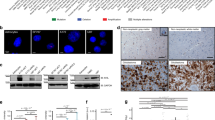
Abstract
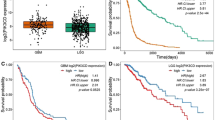
Glioblastoma is the most aggressive primary brain tumor in adults. Although the rapid recurrence of glioblastomas after treatment is a major clinical challenge, the relationships between tumor growth and intracerebral spread remain poorly understood. We have identified the cofilin phosphatase chronophin (gene name: pyridoxal phosphatase, PDXP) as a glial tumor modifier. Monoallelic PDXP loss was frequent in four independent human astrocytic tumor cohorts and increased with tumor grade. We found that aberrant PDXP promoter methylation can be a mechanism leading to further chronophin downregulation in glioblastomas, which correlated with shorter glioblastoma patient survival. Moreover, we observed an inverse association between chronophin protein expression and cofilin phosphorylation levels in glioma tissue samples. Chronophin-deficient glioblastoma cells showed elevated cofilin phosphorylation, an increase in polymerized actin, a higher directionality of cell migration, and elevated in vitro invasiveness. Tumor growth of chronophin-depleted glioblastoma cells xenografted into the immunodeficient mouse brain was strongly impaired. Our study suggests a mechanism whereby the genetic and epigenetic alterations of PDXP resulting in altered chronophin expression may regulate the interplay between glioma cell proliferation and invasion.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout












Similar content being viewed by others
Accession codes
References
Dolecek TA, Propp JM, Stroup NE, Kruchko C . CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005-2009. Neuro-oncology 2012; 14: v1–49.
Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK et al. Malignant glioma: genetics and biology of a grave matter. Genes Dev 2001; 15: 1311–1333.
Carlsson SK, Brothers SP, Wahlestedt C . Emerging treatment strategies for glioblastoma multiforme. EMBO Mol Med 2014; 6: 1359–1370.
Huse JT, Holland EC . Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer 2010; 10: 319–331.
Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR et al. The somatic genomic landscape of glioblastoma. Cell 2013; 155: 462–477.
Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 2012; 22: 425–437.
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010; 17: 98–110.
Sturm D, Bender S, Jones DT, Lichter P, Grill J, Becher O et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer 2014; 14: 92–107.
Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet 2013; 45: 1141–1149.
Diaz de Stahl T, Hartmann C, de Bustos C, Piotrowski A, Benetkiewicz M, Mantripragada KK et al. Chromosome 22 tiling-path array-CGH analysis identifies germ-line- and tumor-specific aberrations in patients with glioblastoma multiforme. Genes Chrom Cancer 2005; 44: 161–169.
Hartmann C, Numann A, Mueller W, Holtkamp N, Simon M, von Deimling A . Fine mapping of chromosome 22q tumor suppressor gene candidate regions in astrocytoma. Int J Cancer 2004; 108: 839–844.
Ino Y, Silver JS, Blazejewski L, Nishikawa R, Matsutani M, von Deimling A et al. Common regions of deletion on chromosome 22q12.3–q13.1 and 22q13.2 in human astrocytomas appear related to malignancy grade. J Neuropathol Exp Neurol 1999; 58: 881–885.
Muhammad AK, Yoshimine T, Maruno M, Tokiyoshi K, Takemoto O, Hayakawa T . Chromosome 22q allelic losses at microsatellite loci in human astrocytic tumors. Neurol Medico-Chirurg 1997; 37: 606–610; discussion 611.
Oskam NT, Bijleveld EH, Hulsebos TJ . A region of common deletion in 22q13.3 in human glioma associated with astrocytoma progression. Int J Cancer 2000; 85: 336–339.
Seng TJ, Ichimura K, Liu L, Tingby O, Pearson DM, Collins VP . Complex chromosome 22 rearrangements in astrocytic tumors identified using microsatellite and chromosome 22 tile path array analysis. Genes Chromosomes Cancer 2005; 43: 181–193.
Seifried A, Schultz J, Gohla A . Human HAD phosphatases: structure, mechanism, and roles in health and disease. FEBS J 2013; 280: 549–571.
