Abstract
The transcription factor Snail is a master regulator of cellular identity and epithelial-to-mesenchymal transition (EMT) directly repressing a broad repertoire of epithelial genes. How chromatin modifiers instrumental to its activity are recruited to Snail-specific binding sites is unclear. Here we report that the long non-coding RNA (lncRNA) HOTAIR (for HOX Transcript Antisense Intergenic RNA) mediates a physical interaction between Snail and enhancer of zeste homolog 2 (EZH2), an enzymatic subunit of the polycomb-repressive complex 2 and the main writer of chromatin-repressive marks. The Snail-repressive activity, here monitored on genes with a pivotal function in epithelial and hepatic morphogenesis, differentiation and cell-type identity, depends on the formation of a tripartite Snail/HOTAIR/EZH2 complex. These results demonstrate an lncRNA-mediated mechanism by which a transcriptional factor conveys a general chromatin modifier to specific genes, thereby allowing the execution of hepatocyte transdifferentiation; moreover, they highlight HOTAIR as a crucial player in the Snail-mediated EMT.
Similar content being viewed by others
Introduction
Epithelial cells may undergo an epithelial-to-mesenchymal transition (EMT) with acquisition of a mesenchymal phenotype and a migratory capacity. Remarkably, these changes appear to be signs of cellular plasticity as EMT is often reversed, in secondary anatomical sites, by a mesenchymal-to-epithelial transition. The EMT/mesenchymal-to-epithelial transition dynamics characterize physiological processes (i.e. organogenesis, development, wound healing and regeneration) and drives epithelial tumor metastasis (for a review see Thiery et al.1).
Various repressors are sufficient to trigger and orchestrate EMT: ZEB1, SIP1/ZEB2, Twist1, Twist2, E12/E47, Tbx3, Slug, Smuc and, in particular, Snail1 (Snail).2, 3, 4, 5, 6 While chromatin modifiers and epigenetic modifications are required in EMT,7, 8, 9, 10, 11 molecular mechanisms linking, in a site-specific manner, master transcription factors and epigenetic machineries are still largely unexplored.
The current knowledge identifies as a key chromatin-repressive modification the trimethylation of histone H3 lysine 27 (H3K27me3), accomplished by the member of the polycomb-repressive complex 2 (PRC2) named enhancer of zeste Homolog 2 (EZH2).12, 13, 14 The mechanism in cell reprogramming that confers specificity to polycomb targeting is unknown,15 even if specific determinants must exist, and the early binding of DNA-binding factors facilitating PRC complexes recruitment has been demonstrated in few cases.16, 17, 18, 19
The long non-coding RNA (lncRNA) HOTAIR (for HOX Transcript Antisense Intergenic RNA)20 is an assembling scaffold for EZH2 targeting H3K27 methylation to discrete regions of the genome.21, 22 If the specificity of PRC2–RNA interactions has been recently questioned as promiscuous in vitro,23, 24 in vivo HOTAIR is an excellent predictor of epithelial tumor metastasis causing genome-wide PRC2 retargeting.21, 25, 26 Once again, the mechanism of in trans HOTAIR-mediated recruitment of PRCs is not established, while clearly involving more than direct RNA-DNA complementarity21 and specific partners.
Here we focused on Snail starting from the hypothesis that a repressor sufficient to drive EMT must directly guide the recruitment of enzymes locally impacting chromatin modifications. We demonstrated the role of Snail as an organizer, on a number of epithelial genes, of a molecular platform that includes PRC2: specifically, Snail was found to regulate histone modifications through the enrollment of HOTAIR.
Notably, we concentrated on in vivo studies: inspired by the innovative approach described by McHugh et al.,27 we used the RNA pull-down after UV crosslinking, coupled to RNA immunoprecipitation (RIP) experiments, to demonstrate the direct and specific interaction between HOTAIR and Snail and to validate in vivo the binding between HOTAIR and EZH2. Furthermore, the interaction among Snail, HOTAIR and EZH2 on epithelial genes was found instrumental for the execution of the EMT. Chromatin isolation by RNA purification (ChIRP) and chromatin immunoprecipitation (ChIP) approaches highlighted the HOTAIR/SNAIL/EZH2 complex specifically localized at Snail binding sites on the promoters of a number of epithelial gene targets of this repressor only if Snail is present. Further, ChIP analysis for EZH2 and H3K27 methylation demonstrated that the repression of these epithelial genes needs the enrollment of HOTAIR/EZH2 by Snail.
Results
HOTAIR behaves as a mesenchymal gene with a functional role in the Snail-dependent epithelial gene repression
Evidence correlating HOTAIR expression to the acquisition of invasive properties by epithelial tumor cells21, 25, 26 prompted us to investigate the role of this lncRNA in differentiated hepatocytes responsive to EMT dynamics (Figure 1 and Supplementary Figure 1). To this aim, HOTAIR levels were first monitored in a hepatocyte cell line well characterized to undergo into a transforming growth factor-β (TGFβ)-induced EMT:28, 29, 30, 31, 32 HOTAIR was positively regulated in EMT, in parallel to the induction of Snail and of the mesenchymal genes vimentin and fibronectin, whereas the epithelial targets E-cadherin and HNF4α were repressed (Figure 1a; the modulation of these genes is well known to be causative of the transition33, 28). The levels of EZH2 were not significantly modulated (Figure 1a).
