Abstract
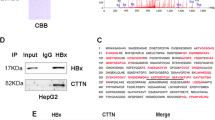
Chronic hepatitis B virus (HBV) infection is a major risk factor for developing hepatocellular carcinoma (HCC), and HBV X protein (HBx) acts as cofactor in hepatocarcinogenesis. In liver tumors from animals modeling HBx- and HBV-mediated hepatocarcinogenesis, downregulation of chromatin regulating proteins SUZ12 and ZNF198 induces expression of several genes, including epithelial cell adhesion molecule (EpCAM). EpCAM upregulation occurs in HBV-mediated HCCs and hepatic cancer stem cells, by a mechanism not understood. Herein we demonstrate HBx induces EpCAM expression via active DNA demethylation. In hepatocytes, EpCAM is silenced by polycomb repressive complex 2 (PRC2) and ZNF198/LSD1/Co-REST/HDAC1 chromatin-modifying complexes. Cells with stable knockdown of SUZ12, an essential PRC2 subunit, upon HBx expression demethylate a CpG dinucleotide located adjacent to NF-κB/RelA half-site. This NF-κB/RelA site is in a CpG island downstream from EpCAM transcriptional start site (TSS). Chromatin immunoprecipitation (ChIP) assays demonstrate HBx-dependent RelA occupancy of NF-κB half-site, whereas RelA knockdown suppresses CpG demethylation and EpCAM expression. Tumor necrosis factor-α activates RelA, propagating demethylation to nearby CpG sites, shown by sodium bisulfite sequencing. RelA-dependent demethylation occurring upon HBx expression requires methyltrasferase EZH2, TET2 a key factor in cytosine demethylation and inactive DNMT3L, shown by knockdown assays and sodium bisulfite sequencing. Co-immunoprecipitations and sequential ChIP assays demonstrate that RelA in the presence of HBx forms a complex with EZH2, TET2 and DNMT3L, although the role of DNMT3L remains to be understood. Interestingly, the human EpCAM gene also has a CpG island downstream from its TSS, and a NF-κB-binding site flanked by CpGs. HepG2 cells derived from human HCC exhibit demethylation of these NF-κB-flanking CpG sites, and HBV replication propagates demethylation to nearby CpG sites. DLK1, another PRC2 target gene, also upregulated in HBV-mediated HCCs, is demethylated in liver tumors at CpG dinucleotides flanking the NF-κB-binding sequence, supporting that this active DNA demethylation mechanism functions during oncogenic transformation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Accession codes
References
Beasley RP, Hwang LY, Lin CC, Chien CS . Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet 1981; 2: 1129–1133.
Andrisani OM, Barnabas S . The transcriptional function of the hepatitis B virus X protein and its role in hepatocarcinogenesis (review). Int J Oncol 1999; 15: 373–379.
Bouchard MJ, Schneider RJ . The enigmatic X gene of hepatitis B virus. J Virol 2004; 78: 12725–12734.
Zoulim F, Saputelli J, Seeger C . Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol 1994; 68: 2026–2030.
Madden CR, Finegold MJ, Slagle BL . Hepatitis B virus X protein acts as a tumor promoter in development of diethylnitrosamine-induced preneoplastic lesions. J Virol 2001; 75: 3851–3858.
Terradillos O, Billet O, Renard CA, Levy R, Molina T, Briand P et al. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene 1997; 14: 395–404.
Studach L, Wang WH, Weber G, Tang J, Hullinger RL, Malbrue R et al. Polo-like kinase 1 activated by the hepatitis B virus X protein attenuates both the DNA damage checkpoint and DNA repair resulting in partial polyploidy. J Biol Chem 2010; 285: 30282–30293.
Studach LL, Rakotomalala L, Wang WH, Hullinger RL, Cairo S, Buendia MA et al. Polo-like kinase 1 inhibition suppresses hepatitis B virus X protein-induced transformation in an in vitro model of liver cancer progression. Hepatology 2009; 50: 414–423.
Wang WH, Studach LL, Andrisani OM . Proteins ZNF198 and SUZ12 are down-regulated in hepatitis B virus (HBV) X protein-mediated hepatocyte transformation and in HBV replication. Hepatology 2011; 53: 1137–1147.
Margueron R, Reinberg D . The Polycomb complex PRC2 and its mark in life. Nature 2011; 469: 343–349.
Gocke CB, Yu H . ZNF198 stabilizes the LSD1-CoREST-HDAC1 complex on chromatin through its MYM-type zinc fingers. PloS One 2008; 3: e3255.
Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329: 689–693.
