Abstract
Gliomas are highly malignant brain tumors that are highly invasive and resistant to conventional therapy. Receptor tyrosine kinases (RTKs) such as PDGFRα (platelet-derived growth factor receptor-α), which show frequent aberrant activation in gliomas, are associated with a process of epithelial–mesenchymal transition (EMT), a cellular alteration that confers a more invasive and drug-resistant phenotype. Although this phenomenon is well documented in human cancers, the processes by which RTKs including PDGFRα mediate EMT are largely unknown. Here, we report that SHP-2 (encoded by PTPN11) upregulates an EMT inducer, ZEB1, to mediate PDGFRα-driven glioma EMT, invasion and growth in glioma cell lines and patient-derived glioma stem cells (GSCs) using cell culture and orthotopic xenograft models. ZEB1 and activated PDGFRα were coexpressed in invasive regions of mouse glioma xenografts and clinical glioma specimens. Glioma patients with high levels of both phospho-PDGFRα (p-PDGFRα) and ZEB1 had significantly shorter overall survival compared with those with low expression of p-PDGFRα and ZEB1. Knockdown of ZEB1 inhibited PDGFA/PDGFRα-stimulated glioma EMT, tumor growth and invasion in glioma cell lines and patient-derived GSCs. PDGFRα mutant deficient of SHP2 binding (PDGFRα-F720) or phosphoinositide 3-kinase (PI3K) binding (PDGFRα-F731/42), knockdown of SHP2 or treatments of pharmacological inhibitor for PDGFRα-signaling effectors attenuated PDGFA/PDGFRα-stimulated ZEB1 expression, cell migration and GSC proliferation. Importantly, SHP-2 acts together with PI3K/AKT to regulate a ZEB1-miR-200 feedback loop in PDGFRα-driven gliomas. Taken together, our findings uncover a new pathway in which ZEB1 functions as a key regulator for PDGFRα-driven glioma EMT, invasiveness and growth, suggesting that ZEB1 is a promising therapeutic target for treating gliomas with high PDGFRα activation.
Similar content being viewed by others
Introduction
Aberrant activation of oncogenic signaling pathways facilitates brain tumor malignancy and treatment resistance.1, 2 PDGFRα (platelet-derived growth factor receptor-α), a receptor tyrosine kinase (RTK), is commonly overexpressed and amplified in glioblastoma (GBM).3 Expression of PDGFRα and its ligand, PDGFA, enhances GBM tumor growth and invasion in the brain.4, 5 Although it was reported that PDGFA/PDGFRα activates phosphoinositide 3-kinase (PI3K), SHP-2 and Src signaling to promote tumor proliferation and motility,4, 5 other downstream effectors that mediate tumor growth, invasiveness and treatment resistance are also likely involved.
Epithelial–mesenchymal transition (EMT), a cellular process typified by loss of polarized epithelial features toward a more motile mesenchymal (MES) phenotype, is frequently observed in malignant and invasive human cancers.6 ZEB1, a zinc-finger protein, is an inducer of EMT,7, 8 through downregulation of E-cadherin and upregulation of MES molecular markers such as vimentin.7, 8 Cancer cells undergoing EMT are thought to acquire stem cell traits and become more resistant to therapies.7, 9 ZEB1 was found preferentially expressed in epithelial cancers, and its expression is correlated to a shorter survival and poor therapy response.7, 9, 10 ZEB1 is an transcriptional repressor of cell-adhesion genes and several microRNAs, particularly members of miR-200 family (including miR-200a, miR200b, miR-200c, miR-141 and miR-429), which function not only as strong inducers of mesenchymal–epithelial transition (MET) but also inhibit undifferentiated stem cell properties.11, 12 Reciprocally, members of miR-200 family downregulate ZEB1 by targeting its 3′-untranslated region, thus establishing a double-negative feedback loop between ZEB1 and members of miR-200 family.13
Cell transformation promoted by PDGFRα and PDGFRβ signaling has been observed in multiple types of cancers.14, 15, 16 PDGFRα is preferentially amplified in the clinically relevant proneural (PN) GBM subtype.3 We previously reported that tumors derived from Ink4a/Arf−/− PDGFRα or PDGFA/PDGFRα mouse astrocytes are highly invasive and express higher levels of the neural progenitor marker nestin, suggesting that PDGFRα signaling contributes to a stem cell phenotype.4 However, the mechanism by which PDGFRα signaling causes GBM tumor transformation and invasion is still unclear. As ZEB1 is involved in glioma initiation and invasion,9 we hypothesize that EMT inducer ZEB1 has important roles in PDGFA/PDGFRα signaling-mediated glioma EMT and invasion. In this study, we determined the expression of ZEB1 and phospho-PDGFRα (p-PDGFRα) in mouse glioma brain xenografts and clinical GBM specimens. We then examined the roles of ZEB1 along with its downstream targets in PDGFA/PDGFRα-activated glioma EMT, invasion and glioma stem cell (GSC) growth. Finally, we used genetic and pharmacological approaches targeting individual downstream signaling effectors and determined which specific pathways emanating from PDGFRα are critical in ZEB1-induced glioma EMT.
Results
ZEB1 and PDGFRα are coexpressed in invasive areas of glioblastoma in mice and humans
We first examined ZEB1 mRNA expression in clinical GBM using The Cancer Genome Atlas dataset.17 We found that the levels of ZEB1 expression are significantly higher in classical and PN subtypes of GBM compared with MES and neural subtypes (Figure 1A). Moreover, compared with the MES GSCs and glioma cells, the levels of ZEB1 mRNA expression were the highest in PN GSCs (Figure 1B).18 This result suggests that ZEB1 may be involved in PDGFRα signaling that is aberrantly activated in the PN GBM subtype.17
ZEB1 and PDGFRα are coexpressed in invasive areas of glioblastomas in mice and humans. (A) Expression levels of ZEB1 mRNA are significantly higher in classical (CL) and PN GBM subtypes compared with MES and neural (NL) GBM. Expression data of ZEB1 mRNA in the four GBM subtypes were downloaded from The Cancer Genome Atlas (TCGA) dataset17 and analyzed. (B) Analysis of ZEB1 mRNA expression in PN GSCs, MES GSCs and glioma cells. ZEB1 mRNA expression level in various cells was determined with gene expression profiling as described previously.18 (C) ZEB1 and PDGFRα are coexpressed in invasive areas of PDGFRα-driven glioma brain tumor xenografts in mice. (a and d) Representative hematoxylin and eosin (H&E) staining images of LN444/PDGFA brain tumor sections. Brains were harvested at 6–7 weeks post-transplantation. (b and e) Representative images of GBM sections that were stained for ZEB1protein. (c and f) Representative images of sister sections of panels a and e that were stained for p-PDGFRα protein. (d–f) Enlarged areas of square marks in (a–c). Insets show isotype-matched immunoglobulin G (IgG) controls of the identical areas (original magnification, x400). Arrows show positive staining. Scale bars: 100 μm. Data were from two independent experiments with at least six mice per group with similar results. (D) Representative IHC images of ZEB1 and p-PDGFRα within invasive areas of sister sections of two representative clinical GBM tumor specimens, RJ18 and RJ13. Scale bars: 50 μm. Insets show isotype-matched IgG controls of the identical areas (original magnification, x400). Arrows show positive staining. Data of IHC staining of individual GBM tumor specimens are shown in Supplementary Table S1. (E) Kaplan–Meier analysis of patients with high p-PDGFRα and high ZEB1-expressing glioma tumors versus low p-PDGFRα and low ZEB1-expressing tumors in IHC staining (D) assays. Median survival (in months): low, 16.87; high, 10.43. P-values were calculated by log-rank test. Black bars, censored data. Data in (C and D) represent two independent experiments with similar results.