Gohla A, Birkenfeld J, Bokoch GM . Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat Cell Biol 2005; 7: 21–29.
Bernstein BW, Bamburg JR . ADF/cofilin: a functional node in cell biology. Trends Cell Biol 2010; 20: 187–195.
Mizuno K . Signaling mechanisms and functional roles of cofilin phosphorylation and dephosphorylation. Cell Signal 2013; 25: 457–469.
Bugyi B, Carlier MF . Control of actin filament treadmilling in cell motility. Annu Rev Biophys 2010; 39: 449–470.
Hall A . The cytoskeleton and cancer. Cancer Metast Rev 2009; 28: 5–14.
Pollard TD, Borisy GG . Cellular motility driven by assembly and disassembly of actin filaments. Cell 2003; 112: 453–465.
Collazo J, Zhu B, Larkin S, Martin SK, Pu H, Horbinski C et al. Cofilin drives cell-invasive and metastatic responses toTGF-beta in prostate cancer. Cancer Res 2014; 74: 2362–2373.
Park JB, Agnihotri S, Golbourn B, Bertrand KC, Luck A, Sabha N et al. Transcriptional profiling of GBM invasion genes identifies effective inhibitors of the LIM kinase-Cofilin pathway. Oncotarget 2014; 5: 9382–9395.
Wang W, Mouneimne G, Sidani M, Wyckoff J, Chen X, Makris A et al. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J Cell Biol 2006; 173: 395–404.
Wang W, Eddy R, Condeelis J . The cofilin pathway in breast cancer invasion and metastasis. Nat Rev Cancer 2007; 7: 429–440.
Yoshioka K, Foletta V, Bernard O, Itoh K . A role for LIM kinase in cancer invasion. Proc Natl Acad Sci USA 2003; 100: 7247–7252.
Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008; 455: 1061–1068.
Di Tomaso E, Pang JC, Lam HK, Tian XX, Suen KW, Hui AB et al. Establishment and characterization of a human cell line from paediatric cerebellar glioblastoma multiforme. Neuropathol Appl Neurobiol 2000; 26: 22–30.
Kruse CA, Mitchell DH, Kleinschmidt-DeMasters BK, Franklin WA, Morse HG, Spector EB et al. Characterization of a continuous human glioma cell line DBTRG-05MG: growth kinetics, karyotype, receptor expression, and tumor suppressor gene analyses. In vitro cellular & developmental biology: journal of the Tissue Culture Association. In vitro Cell Dev Biol 1992; 28A: 609–614.
Clark MJ, Homer N, O'Connor BD, Chen Z, Eskin A, Lee H et al. U87MG decoded: the genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet 2010; 6: e1000832.
Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T . Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteom 2006; 5: 749–757.
Jain RK, di Tomaso E, Duda DG, Loeffler JS, Sorensen AG, Batchelor TT . Angiogenesis in brain tumours. Nat Rev Neurosci 2007; 8: 610–622.
Lu KV, Chang JP, Parachoniak CA, Pandika MM, Aghi MK, Meyronet D et al. VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell 2012; 22: 21–35.
Floyd D, Purow B . Micro-masters of glioblastoma biology and therapy: increasingly recognized roles for microRNAs. Neuro-oncology 2014; 16: 622–627.
Nagai S, Moreno O, Smith CA, Ivanchuk S, Romagnuolo R, Golbourn B et al. Role of the cofilin activity cycle in astrocytoma migration and invasion. Genes Cancer 2011; 2: 859–869.
Binda E, Visioli A, Giani F, Lamorte G, Copetti M, Pitter KL et al. The EphA2 receptor drives self-renewal and tumorigenicity in stem-like tumor-propagating cells from human glioblastomas. Cancer Cell 2012; 22: 765–780.
Horing E, Harter PN, Seznec J, Schittenhelm J, Buhring HJ, Bhattacharyya S et al. The 'go or grow' potential of gliomas is linked to the neuropeptide processing enzyme carboxypeptidase E and mediated by metabolic stress. Acta Neuropathol 2012; 124: 83–97.
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674.
