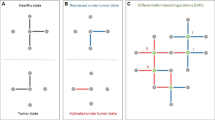
Abstract
Multiple clinical studies have correlated gene expression with survival outcome in cancer on a genome-wide scale. However, in many cases, no obvious correlation between expression of well-known tumour-related genes (that is, p53, p73 and p21) and survival rates of patients has been observed. This can be mainly explained by the complex molecular mechanisms involved in cancer, which mask the clinical relevance of a gene with multiple functions if only gene expression status is considered. As we demonstrate here, in many such cases, the expression of the gene interaction partners (gene ‘interactome’) correlates significantly with cancer survival and is indicative of the role of that gene in cancer. On the basis of this principle, we have implemented a free online datamining tool (http://www.bioprofiling.de/PPISURV). PPISURV automatically correlates expression of an input gene interactome with survival rates on >40 publicly available clinical expression data sets covering various tumours involving about 8000 patients in total. To derive the query gene interactome, PPISURV employs several public databases including protein–protein interactions, regulatory and signalling pathways and protein post-translational modifications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res 2009; 37: D885–D890.
Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA 2005; 102: 13550–13555.
Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010; 123: 725–731.
Gyorffy B, Lanczky A, Szallasi Z . Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer 2012; 19: 197–208.
Antonov AV, Knight RA, Melino G, Barlev NA, Tsvetkov PO . MIRUMIR: an online tool to test microRNAs as biomarkers to predict survival in cancer using multiple clinical data sets. Cell Death Differ 2013; 20: 367.
Na II, Rho JK, Choi YJ, Kim CH, Koh JS, Ryoo BY et al. Clinical features reflect exon sites of EGFR mutations in patients with resected non-small-cell lung cancer. J Korean Med Sci 2007; 22: 393–399.
Tandon S, Tudur-Smith C, Riley RD, Boyd MT, Jones TM . A systematic review of p53 as a prognostic factor of survival in squamous cell carcinoma of the four main anatomical subsites of the head and neck. Cancer Epidemiol Biomarkers Prev 2010; 19: 574–587.
Xia W, Chen JS, Zhou X, Sun PR, Lee DF, Liao Y et al. Phosphorylation/cytoplasmic localization of p21Cip1/WAF1 is associated with HER2/neu overexpression and provides a novel combination predictor for poor prognosis in breast cancer patients. Clin Cancer Res 2004; 10: 3815–3824.
Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res 2012; 40: D841–D846.
Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res 2011; 39: D691–D697.
Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res 2012; 40: D261–D270.
Dietmann S, Lee W, Wong P, Rodchenkov I, Antonov AV . CCancer: a bird’s eye view on gene lists reported in cancer-related studies. Nucleic Acids Res 2010; 38: W118–W123.
Antonov AV, Schmidt EE, Dietmann S, Krestyaninova M, Hermjakob H . R Spider: a network-based analysis of gene lists by combining signaling and metabolic pathways from reactome and KEGG databases. Nucleic Acids Res 2010; 38: W78–W83.
Antonov AV . BioProfiling.de: analytical web portal for high-throughput cell biology. Nucleic Acids Res 2011; 39: W323–W327.
Lee JT, Gu W . The multiple levels of regulation by p53 ubiquitination. Cell Death Differ 2010; 17: 86–92.
Tomasini R, Mak TW, Melino G . The impact of p53 and p73 on aneuploidy and cancer. Trends Cell Biol 2008; 18: 244–252.
Rufini A, Agostini M, Grespi F, Tomasini R, Sayan BS, Niklison-Chirou MV et al. p73 in Cancer. Genes Cancer 2011; 2: 491–502.
Levine AJ, Tomasini R, McKeon FD, Mak TW, Melino G . The p53 family: guardians of maternal reproduction. Nat Rev Mol Cell Biol 2011; 12: 259–265.
Suzuki A, Tsutomi Y, Akahane K, Araki T, Miura M . Resistance to Fas-mediated apoptosis: activation of caspase 3 is regulated by cell cycle regulator p21WAF1 and IAP gene family ILP. Oncogene 1998; 17: 931–939.
Gartel AL, Radhakrishnan SK . Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res 2005; 65: 3980–3985.
Callagy GM, Pharoah PD, Pinder SE, Hsu FD, Nielsen TO, Ragaz J et al. Bcl-2 is a prognostic marker in breast cancer independently of the Nottingham Prognostic Index. Clin Cancer Res 2006; 12: 2468–2475.
Castiglione F, Sarotto I, Fontana V, Destefanis M, Venturino A, Ferro S et al. Bcl2, p53 and clinical outcome in a series of 138 operable breast cancer patients. Anticancer Res 1999; 19: 4555–4563.
Dawson SJ, Makretsov N, Blows FM, Driver KE, Provenzano E, Le QJ et al. BCL2 in breast cancer: a favourable prognostic marker across molecular subtypes and independent of adjuvant therapy received. Br J Cancer 2010; 103: 668–675.
Ciardiello F, Tortora G . Inhibition of bcl-2 as cancer therapy. Ann Oncol 2002; 13: 501–502.
Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 2004; 32: D258–D261.
Wu H, Zeinab RA, Flores ER, Leng RP . Pirh2, a ubiquitin E3 ligase, inhibits p73 transcriptional activity by promoting its ubiquitination. Mol Cancer Res 2011; 9: 1780–1790.
Malatesta M, Peschiaroli A, Memmi EM, Zhang J, Antonov A, Green DR et al. The Cul4A–DDB1 E3 ubiquitin ligase complex represses p73 transcriptional activity. Oncogene 2013; 32: 4721–4726.
Hochberg Y, Benjamini Y . More powerful procedures for multiple significance testing. Stat Med 1990; 9: 811–818.
Harrington F . A class of rank test procedures for censored survival data. Biometrika 1982; 69: 553–566.
Acknowledgements
This work was supported by the UK Medical Research Council (MRC) and funding from Russian Federal grants 14.B37.21.1967 (to AA) 16.740.11.036 (to NB) and 11.G34.31.0069 (to GM).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflicts of interest.
Rights and permissions
About this article
Cite this article
Antonov, A., Krestyaninova, M., Knight, R. et al. PPISURV: a novel bioinformatics tool for uncovering the hidden role of specific genes in cancer survival outcome. Oncogene 33, 1621–1628 (2014). https://doi.org/10.1038/onc.2013.119
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2013.119
Keywords
This article is cited by
-
GNA13 regulates BCL2 expression and the sensitivity of GCB-DLBCL cells to BCL2 inhibitors in a palmitoylation-dependent manner
Cell Death & Disease (2021)
-
The RNA-binding protein HuR is a novel target of Pirh2 E3 ubiquitin ligase
Cell Death & Disease (2021)
-
New immunological potential markers for triple negative breast cancer: IL18R1, CD53, TRIM, Jaw1, LTB, PTPRCAP
Discover Oncology (2021)
-
The oncogene AAMDC links PI3K-AKT-mTOR signaling with metabolic reprograming in estrogen receptor-positive breast cancer
Nature Communications (2021)
-
SEMG1/2 augment energy metabolism of tumor cells
Cell Death & Disease (2020)