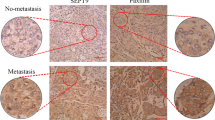
srGAP3, a member of the Slit-Robo sub-family of Rho GTPase-activating proteins (Rho GAPs), controls actin and microtubule dynamics through negative regulation of Rac. Here, we describe a potential role for srGAP3 as a tumor suppressor in mammary epithelial cells. We show that RNAi-mediated depletion of srGAP3 promotes Rac dependent, anchorage-independent growth of partially transformed human mammary epithelial cells (HMECs). Furthermore, srGAP3 expression is absent, or significantly reduced in 7/10 breast cancer cell lines compared with normal HMECs. Re-expression of srGAP3 in a subset of these cell lines inhibits both anchorage-independent growth and cell invasion in a GAP-dependent manner, and this is accompanied by an increase in phosphorylation of the ezrin/radixin/moesin (ERM) family proteins and myosin light chain 2 (MLC2). Inhibition of the Rho regulated kinase, ROCK, reduces ERM and MLC2 phosphorylation and restores invasion. We conclude that srGAP3 has tumor suppressor-like activity in HMECs, likely through its activity as a negative regulator of Rac1.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Ellenbroek SI, Collard JG . Rho GTPases: functions and association with cancer. Clin Exp Metastasis 2007; 24: 657–672.
Vega FM, Ridley AJ . Rho GTPases in cancer cell biology. FEBS Lett 2008; 582: 2093–2101.
Jaffe AB, Hall A . Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol 2005; 21: 247–269.
Lazer G, Katzav S . Guanine nucleotide exchange factors for RhoGTPases: good therapeutic targets for cancer therapy? Cell Signal 2011; 23: 969–979.
Xue W, Krasnitz A, Lucito R, Sordella R, Vanaelst L, Cordon-Cardo C et al. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev 2008; 22: 1439–1444.
Yang C, Kazanietz MG . Chimaerins: GAPs that bridge diacylglycerol signalling and the small G-protein Rac. Biochem J 2007; 403: 1–12.
Aspenstrom P . Roles of F-BAR/PCH proteins in the regulation of membrane dynamics and actin reorganization. Int Rev Cell Mol Biol 2009; 272: 1–31.
Wong K, Ren XR, Huang YZ, Xie Y, Liu G, Saito H et al. Signal transduction in neuronal migration: roles of GTPase activating proteins and the small GTPase Cdc42 in the slit-robo pathway. Cell 2001; 107: 209–221.
Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K et al. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell 2009; 138: 990–1004.
Madura T, Yamashita T, Kubo T, Tsuji L, Hosokawa K, Tohyama M . Changes in mRNA of Slit-Robo GTPase-activating protein 2 following facial nerve transection. Brain Res Mol Brain Res 2004; 123: 76–80.
Endris V, Wogatzky B, Leimer U, Bartsch D, Zatyka M, Latif F et al. The novel Rho-GTPase activating gene MEGAP/srGAP3 has a putative role in severe mental retardation. Proc Natl Acad Sci USA 2002; 99: 11754–11759.
Endris V, Haussmann L, Buss E, Bacon C, Bartsch D, Rappold G . SrGAP3 interacts with lamellipodin at the cell membrane and regulates Rac-dependent cellular protrusions. J Cell Sci 2011; 124 (Pt 23): 3941–3955.
Yang Y, Marcello M, Endris V, Saffrich R, Fischer R, Trendelenburg MF et al. MEGAP impedes cell migration via regulating actin and microtubule dynamics and focal complex formation. Exp Cell Res 2006; 312: 2379–2393.
Carlson BR, Lloyd KE, Kruszewski A, Kim IH, Rodriguiz RM, Heindel C et al. WRP/srGAP3 facilitates the initiation of spine development by an inverse F-BAR domain, and its loss impairs long-term memory. J Neurosci 2011; 31: 2447–2460.
Soderling SH, Binns KL, Wayman GA, Davee SM, Ong SH, Pawson T et al. The WRP component of the WAVE-1 complex attenuates Rac-mediated signalling. Nat Cell Biol 2002; 4: 970–975.
Soderling SH, Guire ES, Kaech S, White J, Zhang F, Schutz K et al. A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. J Neurosci 2007; 27: 355–365.
Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B et al. A genetic screen for candidate tumor suppressors identifies REST. Cell 2005; 121: 837–848.
Zhao JJ, Gjoerup OV, Subramanian RR, Cheng Y, Chen W, Roberts TM et al. Human mammary epithelial cell transformation through the activation of phosphatidylinositol 3-kinase. Cancer Cell 2003; 3: 483–495.
Chen K, Mi YJ, Ma Y, Fu HL, Jin WL . The mental retardation associated protein, srGAP3 negatively regulates VPA-induced neuronal differentiation of Neuro2A cells. Cell Mol Neurobiol 2011; 31: 675–686.