Next, we determined the roles of ZEB1 in glioma tumorigenesis using LN444 cells with a stably expressed PDGFA (LN444/PDGFA) or a control (LN444/GFP). We observed that PDGFA overexpression promoted tumor growth and invasion (Figures 1Ca and d and Supplementary Figures S1Ab and d) with a shorter survival (Supplementary Figure S1B) compared with the control xenograft models (Supplementary Figures S1Aa and c) as reported previously.4, 5, 19 ZEB1 was detected at the invasive areas in xenograft glioma tumors, associating its role in regulating glioma cell invasion.9 We determined whether ZEB1 is coexpressed with PDGFRα in the invasive front of LN444/PDGFA glioma brain xenografts. As shown in Figure 1C, ZEB1 (panels b and e) and p-PDGFRα (panels c and f) were found coexpressed at high levels in the invasive areas of PDGFA/PDGFRα-driven LN444 orthotopic xenograft tumors. These data suggest that ZEB1 may be dependent on PDGFA/PDGFRα signaling to promote glioma tumor invasion.
We then sought clinical evidence for a link between ZEB1 and PDGFA/PDGFRα signaling. We analyzed a total of 86 clinical GBM tumor samples using anti-p-PDGFRα and anti-ZEB1 antibodies. High levels of p-PDGFRα expression were detected in 21 of 86 GBM specimens (24.4%), corresponding to the frequencies of PDGFRα activation in clinical gliomas,17 and high levels of ZEB1 expression were detected in 33 GBMs (38.4%) (Figure 1D and Supplementary Table S1). Nineteen GBM samples (22.1%) were found to coexpress high levels of ZEB1 and p-PDGFRα, but only two patient samples with high p-PDGFRα contained low ZEB1 expression (Supplementary Table S1). We used a two-tailed Fisher’s exact test to show the frequency distribution pattern of p-PDGFRα and ZEB1 expression in patient samples is unlikely the results of chance (Supplementary Table S2). Fourteen GBM samples with high ZEB1 levels showed weak or no detectable p-PDGFRα signal (Supplementary Figure S2 and Supplementary Table S1). p-PDGFRα was detected in invasive and center regions in the GBM tumors (Figures 1Da and b and Supplementary Figures S3a and b). ZEB1 was expressed in the majority of p-PDGFRα-positive tumor cells in invasive areas of clinical GBM specimens (Figures 1Dc and d), but the tumor core tended to contain little to no detectable ZEB1 expression (Supplementary Figures S3c and d).
We examined the relationship between p-PDGFRα and ZEB1 expression on survival using Kaplan–Meier survival probability estimates. When combining the expression status of p-PDGFRα and ZEB1 in the analyses, a statistically significant worse prognosis was apparent in GBM with high expression of both proteins compared with those with low p-PDGFRα and low ZEB1 expression (Figure 1E), with median patient survival times of 10.43 and 16.87 months, respectively (P=0.0197 with a hazard ratio of 2.193, 95% confidence interval 1.133–4.242; Supplementary Table S2). Survival analysis based on the expression of ZEB1 alone showed a nonsignificant trend toward a modest decrease in median survival for patients with high ZEB1-expressing tumors compared with ZEB1 low tumors (15.53 and 17.97 months, respectively, P=0.0608; Supplementary Figure S4A). Evaluating patient survival based on p-PDGFRα staining alone (Supplementary Figure S4B) reveals a statistically significant increase in survival for patients with low p-PDGFRα tumors, with a median survival of 19.97 months, compared with 12.97 months for patients with high p-PDGFRα tumors (P=0.0492). However, when median survival is calculated for patients with tumors expressing high ZEB1 and low p-PDGFRα, the median survival increases to 26.8 months (Supplementary Table S2), justifying the use of these markers together and supporting our results showing a novel mechanism by which PDGFRα regulates tumor growth and invasiveness through ZEB1.
Taken together, these data demonstrate that ZEB1 is coexpressed with activated PDGFRα in invasive regions of mouse glioma xenograft tumors and clinical GBM specimens. These results also suggest that PDGFRα-driven glioma tumorigenesis and invasion involves ZEB1 and that the utilization of p-PDGFRα and ZEB1 serves as a prognostic marker for glioma patients.
PDGFA-promoted ZEB1 regulates glioma EMT, migration and colony formation
To determine the relationship between ZEB1 and PDGFRα signaling, we examined the expression of ZEB1 after PDGFA treatment of glioma cells. PDGFA stimulation significantly enhanced the expression of ZEB1 and vimentin (a mesenchymal protein; Figure 2a) and a fibroblast-like morphology (Figure 2d and data not shown). Moreover, decreased levels of E-cadherin (an epithelial marker) were found in glioma LN18 and LN444 cells with high endogenous levels of PDGFRα and p-PDGFRα, but not in glioma T98G and LN235 cells that lacked detection of endogenous PDGFRα (Figure 2a). PDGFA stimulation also markedly upregulated ZEB1 mRNA in glioma LN18 and LN444 cells but not in T98G and LN235 cells (Figure 2b). Additionally, expression of ZEB2, Snai1, Snail2 and Twist1 were not affected in these treated glioma cells (Supplementary Figure S5). These data suggest that PDGFA/PDGFRα signaling specifically regulates ZEB1 transcription in glioma cells.