Salhia B, Rutten F, Nakada M, Beaudry C, Berens M, Kwan A et al. Inhibition of Rho-kinase affects astrocytoma morphology, motility, and invasion through activation of Rac1. Cancer Res 2005; 65: 8792–8800.
Wiggan O, Shaw AE, DeLuca JG, Bamburg JR . ADF/cofilin regulates actomyosin assembly through competitive inhibition of myosin II binding to F-actin. Dev Cell 2012; 22: 530–543.
Ridley AJ . Life at the leading edge. Cell 2011; 145: 1012–1022.
Poincloux R, Collin O, Lizarraga F, Romao M, Debray M, Piel M et al. Contractility of the cell rear drives invasion of breast tumor cells in 3D Matrigel. Proc Natl Acad Sci USA 2011; 108: 1943–1948.
Bellail AC, Hunter SB, Brat DJ, Tan C, Van Meir EG . Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int J Biochem Cell Biol 2004; 36: 1046–1069.
Galluzzi L, Vitale I, Senovilla L, Olaussen KA, Pinna G, Eisenberg T et al. Prognostic impact of vitamin B6 metabolism in lung cancer. Cell Rep 2012; 2: 257–269.
Huang TY, Minamide LS, Bamburg JR, Bokoch GM . Chronophin mediates an ATP-sensing mechanism for cofilin dephosphorylation and neuronal cofilin-actin rod formation. Dev Cell 2008; 15: 691–703.
Bernstein BW, Chen H, Boyle JA, Bamburg JR . Formation of actin-ADF/cofilin rods transiently retards decline of mitochondrial potential and ATP in stressed neurons. Am J Physiol Cell Physiol 2006; 291: C828–C839.
Ren S, Ouyang DY, Saltis M, Xu LH, Zha QB, Cai JY et al. Anti-proliferative effect of 23,24-dihydrocucurbitacin F on human prostate cancer cells through induction of actin aggregation and cofilin-actin rod formation. Cancer Chemother Pharmacol 2012; 70: 415–424.
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007; 114: 97–109.
Chopra M, Kraus S, Schwinn S, Ritz M, Mattenheimer K, Mottok A et al. Non-invasive bioluminescence imaging to monitor the immunological control of a plasmablastic lymphoma-like B cell neoplasia after hematopoietic cell transplantation. PLoS One 2013; 8: e81320.
Seifried A, Knobloch G, Duraphe PS, Segerer G, Manhard J, Schindelin H et al. Evolutionary and structural analyses of mammalian haloacid dehalogenase-type phosphatases AUM and chronophin provide insight into the basis of their different substrate specificities. J Biol Chem 2014; 289: 3416–3431.
Sauzeau V, Berenjeno IM, Citterio C, Bustelo XR . A transcriptional cross-talk between RhoA and c-Myc inhibits the RhoA/Rock-dependent cytoskeleton. Oncogene 2010; 29: 3781–3792.
Acknowledgements
We thank Andrea Odersky and Beate Vogt for excellent technical assistance and Miriam Ritz, Carolin Graf, Thorsten Winter and Katja Ottmüller for help with bioluminescence imaging. We also thank Stephan Kissler for the gift of viral vectors, Christian Linden for FACS sorting and Michael Krause and Lukas Rycak, Marburg University, for microarray analysis. This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB728 to AG and GR; SFB688 and FZ82 to AG), the Forschungskommission of the Medical Faculty of Heinrich Heine University Düsseldorf (to AG) and by an IZKF Würzburg research grant (to AB). TGZ was supported by a predoctoral fellowship from the Medical Faculty of the University of Würzburg (Graduate School of Life Sciences) and by the Deutsche Forschungsgemeinschaft TR17. SK was supported by a fellowship of the Else Kröner-Fresenius-Stiftung, and CBKT is supported by the Max Eder Program of the German Cancer Aid (Deutsche Krebshilfe).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
GR has received grants from Roche and Merck, as well as honoraria for advisory boards or lectures from Amgen, Merck and Roche. The other authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Schulze, M., Fedorchenko, O., Zink, T. et al. Chronophin is a glial tumor modifier involved in the regulation of glioblastoma growth and invasiveness. Oncogene 35, 3163–3177 (2016). https://doi.org/10.1038/onc.2015.376
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.376