Studach LL, Menne S, Cairo S, Buendia MA, Hullinger RL, Lefrancois L et al. Subset of Suz12/PRC2 target genes is activated during hepatitis B virus replication and liver carcinogenesis associated with HBV X protein. Hepatology 2012; 56: 1240–1251.
de Boer CJ, van Krieken JH, Janssen-van Rhijn CM, Litvinov SV . Expression of Ep-CAM in normal, regenerating, metaplastic, and neoplastic liver. J Pathol 1999; 188: 201–206.
Schmelzer E, Zhang L, Bruce A, Wauthier E, Ludlow J, Yao HL et al. Human hepatic stem cells from fetal and postnatal donors. J Exp Med 2007; 204: 1973–1987.
Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology 2009; 136: 1012–1024.
Tarn C, Bilodeau ML, Hullinger RL, Andrisani OM . Differential immediate early gene expression in conditional hepatitis B virus pX-transforming versus nontransforming hepatocyte cell lines. J Biol Chem 1999; 274: 2327–2336.
Schnell U, Cirulli V, Giepmans BN . EpCAM: structure and function in health and disease. Biochimica et Biophysica Acta 2013; 1828: 1989–2001.
Went P, Vasei M, Bubendorf L, Terracciano L, Tornillo L, Riede U et al. Frequent high-level expression of the immunotherapeutic target Ep-CAM in colon, stomach, prostate and lung cancers. Br J Cancer 2006; 94: 128–135.
Hachmeister M, Bobowski KD, Hogl S, Dislich B, Fukumori A, Eggert C et al. Regulated intramembrane proteolysis and degradation of murine epithelial cell adhesion molecule mEpCAM. PloS One 2013; 8: e71836.
Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol 2009; 11: 162–171.
Chaves-Perez A, Mack B, Maetzel D, Kremling H, Eggert C, Harreus U et al. EpCAM regulates cell cycle progression via control of cyclin D1 expression. Oncogene 2013; 32: 641–650.
Munz M, Kieu C, Mack B, Schmitt B, Zeidler R, Gires O . The carcinoma-associated antigen EpCAM upregulates c-myc and induces cell proliferation. Oncogene 2004; 23: 5748–5758.
Munz M, Baeuerle PA, Gires O . The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res 2009; 69: 5627–5629.
van der Gun BT, de Groote ML, Kazemier HG, Arendzen AJ, Terpstra P, Ruiters MH et al. Transcription factors and molecular epigenetic marks underlying EpCAM overexpression in ovarian cancer. Br J Cancer 2011; 105: 312–319.
Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K . Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 2006; 20: 1123–1136.
Lee HJ, Hore TA, Reik W . Reprogramming the methylome: erasing memory and creating diversity. Cell Stem Cell 2014; 14: 710–719.
Denis H, Ndlovu MN, Fuks F . Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO Rep 2011; 12: 647–656.
Neri F, Krepelova A, Incarnato D, Maldotti M, Parlato C, Galvagni F et al. Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell 2013; 155: 121–134.
Delatte B, Deplus R, Fuks F . Playing TETris with DNA modifications. EMBO J 2014; 33: 1198–1211.
de la Rica L, Rodriguez-Ubreva J, Garcia M, Islam AB, Urquiza JM, Hernando H et al. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol 2013; 14: R99.
Fujiki K, Shinoda A, Kano F, Sato R, Shirahige K, Murata M . PPARgamma-induced PARylation promotes local DNA demethylation by production of 5-hydroxymethylcytosine. Nat Commun 2013; 4: 2262.
Ko M, An J, Bandukwala HS, Chavez L, Aijo T, Pastor WA et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature 2013; 497: 122–126.
Wang VY, Huang W, Asagiri M, Spann N, Hoffmann A, Glass C et al. The transcriptional specificity of NF-kappaB dimers is coded within the kappaB DNA response elements. Cell Rep 2012; 2: 824–839.
Chen YQ, Ghosh S, Ghosh G . A novel DNA recognition mode by the NF-kappa B p65 homodimer. Nat Struct Biol 1998; 5: 67–73.
Chen YQ, Sengchanthalangsy LL, Hackett A, Ghosh G . NF-kappaB p65 (RelA) homodimer uses distinct mechanisms to recognize DNA targets. Structure 2000; 8: 419–428.
Wu JC, Merlino G, Fausto N . Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc Natl Acad Sci USA 1994; 91: 674–678.
Meissner A . Guiding DNA methylation. Cell Stem Cell 2011; 9: 388–390.
Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008; 454: 766–770.
Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 2007; 39: 457–466.
Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006; 439: 871–874.
Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol Cell 2011; 43: 798–810.
Kriaucionis S, Heintz N . The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 2009; 324: 929–930.
Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009; 324: 930–935.
Cartron PF, Nadaradjane A, Lepape F, Lalier L, Gardie B, Vallette FM . Identification of TET1 partners that control Its DNA-demethylating function. Genes Cancer 2013; 4: 235–241.
Su F, Schneider RJ . Hepatitis B virus HBx protein activates transcription factor NF-kappaB by acting on multiple cytoplasmic inhibitors of rel-related proteins. J Virol 1996; 70: 4558–4566.
Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT et al. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother 1997; 41: 1715–1720.
Boyault S, Rickman DS, de Reynies A, Balabaud C, Rebouissou S, Jeannot E et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 2007; 45: 42–52.
Tanaka M, Okabe M, Suzuki K, Kamiya Y, Tsukahara Y, Saito S et al. Mouse hepatoblasts at distinct developmental stages are characterized by expression of EpCAM and DLK1: drastic change of EpCAM expression during liver development. Mech Dev 2009; 126: 665–676.
Zhang H, Diab A, Fan H, Kumar S, Mani K, Hullinger R et al. PLK1 and HOTAIR accelerate proteasomal degradation of SUZ12 and ZNF198 during hepatitis B virus-induced liver carcinogenesis. Cancer Res, in press.
Lu T, Yang M, Huang DB, Wei H, Ozer GH, Ghosh G et al. Role of lysine methylation of NF-kappaB in differential gene regulation. Proc Natl Acad Sci USA 2013; 110: 13510–13515.
Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 2013; 23: 839–852.
Baud V, Karin M . Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Disc 2009; 8: 33–40.
Nakamoto Y, Guidotti LG, Pasquetto V, Schreiber RD, Chisari FV . Differential target cell sensitivity to CTL-activated death pathways in hepatitis B virus transgenic mice. J Immunol 1997; 158: 5692–5697.
Ning BF, Ding J, Liu J, Yin C, Xu WP, Cong WM et al. Hepatocyte nuclear factor 4alpha-nuclear factor-kappaB feedback circuit modulates liver cancer progression. Hepatology 2014; 60: 1607–1619.
Sun L, Mathews LA, Cabarcas SM, Zhang X, Yang A, Zhang Y et al. Epigenetic regulation of SOX9 by the NF-kappaB signaling pathway in pancreatic cancer stem cells. Stem Cells 2013; 31: 1454–1466.
Zeng SS, Yamashita T, Kondo M, Nio K, Hayashi T, Hara Y et al. The transcription factor SALL4 regulates stemness of EpCAM-positive hepatocellular carcinoma. J Hepatol 2014; 60: 127–134.
Chang CJ, Hung MC . The role of EZH2 in tumour progression. Br J Cancer 2012; 106: 243–247.
Li LC, Dahiya R . MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002; 18: 1427–1431.
Livak KJ, Schmittgen TD . Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 2001; 25: 402–408.
R Core Team R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria 2014; http://www.R-project.org/.
Gautier L, Cope L, Bolstad BM, Irizarry R A . Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004; 20: 307–315.
Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M . Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 2007; 3: 11.
Acknowledgements
This work was supported by NIH grant DK044533 to OA. Shared Resources (flow cytometry and DNA sequencing) are supported by NIH grant P30CA023168 to Purdue Cancer Research Center. The authors thank Dr R Hullinger for critical review of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Fan, H., Zhang, H., Pascuzzi, P. et al. Hepatitis B virus X protein induces EpCAM expression via active DNA demethylation directed by RelA in complex with EZH2 and TET2. Oncogene 35, 715–726 (2016). https://doi.org/10.1038/onc.2015.122
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.122
This article is cited by
-
Epigenetic reprogramming of T cells: unlocking new avenues for cancer immunotherapy
Cancer and Metastasis Reviews (2024)
-
Tazemetostat decreases β-catenin and CD13 protein expression in HEPG-2 and Hepatitis B virus-transfected HEPG-2 with decreased cell viability
Clinical Epigenetics (2023)
-
Environment factors, DNA methylation, and cancer
Environmental Geochemistry and Health (2023)
-
Cancer stem cells in hepatocellular carcinoma — from origin to clinical implications
Nature Reviews Gastroenterology & Hepatology (2022)
-
Bioinformatics analysis on multiple Gene Expression Omnibus datasets of the hepatitis B virus infection and its response to the interferon-alpha therapy
BMC Infectious Diseases (2020)