PDGFA promotes ZEB1 expression and glioma EMT, proliferation, migration and colony formation. (a) Western blotting analyses. Compared with the control (vehicle, phosphate-buffered saline (PBS)), PDGFA stimulation upregulated ZEB1, vimentin and inhibited E-cadherin in LN18 and LN444 cell lines that have high levels of endogenous PDGFRα. In contrast, PDGFA had no effects on T98G and LN235 cells that had non-detectable PDGFRα protein. After starvation, indicated glioma cells were cultured in Dulbecco's modified Eagle's medium (DMEM) plus 0.5% fetal bovine serum (FBS) with or without 50 ng/ml PDGFA for 2 days. β-Actin was used as a control. (b) Quantitative reverse transcription–PCR (QRT–PCR) assays of PDGFA-stimulated ZEB1 mRNA expression in indicated cell lines from (a). ACTB (encoding β-actin) was used as a control. (c) Effect of ZEB1 knockdown with two different shRNAs (shZEB1-1 and shZEB1-2) or control shRNA (shC) on expression of vimentin and E-cadherin in indicated glioma cell lines. (d) Representative images of cell phenotypes of PDGFA stimulation and/or ZEB1 knockdown. After starvation, LN18 cells were cultured in DMEM plus 0.5% FBS with or without 50 ng/ml PDGFA for 7 days. Medium was changed every 2 days. Scale bars: 200 μm. (e) Effect of overexpression of PDGFA on expression of ZEB1, vimentin and E-cadherin in glioma cells. LN18 and LN444 glioma cells stably expressed exogenous PDGFA (LN18/PDGFA and LN444/PDGFA) or GFP (LN18/P and LN444/P). Western blot (WB) or enzyme-linked immunosorbent assay (ELISA) did not detect endogenous PDGFA in GFP controls of both cell lines. β-actin was used as a control. (f) Cell proliferation assays. Various cells were cultured in DMEM medium with 10% FBS for 3 days, and then cell numbers were analyzed. (g) In vitro cell migration assays. Various cells were serum starved for 16 h, and then placed on upper wells of a Boyden chamber. After 12–16 h, migrated cells were counted. (h) Soft agar colony assays. Scale bars: 100 μm. Data in (a–h) represent two or three independent experiments with similar results. Error bars, s.d. *P<0.05.
To further validate that ZEB1 is necessary for PDGFA/PDGFRα signaling-driven glioma EMT, we used short hairpin RNAs (shRNAs) to deplete ZEB1 and assessed the effect of ZEB1 inhibition on PDGFRα-stimulated glioma EMT. Knockdown of endogenous ZEB1 in LN18 and LN444 cells markedly impaired basal and PDGFA-stimulated vimentin expression and rescued E-cadherin expression (Figure 2c) and PDGFA/PDGFRα-driven EMT (Figure 2d). We previously reported that in LN444/PDGFA and LN18/PDGFA cells, autocrine PDGFA activates endogenous PDGFRα signaling and enhanced glioma cell tumorigenesis in mouse orthotopic xenografts.4, 5 ZEB1 knockdown inhibited basal and PDGFA-induced expression of both ZEB1 and vimentin, while increasing expression of E-cadherin (Figure 2e). Moreover, ZEB1 depletion attenuated PDGFA-promoted cell proliferation (Figure 2f), migration (Figure 2g) and colony formation (Figure 2h) of LN18 and LN444 cells in vitro. These data suggest that ZEB1 is involved in PDGFRα-driven glioma EMT, cell proliferation, migration and colony formation.
ZEB1 is important for PDGFRα-driven glioma tumor growth, invasion and GSC renewal
To determine the roles of ZEB1 in PDGFRα-driven glioma tumorigenesis and invasion in vivo, we separately implanted LN444/PDGFA/shC, LN444/PDGFA/shZEB1-1 and LN444/PDGFA/shZEB1-2 cells into the brains of mice. Compared with control LN444/PDGFA/shC, knockdown of ZEB1 markedly suppressed PDGFRα-stimulated glioma tumor growth (Figures 3A and B) and invasion (Figures 3A and C) in mouse brains tumors and resulted in an increase in survival (Figure 3D).
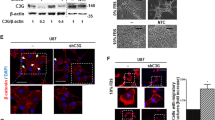
ZEB1 is important for PDGFRα-driven glioma tumor growth, invasion, survival and GSC self-renewal. (A) Representative hematoxylin and eosin (H&E) staining images of various brain sections. Brains were harvested at 6–7 weeks after transplantation. (a and d) LN444/PDGFA/shC tumors with a control shRNA (shC) from mice that developed neuropathological symptoms. (b, c, e and f) LN444/PDGFA/shZEB1 tumors with shZEB1-1 or shZEB1-2 from mice without noticeable neuropathological symptoms. (a–c) H&E staining. (d and f) Enlarged areas in panels a–c marked with squares. Scale bars: 1 mm in panels a–c; 200 μm in panels d–f. Data were from two independent experiments with at least five mice per group with similar results. (B) Quantification of tumor size in (a). Error bars, s.d. **P<0.01. (C) Quantification of tumors showing invasive fingers in (a). Error bars, s.d. **P<0.01. (D) Kaplan–Meier survival curves of mice with LN444/PDGFA/shC, LN444/PDGFA/shZEB1-1 or LN444/PDGFA/shZEB1-2 tumors. (E) Effect of knockdown of ZEB1 on expression of vimentin, E-cadherin, p-PDGFRα, PDGFRα in GSCs, AC17 and 157. (F) Representative images of effect of ZEB1 knockdown on the size of GSC neurospheres. Scale bars: 20 μm. (G) Quantification of relative neurosphere sizes of indicated GSC cells in (F). Error bars, s.d. *P<0.05. (H) Quantification of numbers of indicated cells in (F). Error bars, s.d. *P<0.05. (I) Limiting dilution neurosphere-forming assays of effect of ZEB1 knockdown on AC17 and 157 GSCs. Error bars, s.d. *P<0.05. Data in (D–I) represent two to three independent experiments with similar results.
Recently, we and other investigators have identified and characterized at least two distinct subtypes of patient-derived GSCs: PN and MES GSCs.18, 20 Molecular expression profiling of these GSCs correlated with the corresponding PN and MES subtypes of clinical high-grade gliomas.17, 21 PN GSCs are associated with unique genetic alterations such as PDGFRA amplification.18, 20 To determine the role of ZEB1 in glioma EMT, we tested the expression of ZEB1, vimentin, E-cadherin, PDGFRα and p-PDGFRα in PN GSCs, AC17 and 157.18 As shown in Figure 3E, we found that endogenous PDGFRα is highly expressed and activated in patient-derived AC17 and 157 GSC lines. Moreover, ZEB1 and vimentin expression were high and positively correlated with activated PDGFRα, whereas E-cadherin displayed an inverse correlation (Figure 3E). Depletion of ZEB1 (shZEB1) in two GSC lines resulted in marked reductions in vimentin expression (Figure 3E), neurosphere size (Figures 3F and G), GSC proliferation (Figure 3H) and neurosphere formation (Figure 3I). These results support that ZEB1 is critical for PDGFA/PDGFRα-stimulated glioma tumor growth, invasion in the brain and GSC renewal in vitro.
SHP-2 and PI3K signaling are required for PDGFRα-promoted ZEB1 expression
Using genetic and biochemical methods, we and other investigators have described the roles of signaling molecules in PDGFRα-mediated cellular functions by specific tyrosine-to-phenylalanine (Y-to-F) mutations (Figure 4a).4, 22, 23, 24 To determine the impact of downstream effectors of PDGFRα signaling on ZEB1 expression, we separately expressed PDGFRα wild-type (WT) or mutants in Ink4a/Arf−/− mouse astrocytes as described previously.4 The R627 mutant (PDGFRα-R627) with a lysine-to-arginine (K-to-R) mutation was used as a ‘receptor kinase-dead’ control. The F7 mutant (PDGFRα-F7) harbors seven Y-to-F mutations including Y572/74F, Y720F, Y731/42F, Y988F and Y1018F.22 As shown in Figures 4b and c, stimulation of WT PDGFRα by PDGFA resulted in high phosphorylation of the receptor and promoted cell migration compared with the control. In contrast, PDGFA stimulation of cells expressing PDGFRα-R627 or PDGFRα-F7 mutant did not yield appreciable effects of protein phosphorylation of PDGFRα and cell migration. Consistent with this, PDGFA stimulation induced ZEB1 expression in cells with WT PDGFRα, but not cells expressing R627 or F7 PDGFRα mutations. These data further support that stimulation of PDGFRα signaling upregulates ZEB1 expression.