Radvanyi L, Singh-Sandhu D, Gallichan S, Lovitt C, Pedyczak A, Mallo G et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci USA 2005; 102: 11005–11010.
Zhao H, Langerod A, Ji Y, Nowels KW, Nesland JM, Tibshirani R et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell 2004; 15: 2523–2536.
Shuib S, McMullan D, Rattenberry E, Barber RM, Rahman F, Zatyka M et al. Microarray based analysis of 3p25-p26 deletions (3p- syndrome). Am J Med Genet A 2009; 149A: 2099–2105.
Kristensen LS, Nielsen HM, Hansen LL . Epigenetics and cancer treatment. Eur J Pharmacol 2009; 625: 131–142.
Tang YA, Wen WL, Chang JW, Wei TT, Tan YH, Salunke S et al. A novel histone deacetylase inhibitor exhibits antitumor activity via apoptosis induction, F-actin disruption and gene acetylation in lung cancer. PLoS One 2010; 5: e12417.
Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol 2007; 1: 84–96.
Han J, Chang H, Giricz O, Lee GY, Baehner FL, Gray JW et al. Molecular predictors of 3D morphogenesis by breast cancer cell lines in 3D culture. PLoS Comput Biol 2010; 6: e1000684.
Guo Y, Pakneshan P, Gladu J, Slack A, Szyf M, Rabbani SA . Regulation of DNA methylation in human breast cancer. Effect on the urokinase-type plasminogen activator gene production and tumor invasion. J Biol Chem 2002; 277: 41571–41579.
Zajchowski DA, Bartholdi MF, Gong Y, Webster L, Liu HL, Munishkin A et al. Identification of gene expression profiles that predict the aggressive behavior of breast cancer cells. Cancer Res 2001; 61: 5168–5178.
Ikebe M, Hartshorne DJ . Phosphorylation of smooth muscle myosin at two distinct sites by myosin light chain kinase. J Biol Chem 1985; 260: 10027–10031.
Lorentzen A, Bamber J, Sadok A, Elson-Schwab I, Marshall CJ . An ezrin-rich, rigid uropod-like structure directs movement of amoeboid blebbing cells. J Cell Sci 2011; 124 (Pt 8): 1256–1267.
Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell 2008; 135: 510–523.
Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M et al. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science 1996; 273: 245–248.
Matsui T, Maeda M, Doi Y, Yonemura S, Amano M, Kaibuchi K et al. Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J Cell Biol 1998; 140: 647–657.
Sosa MS, Lopez-Haber C, Yang C, Wang H, Lemmon MA, Busillo JM et al. Identification of the Rac-GEF P-Rex1 as an essential mediator of ErbB signaling in breast cancer. Mol Cell 2010; 40: 877–892.
Sander EE, ten Klooster JP, van Delft S, van der Kammen RA, Collard JG . Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J Cell Biol 1999; 147: 1009–1022.
Zondag GC, Evers EE, ten Klooster JP, Janssen L, van der Kammen RA, Collard JG . Oncogenic Ras downregulates Rac activity, which leads to increased Rho activity and epithelial-mesenchymal transition. J Cell Biol 2000; 149: 775–782.
Counter CM, Meyerson M, Eaton EN, Ellisen LW, Caddle SD, Haber DA et al. Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene 1998; 16: 1217–1222.
Soderling SH, Langeberg LK, Soderling JA, Davee SM, Simerly R, Raber J et al. Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc Natl Acad Sci USA 2003; 100: 1723–1728.
Acknowledgements
We thank William C Hahn, Dana-Farber Cancer Institute, for providing the cell line HMEC/hTERT/LT/c-myc and John D Scott, University of Washington, for the gift of pCDNA3.1-WRP-V5His and pCDNA3.1-WRPΔGAP-V5His plasmids. We also thank Memorial Sloan-Kettering Cancer Center Molecular Cytogenetics Core facility for performing the FISH assays, the Bioinformatic Core facility for the analysis of the microarray data and Overholtzer laboratories for critical reading of the manuscript and helpful discussions. This work was supported by Fundacion Caja Madrid, Spain (AL) and by an MSKCC Geoffrey Beene Cancer Research Center Grant (AH).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Lahoz, A., Hall, A. A tumor suppressor role for srGAP3 in mammary epithelial cells. Oncogene 32, 4854–4860 (2013). https://doi.org/10.1038/onc.2012.489
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2012.489
Keywords
This article is cited by
-
The oncogenic fusion landscape in pediatric CNS neoplasms
Acta Neuropathologica (2022)
-
The emerging roles of srGAPs in cancer
Molecular Biology Reports (2022)
-
A pilot radiogenomic study of DIPG reveals distinct subgroups with unique clinical trajectories and therapeutic targets
Acta Neuropathologica Communications (2021)
-
Estimating the predictive power of silent mutations on cancer classification and prognosis
npj Genomic Medicine (2021)
-
CRAF gene fusions in pediatric low-grade gliomas define a distinct drug response based on dimerization profiles
Oncogene (2017)