SHP-2 and PI3K signaling are required for PDGFRα-promoted ZEB1 expression. (a) Schematics of various PDGFRα mutants. (b) Western blotting analyses of ZEB1 expression in PDGFA-stimulated Ink4a/Arf −/− mouse astrocytes with overexpressing PDGFRα wild-type (WT), kinase-dead mutant (R627), seven-point mutant (F7) or a vector control (control). (c) Cell migration assays from (b). Error bars, s.d. *P<0.05. (d) Effect of individual PDGFRα mutations on ZEB1 expression. (e) Cell migration assays from (d). Error bars, s.d. *P<0.05. (f) Effect of PI3K inhibitor LY294002 (10 μm) and SHP-2 inhibitor PHPS-1 (100 μm) on expression of ZEB1, vimentin, E-cadherin, p-Akt and p-Erk1/2 in LN444/PDGFA cells. (g) Cell migration assays from (f). Error bars, s.d. *P<0.05. Results in (b–g) represent two to three independent experiments with similar results.
Next, we determined which downstream effector of PDGFRα signaling promotes ZEB1 expression in Ink4a/Arf −/− mouse astrocytes with stable expression of WT or various PDGFRα mutants. As shown in Figures 4d and e, compared with WT PDGFRα, PDGFRα Y-to-F mutations at Y720 (PDGFRα-F720, for SHP-2 binding) and Y731/42 (PDGFRα-F731/42, for PI3K binding) inhibited ZEB1 expression, whereas PDGFRα Y-to-F mutations at Y572/74 (PDGFRα-F572/74, for Src binding), Y988 (PDGFRα-F988) and Y1018 (PDGFRα-F1018, for PLCγ binding) did not affect ZEB1 expression (Figure 4d). Corresponding to the regulation of ZEB1 expression, cell migration was impaired by F720 and F731/42 PDGFRα mutants compared with WT PDGFRα, whereas other PDGFRα mutants did not exert any effect (Figure 4e). These results show that PDGFRα signaling downstream effectors, PI3K and SHP-2 induce ZEB1 expression and cell migration in glioma cells.
To further demonstrate that PDGFRα-PI3K/SHP2 signaling regulates ZEB1 expression and glioma cell migration, we used PI3K inhibitor LY294002 and SHP-2 inhibitor PHPS-1 to treat LN444/PDGFA glioma cells as reported previously.4 As shown in Figures 4f and g, compared with the control, treatment with 10 μm LY294002 or 100 μm PHPS-1 blocked PDGFA-enhanced expression of ZEB1, vimentin, p-Akt and cell migration, while increasing E-cadherin expression (Figures 4f and g). PHPS-1 inhibited p-Erk1/2, whereas LY294002 had no effect (Figure 4f). Taken together, these data demonstrate that PDGFA/PDGFRα-regulated ZEB1 expression requires SHP-2 and PI3K signaling in mouse astrocytes and human glioma cells.
SHP-2 signaling is required for ZEB1-miR-200 feedback loop-mediated glioma cell migration and GSC proliferation
Emerging evidence suggests that ZEB1 and miR-200 family members form a feedback loop to regulate cancer cell EMT, cell migration and tumorigenesis.11, 13, 25 Moreover, PI3K-Akt signaling directly or indirectly regulates this ZEB1-miR-200 feedback loop in gastric cancer, bladder cancer and ovarian cancer.26, 27, 28 We hypothesize that SHP-2 also regulates ZEB1-miR-200 feedback loop and mediates PDGFRα-driven glioma EMT and cell migration. As shown in Figure 5a, knockdown of SHP-2 using small interfering RNAs (siRNAs) in LN18 and LN444 glioma cells prevented PDGFA-stimulated phosphorylation of Akt and Erk1/2. SHP-2 knockdown also ablated PDGFA-stimulated ZEB1 and vimentin expression, and rescued PDGFA-inhibited E-cadherin expression. Next, we determined the effect of SHP-2 depletion on expression of miR-200a, a member of miR-200 family that is known to be regulated by ZEB1.29 We found that miR-200a expression was inhibited by PDGFA stimulation in both glioma cell lines compared with the control (Figure 5b). Interestingly, knockdown of SHP-2 markedly increased miR-200a expression with or without PDGFA stimulation compared with the control (Figure 5b). Consistent with our previous work,19 knockdown of SHP-2 inhibited basal and PDGFA-stimulated cell migration (Figure 5c). Similarly, in patient-derived GSCs, depletion of SHP-2 attenuated Akt and Erk1/2 activity, as well as the expression of ZEB1 and vimentin, but promoted E-cadherin expression (Figure 5d). Reciprocally, knockdown of SHP-2 in GSCs increased miR-200a expression, but significantly diminished GSC proliferation (Figures 5e and f). We also found that treatment with MEK inhibitor PD98059 significantly blocked PDGFA-stimulated ZEB1 upregulation (Figure 5g) and cell migration (Figure 5i) without affecting Akt activity (Figure 5g) in LN444 and LN18 cells. Additionally, treatment with PD98059 attenuated PDGFA/PDGFRα inhibition of miR-200a expression (Figure 5h). Moreover, overexpression of miR-200a decreased the expression of ZEB1 and vimentin (Figure 5j) and cell migration (Figure 5k), but increased E-cadherin expression (Figure 5j) in LN18/PDGFA and LN444/PDGFA cells. Overexpression of miR-200a also inhibited PDGFA/PDGFRα-driven EMT in LN18 cells (Figure 5l). Taken together, these findings suggest that glioma cell migration and GSC renewal require SHP-2 activation of the ZEB1-miR-200 feedback loop.
SHP-2 regulates ZEB1-miR-200 feedback loop-mediated glioma cell migration and GSC proliferation. (a) Knockdown of SHP-2 by siRNAs inhibited PDGFA/PDGFRα-promoted Akt and Erk1/2 activity, ZEB1 and vimentin expression but increased E-cadherin expression in LN18 and LN444 cells. (b) Quantitative RT–PCR (QRT–PCR) assays of effect of SHP-2 knockdown on miR-200a expression. U6-snRNA was used as a control. (c) Cell migration assays from (a). Bars, s.d. *P<0.05. (d) Effect of SHP-2 depletion on the expression of ZEB1, vimentin, E-cadherin, p-Akt and p-Erk1/2 in GSC AC17 cell line. (e) QRT–PCR assays of miR-200a expression. U6-snRNA was used as a control. (f) GSC proliferation assays. (g–i). Effects of treatment of MEK inhibitor PD98059 (10 μm) on p-Erk1/2, ZEB1 expression, p-Akt (g), miR-200a expression (h) and cell migration (i) in LN444 and LN18 cells with or without PDGFA stimulation. (j and k) Effects of miR-200a overexpression on the expression of ZEB1, vimentin, E-cadherin (j) and cell migration (k). (l) Representative images of cell phenotypes of effect of miR-200a overexpression on PDGFA/PDGFRα-stimulated LN18 cells. Scale bars: 200 μm. Data in (a–l) represent two independent experiments with similar results. Error bars, s.d. *P<0.05.
SHP-2 acts together with PI3K-Akt to regulate ZEB1-miR-200 feedback loop
SHP-2 has been identified as a critical modulator that connects several signaling pathways including PI3K-Akt in glioma tumorigenesis.4, 30 Previous studies reported that PI3K/AKT activation has important roles in contributing to cancer cell EMT and cancer stem cell properties.31, 32 We then determined the relationship of SHP-2 and PI3K-Akt in ZEB1-miR-200-mediated glioma cell migration by overexpressing a constitutively activated (CA) Akt (Myr-Akt) mutant in Ink4a/Arf −/− mouse astrocytes with stable expression of WT PDGFRα or F720 PDGFRα mutant. As shown in Figure 6a, compared with the control, overexpression of the CA Akt mutant in WT PDGFRα cells promoted ZEB1 expression (Figure 6a) and cell migration (Figure 6c). Interestingly, overexpression of the CA Akt mutant partially rescued PDGFRα F720 inhibition of ZEB1 expression (Figure 6a) and cell migration (Figure 6b) compared with WT PDGFRα. In addition, overexpression of the CA Akt mutant-inhibited miR-200a expression in both WT PDGFRα and F720 PDGFRα mutant cells (Figure 6c). These results suggest that SHP-2 synergizes with PI3K-Akt to mediate ZEB1-miR-200 feedback loop in PDGFRα-stimulated glioma cell migration.
SHP-2 acts together with PI3K-Akt to regulate ZEB1-miR-200 feedback loop. (a) Overexpression of constitutively active AKT (Myr-AKT, CA Akt) partially restored F720 (SHP-2 binding site mutation) PDGFRα mutant-inhibited ZEB1 expression. (b) Cell migration assays from (a). (c) Quantitative RT–PCR (QRT–PCR) assays of the effect of Myr-AKT overexpression on miR-200a expression from (a). (d) Knockdown of AKT1 and AKT2 with siRNAs (siAKT1/2) moderately inhibited Y720 PDGFRα mutant-promoted ZEB1 expression compared with F7 PDGFRα mutant. (e,f) Cell migration assays (e) and QRT–PCR assays of miR-200a expression (f) of glioma cells from (d). (g) Knockdown of AKT1 and AKT2 by siRNAs inhibited PDGFA/PDGFRα-stimulated ZEB1 expression in LN444 cells. (h and i). QRT–PCR assays (h) and cell migration assays (i) from (g). Data in (a–i) represent two to three independent experiments with similar results. Error bars, s.d. *P<0.05.
To further support these observations, we knocked down Akt1 and Akt2 using siRNAs for Akt1 and Akt233 in Ink4a/Arf−/− mouse astrocytes with stable expression of F7 PDGFRα or Y720 PDGFRα mutant (Figure 4a). As shown in Figure 6d, re-expression of Y720 (SHP-2 binding site) in F7 PDGFRα mutant-expressing cells restored Akt activity and promoted ZEB1 expression in comparison with the F7 PDGFRα mutant. Depletion of Akt1 and Akt2 with siRNA (siAkt1/2) attenuated Y720 PDGFRα-restored ZEB1 expression (Figure 6d) and cell migration (Figure 6e). Consistent with this, re-expression of the Y720 PDGFRα mutant-inhibited miR-200a expression compared with F7 PDGFRα mutant, whereas knockdown of Akt1 and Akt2 marginally rescued SHP-2 inhibition of miR-200a in the Y720 cells (Figure 6f). Lastly, depletion of Akt1 and Akt2 with siRNA (siAkt1/2) in LN444/PDGFA cells inhibited ZEB1 expression, while promoting the expression of miR-200a and cell migration (Figure 6g and i). Moreover, knockdown of ZEB1 promoted miR-200a expression, reduced cell migration and marginally affected Akt activity. Taken together, these results demonstrate that SHP-2/PI3K-Akt and SHP-2-Erk1/2 collectively regulates ZEB1-miR-200 feedback loop in PDGFRα-stimulated glioma EMT, cell migration, tumor growth and GSC self-renewal.
Discussion
In this study, our findings suggest that SHP-2 regulation of ZEB1-miR-200 feedback loop is critical for PDGFA/PDGFRα signaling-driven glioma EMT, cell migration, tumor invasion and GSC growth in mice and humans (Figure 7). PDGFA/PDGFRα regulates the ZEB1-miR-200 feedback loop through SHP-2/PI3K-Akt and SHP-2-Erk1/2 in glioma cells, leading to enhanced glioma tumorigenesis, and invasion and GSC self-renewal.
One of the important aspects of this work is that PDGFA/PDGFRα signaling upregulates ZEB1 and induces glioma EMT, migration and GSC self-renewal. The EMT process is vital for morphogenesis during embryonic development and tissue remodeling as well as tumor growth, invasion and metastasis.34, 35 The processes of EMT could be triggered by a number of cell signaling pathways, such as EGF (epidermal growth factor), TGF-β (tumor frowth factor-β), HGF (hepatocyte growth factor) and PDGF ligands.16, 34, 36, 37 These growth factors act through their RTKs to activate a series of downstream targets. ZEB1 is a known driver of EMT, and targeting ZEB1 represents a potential therapeutic strategy for metastasis and invasion suppression in various cancers.9, 38, 39 In the present study, we described a new mechanism by which ZEB1 mediates PDGFA/PDGFRα-stimulated glioma tumorigenesis. In glioma cells, we examined PDGFA stimulation, or activation of PDGFRα by exogenously expressed PDGFA, led to specific upregulation of ZEB1 and acquisition of EMT-like phenotype. This process was consistent with increased cell growth, cell migration and GSC self-renewal in vitro, and glioma tumor growth and invasion in vivo. Moreover, knockdown of ZEB1 inhibited GSC growth. Taken together, this study identifies a previously unrecognized mechanism, in which PDGFA/PDGFRα signaling induces EMT through stimulation of ZEB1, thereby promoting glioma tumor growth and invasion and GSC stemness.
The second important finding in this study is that our data reveal that SHP-2 mediates ZEB1-miR-200 feedback loop in glioma EMT. SHP-2 is a critical mediator of oncogenic Ras/MAPK signaling and is activated through multiple mechanisms in various types of cancers.40, 41 We previously reported that SHP-2 is a downstream effector for PDGFA/PDGFRα-driven glioma tumorigenesis and dynamin 2-mediated cell migration.4, 19 In this study, we showed that inhibition of SHP-2 function by mutation of its binding site in PDGFRα (F720 mutant), gene knockdown or pharmacological inhibitors markedly impaired PDGFRα stimulation of ZEB1 expression and glioma cell migration and GSC renewal in mice and humans. Collectively, these findings suggest that PDGFA/PDGFRα-SHP-2 activation of the ZEB1-miR-200 feedback loop is another essential signaling for PDGFRα-driven glioma tumorigenesis and invasion.
The last important finding in this study is that our data demonstrate that SHP-2 acts together with PI3K/Akt to regulate the ZEB1-miR-200 loop in glioma cells. Accumulated evidence show that activation of the PI3K/AKT pathway has important roles in contributing to cancer cell EMT and cancer stem cell features, a common mechanism in multiple cancers.32, 42, 43, 44 Integrative genomic analysis of The Cancer Genome Atlas data has found that PTPN11 is one of six ‘linker’ genes that connect major ‘nodes’ of commonly altered cancer GBM genes, suggesting that SHP-2 is critical for glioma tumorigenesis.30 Recently, using gene knockdown and pharmacological inhibitor treatments, we demonstrated that both SHP-2 and PI3K/Akt are important for PDGFRα-driven glioma tumor growth and invasion in mice and humans, in which SHP-2 recruits PI3K to activate Akt/mTOR pathway.4 Here, our data not only validates the critical roles of SHP-2 and PI3K/AKT in glioma proliferation and survival but also establishes them as collective modulators that regulate glioma EMT and cancer stem cell malignancy. We showed that overexpression of the CA Akt mutant in F720 (SHP-2 binding site mutation) PDGFRα mutant-expressing cells, or knockdown of Akt1/2 in Y720 (SHP-2 binding site mutation) PDGFRα mutant cells, altered the SHP-2-mediated ZEB1-miR-200 feedback loop, suggesting that SHP-2 and PI3K/Akt collectively modulate ZEB1-miR-200 in PDGFRα-driven glioma tumorigenesis.
In summary, our findings identify ZEB1 as a potential target for treatment of highly invasive clinical GBMs. This study demonstrates a previously unknown signal relay by which ZEB1 mediates EMT stimulated by PDGFA/PDGFRα-SHP-2/PI3K-Akt signaling, thereby enhancing glioma tumor growth and invasion. The newly elucidated roles of ZEB1 in PDGFRα-driven glioma EMT and invasion also provide a strong rationale for targeting this EMT-related molecule in clinical treatment of human gliomas with high levels of PDGFRα activity.
Materials and methods
Cell lines
Human HEK293T, glioma LN18 and T98G cells were from ATCC (Manassas, VA, USA). The LN444 cell line was a gift from Dr E Van Meir at Emory University (Atlanta, GA, USA), and was also recently authenticated using short tandem repeat DNA fingerprinting by RADIL (Columbia, MO, USA). LN235 cell line was cultured as reported previously.5 Patient-derived GSC lines, GSC157 and GSC AC17, were recently characterized.18 All cells and GSC cells were maintained and cultured as described previously.4, 5, 18 Cell transfections were performed as described previously.5 Mouse astrocytes with various WT or mutant PDGFRα, human LN444/PDGFA and LN18/PDGFA cell lines with overexpression of exogenous PDGFA were established and characterized as described previously.4, 5
Antibodies and reagents
The following antibodies were used in this study: anti-ZEB1 (E-20), anti-vmentin (V9), anti-E-cadherin (H-108), anti-PDGFRα (C-20), anti-p-PDGFRα (Y754) and anti-β-actin (I-19) antibodies (Santa Cruz Biotechnology, Dallas, TX, USA); an anti-SHP-2 antibody (no. 610621; BD Biosciences, San Jose, CA, USA); anti-ZEB1 (D80D3), anti-p-p44/42 MAP kinase (Thr202/Tyr204, no. 9101), rabbit anti-p44/42 MAP kinase (no. 9102), anti-p-Akt (S473, no. 4060) and anti-Akt (no. 9272) antibodies (Cell Signaling Technology, Danvers, MA, USA). The secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA, USA). Peroxidase blocking reagent was from DAKO (Carpinteria, CA, USA); AquaBlock was from East Coast Biologics Inc. (North Berwick, ME, USA). Inhibitors, PD98059, PHPS-1 and LY294002 were from Sigma (St. Louis, MO, USA). Cell culture media and other reagents were from Invitrogen (Carlsbad, CA, USA), Sigma-Aldrich or Peprotech (Rocky Hill, NJ, USA).
Plasmids
ZEB1 shRNAs and control shRNAs were purchased from Shanghai GeneChem Co. Ltd (Shanghai, China). ATK1/2 siRNAs and control siRNAs were purchased from Thermo Fisher Scientific (Waltham, MA, USA). pLenti 4.1 Ex miR200b-200a-429 (plasmid 35533)29 and pLNCX-Myr-Akt (plasmid 17245)45 were purchased from Addgene Inc. (Cambridge, MA, USA). pcDNA3-miR-200a was derived from pLenti 4.1 Ex miR200b-200a-429.
Western blotting assay
Western blotting assay was performed as described previously.46 Briefly, cells were lysed in a buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 2 mm Na3VO4, 5 mm NaF, 1% Triton X-100 and protease inhibitor cocktail) at 4 °C for 30 min, and then the lysates were centrifuged for 20 min at 12 000 g to remove debris. Protein concentrations were determined with a BCA Protein Assay Kit (Thermo Fisher Scientific). Equal amounts of cell lysates were resolved in a 2 × sodium dodecyl sulfate lysis buffer and analyzed.
Cell proliferation and cell migration assays
Cell proliferation assay was performed using a WST-1 Assay Kit (BioVision, Inc., Milpitas, CA, USA), and cell migration was performed using a Boyden chamber assay as described previously.5
Real-time PCR analysis
Total RNA was extracted using Trizol (Invitrogen), according to the manufacturer's instructions. Real-time PCR was performed in triplicate using the QuantiTect SYBR Green PCR Kit (Qiagen, Valencia, CA, USA) on a Rotorgene 6000 series PCR machine (Corbett Research, Valencia, CA, USA). All mRNA quantification data were normalized to ACTB, which was used as an internal control. The following ZEB1 primer sets were used: 5′-TTCAAACCCATAGTGGTTGCT-3′ and 5′-TGGGAGATACCAAACCAACTG-3′.
MicroRNA real-time PCR analysis
The abundance of miR-200a was determined by microRNA real-time PCR assay using the mirVana qRT–PCR miRNA Detection Kit (Thermo Fisher Scientific) and QRT–PCR Primer Sets, according to the manufacturer's instructions (Ambion Inc., Austin, TX, USA). The expression of U6-snRNA was used as an internal control.
Tumorigenesis studies
Athymic (Ncr nu/nu) female mice at an age of 6–8 weeks (SLAC, Shanghai, China) were used for all animal experiments. All experiments using animals were performed in accordance to a protocol approved by Shanghai Jiao Tong University Institutional Animal Care and Use Committee (IACUC). Mice were randomly divided into 5–6 per group. Various human glioma cells (1 × 106 in 5 μl phosphate-buffered saline) were stereotactically implanted into the brain of individual mice. Animals were killed when neuropathological symptoms developed. The brains were removed, processed and analyzed as described previously.46 Tumor volumes were measured and estimated as (a2 × b)/2, a<b.4 The number of invasive tumors was quantitated, and these tumors did not have a clear border with more than two invasive fingers.
Mice group allocation, surgery and assessing the outcome of mice were performed independently by different investigators.
Colony formation and western blot assays
Soft agar colony formation assay and western blot assay were performed as described previously.47
Limiting dilution neurosphere-forming assay
Limiting dilution neurosphere-forming assay was performed as reported previously.18 Briefly, GSC spheres from individual samples were dissociated and seeded into 96-well plates from 50, 100, 500 or 1000 cells per well. Spheres (>50 μm in diameter) were counted at day 4.
IHC of human and mouse glioma specimens
Immunohistochemical (IHC) staining was performed as described previously.5 All experiments using clinical patient samples were performed in accordance with a protocol approved by Shanghai Jiao Tong University Institutional Clinical Care and Use Committee. Informed consent was obtained from all subjects. OCT-embedded mouse brain sister sections containing glioma xenograft tumors or paraffin-embedded human GBM tumor sister sections were separately stained with antibodies against ZEB1 (1:50) or p-PDGFRα-Y754 (1:30). Nonspecific immunoglobulin Gs were used as negative controls. A total of 86 primary human GBM specimens and five normal brain tissues without notable pathological lesions or history were collected from 2002 to 2012 at Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University. These clinical cancer specimens were examined and diagnosed by pathologists at Ren Ji Hospital. IHC staining was quantified by two persons independently as described previously:5 3+, positively signals in ~50% tumor cells; 2+, signals in ~25% tumor cells; 1+,signals in ~5–25% tumor cells; ±, low or no signals in <1% tumor cells; –, no detectable signals in all tumor cells (0%). Tumors with – or ± staining were considered as low expressing, and tumors with 1+ to 3+ scores were considered high expressing. Kaplan–Meier survival assay was performed as described previously.5
Statistical analysis
Statistical analyses were performed in a GraphPad Prism version 5.0 for Windows (GraphPad Software Inc., San Diego, CA, USA). Survival analysis was carried out using log-rank tests, and a Mantel–Haneszel approach was used to determine hazard ratio. A two-tailed Fisher’s exact test was performed to determine if the frequency distribution of the variables were statistically significant. Comparison of treatments was analyzed using one-way analysis of variance with Newman–Keuls post-test or a paired two-way Student’s t-test as described previously.19, 48 P-values <0.05 were considered significant.
References
Wen PY, Kesari S . Malignant gliomas in adults. N Engl J Med 2008; 359: 492–507.
Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 2007; 21: 2683–2710.
Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008; 455: 1061–1068.
Liu KW, Feng H, Bachoo R, Kazlauskas A, Smith EM, Symes K et al. SHP-2/PTPN11 mediates gliomagenesis driven by PDGFRA and INK4A/ARF aberrations in mice and humans. J Clin Invest 2011; 121: 905–917.
Feng H, Hu B, Liu KW, Li Y, Lu X, Cheng T et al. Activation of Rac1 by Src-dependent phosphorylation of Dock180Y1811 mediates PDGFRalpha-stimulated glioma tumorigenesis in mice and humans. J Clin Invest 2011; 121: 4670–4684.
Gonzalez DM, Medici D . Signaling mechanisms of the epithelial–mesenchymal transition. Sci Signal 2014; 7: re8.
Singh A, Settleman J . EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 2010; 29: 4741–4751.
Chaffer CL, Weinberg RA . A perspective on cancer cell metastasis. Science 2011; 331: 1559–1564.
Siebzehnrubl FA, Silver DJ, Tugertimur B, Deleyrolle LP, Siebzehnrubl D, Sarkisian MR et al. The ZEB1 pathway links glioblastoma initiation, invasion and chemoresistance. EMBO Mol Med 2013; 5: 1196–1212.
Schmalhofer O, Brabletz S, Brabletz T . E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metast Rev 2009; 28: 151–166.
Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 2009; 11: 1487–1495.
Korpal M, Lee ES, Hu G, Kang Y . The miR-200 family inhibits epithelial–mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 2008; 283: 14910–14914.
Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial–mesenchymal transition. Cancer Res 2008; 68: 7846–7854.
Meng F, Speyer CL, Zhang B, Zhao Y, Chen W, Gorski DH et al. PDGFRalpha and beta play critical roles in mediating Foxq1-driven breast cancer stemness and chemoresistance. Cancer Res 2015; 75: 584–593.
Jechlinger M, Sommer A, Moriggl R, Seither P, Kraut N, Capodiecci P et al. Autocrine PDGFR signaling promotes mammary cancer metastasis. J Clin Invest 2006; 116: 1561–1570.
Kong D, Wang Z, Sarkar SH, Li Y, Banerjee S, Saliganan A et al. Platelet-derived growth factor-D overexpression contributes to epithelial-mesenchymal transition of PC3 prostate cancer cells. Stem Cells 2008; 26: 1425–1435.
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010; 17: 98–110.
Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci USA 2013; 110: 8644–8649.
Feng H, Liu KW, Guo P, Zhang P, Cheng T, McNiven MA et al. Dynamin 2 mediates PDGFRalpha-SHP-2-promoted glioblastoma growth and invasion. Oncogene 2012; 31: 2691–2702.
Bhat KP, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell 2013; 24: 331–346.
Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006; 9: 157–173.
Rosenkranz S, DeMali KA, Gelderloos JA, Bazenet C, Kazlauskas A . Identification of the receptor-associated signaling enzymes that are required for platelet-derived growth factor-AA-dependent chemotaxis and DNA synthesis. J Biol Chem 1999; 274: 28335–28343.
Van Stry M, Kazlauskas A, Schreiber SL, Symes K . Distinct effectors of platelet-derived growth factor receptor-alpha signaling are required for cell survival during embryogenesis. Proc Natl Acad Sci USA 2005; 102: 8233–8238.
Klinghoffer RA, Hamilton TG, Hoch R, Soriano P . An allelic series at the PDGFalphaR locus indicates unequal contributions of distinct signaling pathways during development. Dev Cell 2002; 2: 103–113.
Park SM, Gaur AB, Lengyel E, Peter ME . The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008; 22: 894–907.
Zang M, Zhang B, Zhang Y, Li J, Su L, Zhu Z et al. CEACAM6 promotes gastric cancer invasion and metastasis by inducing epithelial-mesenchymal transition via PI3K/AKT signaling pathway. PLoS One 2014; 9: e112908.
Wu K, Fan J, Zhang L, Ning Z, Zeng J, Zhou J et al. PI3K/Akt to GSK3beta/beta-catenin signaling cascade coordinates cell colonization for bladder cancer bone metastasis through regulating ZEB1 transcription. Cell Signal 2012; 24: 2273–2282.
Wang Y, Sheng Q, Spillman MA, Behbakht K, Gu H . Gab2 regulates the migratory behaviors and E-cadherin expression via activation of the PI3K pathway in ovarian cancer cells. Oncogene 2012; 31: 2512–2520.
Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008; 10: 593–601.
Cerami E, Demir E, Schultz N, Taylor BS, Sander C . Automated network analysis identifies core pathways in glioblastoma. PLoS One 2010; 5: e8918.
Dong P, Konno Y, Watari H, Hosaka M, Noguchi M, Sakuragi N . The impact of microRNA-mediated PI3K/AKT signaling on epithelial-mesenchymal transition and cancer stemness in endometrial cancer. J Transl Med 2014; 12: 231.
Grille SJ, Bellacosa A, Upson J, Klein-Szanto AJ, van Roy F, Lee-Kwon W et al. The protein kinase Akt induces epithelial mesenchymal transition and promotes enhanced motility and invasiveness of squamous cell carcinoma lines. Cancer Res 2003; 63: 2172–2178.
Imanishi Y, Hu B, Xiao G, Yao X, Cheng SY . Angiopoietin-2, an angiogenic regulator, promotes initial growth and survival of breast cancer metastases to the lung through the integrin-linked kinase (ILK)-AKT-B cell lymphoma 2 (Bcl-2) pathway. J Biol Chem 2011; 286: 29249–29260.
Yang L, Lin C, Liu ZR . P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from beta-catenin. Cell 2006; 127: 139–155.
Zheng S, El-Naggar AK, Kim ES, Kurie JM, Lozano G . A genetic mouse model for metastatic lung cancer with gender differences in survival. Oncogene 2007; 26: 6896–6904.
Grande M, Franzen A, Karlsson JO, Ericson LE, Heldin NE, Nilsson M . Transforming growth factor-beta and epidermal growth factor synergistically stimulate epithelial to mesenchymal transition (EMT) through a MEK-dependent mechanism in primary cultured pig thyrocytes. J Cell Sci 2002; 115: 4227–4236.
Lu Z, Ghosh S, Wang Z, Hunter T . Downregulation of caveolin-1 function by EGF leads to the loss of E-cadherin, increased transcriptional activity of beta-catenin, and enhanced tumor cell invasion. Cancer Cell 2003; 4: 499–515.
Kong D, Li Y, Wang Z, Banerjee S, Ahmad A, Kim HR et al. miR-200 regulates PDGF-D-mediated epithelial–mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells 2009; 27: 1712–1721.
Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev 2009; 23: 2140–2151.
Matozaki T, Murata Y, Saito Y, Okazawa H, Ohnishi H . Protein tyrosine phosphatase SHP-2: a proto-oncogene product that promotes Ras activation. Cancer Sci 2009; 100: 1786–1793.
Chan G, Kalaitzidis D, Neel BG . The tyrosine phosphatase Shp2 (PTPN11) in cancer. Cancer Metast Rev 2008; 27: 179–192.
Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME et al. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol Biol Cell 2001; 12: 27–36.
Du R, Xia L, Ning X, Liu L, Sun W, Huang C et al. Hypoxia-induced Bmi1 promotes renal tubular epithelial cell-mesenchymal transition and renal fibrosis via PI3K/Akt signal. Mol Biol Cell 2014; 25: 2650–2659.
Chen W, Wu S, Zhang G, Wang W, Shi Y . Effect of AKT inhibition on epithelial-mesenchymal transition and ZEB1-potentiated radiotherapy in nasopharyngeal carcinoma. Oncol Lett 2013; 6: 1234–1240.
Schmelzle T, Mailleux AA, Overholtzer M, Carroll JS, Solimini NL, Lightcap ES et al. Functional role and oncogene-regulated expression of the BH3-only factor Bmf in mammary epithelial anoikis and morphogenesis. Proc Natl Acad Sci USA 2007; 104: 3787–3792.
Feng H, Hu B, Jarzynka MJ, Li Y, Keezer S, Johns TG et al. Phosphorylation of dedicator of cytokinesis 1 (Dock180) at tyrosine residue Y722 by Src family kinases mediates EGFRvIII-driven glioblastoma tumorigenesis. Proc Natl Acad Sci USA 2012; 109: 3018–3023.
Yoshida S, Shimizu E, Ogura T, Takada M, Sone S . Stimulatory effect of reconstituted basement membrane components (matrigel) on the colony formation of a panel of human lung cancer cell lines in soft agar. J Cancer Res Clin Oncol 1997; 123: 301–309.
Feng H, Lopez GY, Kim CK, Alvarez A, Duncan CG, Nishikawa R et al. EGFR phosphorylation of DCBLD2 recruits TRAF6 and stimulates AKT-promoted tumorigenesis. J Clin Invest 2014; 124: 3741–3756.
Acknowledgements
We thank E Van Meir for providing glioma cell lines, R Pangeni and N Sastry for editing the manuscript. This work was supported, in part, by National Natural Science Foundation of China (Nos. 81372704 and 81572467) to H Feng and (No. 81470315) (to YL); the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning, Innovation Program of Shanghai Municipal Education Commission (No. 14ZZ111), the State Key Laboratory of Oncogenes and Related Genes in China (No. 90-14-04) (to HF); the Pujiang Talent Plan of Shanghai City, China (No. 14PJ1406500), Natural Science Foundation of Tianjin City, China (No. 13JCYBJC39400), Shanghai Jiao Tong University School of Medicine Hospital Fund (No. 14XJ10069) (to YL); Science and Technology Commission of Shanghai Municipality, China (15ZR1425700) (to ZL); US NIH Grants (CA158911, NS093843 and NS95634), a Zell Scholar Award from the Zell Family Foundation and funds from Northwestern Brain Tumor Institute at Northwestern University (to S-YC); a Brain Cancer Research Award from the James S McDonnell Foundation (to BH) and a NIH/NCI training grant T32 CA070085 (to AA).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Zhang, L., Zhang, W., Li, Y. et al. SHP-2-upregulated ZEB1 is important for PDGFRα-driven glioma epithelial–mesenchymal transition and invasion in mice and humans. Oncogene 35, 5641–5652 (2016). https://doi.org/10.1038/onc.2016.100
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.100
This article is cited by
-
Valtrate, an iridoid compound in Valeriana, elicits anti-glioblastoma activity through inhibition of the PDGFRA/MEK/ERK signaling pathway
Journal of Translational Medicine (2023)
-
Platelets involved tumor cell EMT during circulation: communications and interventions
Cell Communication and Signaling (2022)
-
MYCN protein stability is a better prognostic indicator in neuroblastoma
BMC Pediatrics (2022)
-
Gliosarcoma vs. glioblastoma: a retrospective case series using molecular profiling
BMC Neurology (2021)
-
Novel insights into the interaction between long non-coding RNAs and microRNAs in glioma
Molecular and Cellular Biochemistry (2021)