HOTAIR expression correlates to mesenchymal genes. (a) RT–qPCR analysis for HOTAIR, EZH2, the indicated epithelial (E-cadherin and HNF4α) and mesenchymal (Snail, fibronectin and vimentin) genes on hepatocytes treated with TGFβ or not treated (NT) with TGFβ for 24 h. The values are calculated by the 2(−ΔCt) method, expressed as fold of expression versus the control (arbitrary value=1) and shown as means±s.e.m. Statistically significant differences are reported (*P<0.05, **P<0.01) for five independent experiments (NS=no significance). (b) RT–qPCR analysis for the same genes as in (a) in hepatocytes overexpressing HOTAIR (HOTAIR) or an empty vector (Ctr). The values are calculated by the 2(−ΔCt) method, expressed as fold of expression versus the control (arbitrary value=1) and shown as means±s.e.m. No statistically significant differences were evaluated for five independent experiments.
These data showing HOTAIR induction when the epithelial identity is lost might suggest a causal role for HOTAIR in EMT. Consequently, to move from correlative evidence to a functional investigation, we analyzed the response of the hepatocytes to HOTAIR overexpression or its silencing.
As shown in Figure 1b, ectopic expression of HOTAIR in these epithelial cells did not induce statistically significant modulation of the mesenchymal and epithelial genes analyzed. Conversely, the response of the TGFβ-induced transitional hepatocytes when HOTAIR is silenced (Figure 2a) highlighted a functional role for this RNA: in HOTAIR-interfered cells, the TGFβ-induced upregulation of the mesenchymal genes Snail, fibronectin and vimentin did not correlate with the repression of the epithelial genes HNF4α, E-cadherin and HNF1α (Figure 2a and Supplementary Figure 2). Notably, the TGFβ-induced upregulation of Snail, verified also at the protein level, did not result in the full repression of its main targets E-cadherin and HNF4α, suggesting a Snail activity functional impairment when HOTAIR was silenced (Figure 2a and Supplementary Figure 2).
HOTAIR has a functional role in EMT. (a) RT–qPCR analysis for the indicated markers on HOTAIR-silenced hepatocytes (siHotair), compared with control siGFP cells (siCtr), treated (TGFβ) or not (Ctr) with TGFβ for 24 h. The values are calculated by the 2(−ΔCt) method, expressed as fold of expression versus the control (arbitrary value=1) and shown as means±s.e.m. Statistically significant differences are reported (*P<0.05, **P<0.01) for five independent experiments (NS=no significance). (b) RT–qPCR analysis for the same markers as in (a) on hepatocytes overexpressing Snail (Snail), or a control vector (Ctr), and both HOTAIR (siHotair) or GFP (siCtr) silenced. RNAs were collected 72 h after infection and 48 h after transfection. The values are calculated as in (a). Statistically significant differences are reported (*P<0.05, **P<0.01) for five independent experiments. (c) Phase-contrast and immunofluorescence analysis for the indicated markers in cells as in (b) (magnification × 20). Blue DAPI staining shows the nuclei (DNA).
Taking into account the pleiotropy of the TGFβ-mediated effects, involving the induction of both Snail and other EMT transcriptional factors, we considered necessary to focus on the sole Snail-mediated transcriptional repression activity.1 We therefore analyzed the EMT of hepatocytes both ectopically expressing Snail and interfered for HOTAIR. Although HOTAIR silencing did not interfere with the Snail-mediated mesenchymal marker induction, it profoundly impacted the repression of the epithelial genes HNF4α, E-cadherin and HNF1α (Figure 2b and Supplementary Figure 2), previously experimentally validated as Snail targets.1, 29, 34 Coherently, hepatocytes that ectopically expressed Snail did not undergo a complete morphological EMT when silenced for HOTAIR, still retaining an epithelial morphology with membrane-bound E-cadherin (Figure 2c).
Overall, the loss of the cellular response to TGFβ observed when HOTAIR is silenced can be ascribed to a Snail functional impairment, as monitored by the regulation of the epithelial genes considered. Moreover, the evidence described indicates for the first time a functional HOTAIR requirement in Snail-mediated epithelial target repression during both TGFβ- and Snail-induced EMT.
HOTAIR has a bridging role in the Snail/EZH2 complex
The effect of HOTAIR on the Snail-mediated repression suggests the possibility that this lncRNA might interact with Snail, forming a complex with a function in EMT. We first explored by RIP assays the possible HOTAIR–Snail interaction and analyzed whether the previously reported in vitro interaction between EZH2 and HOTAIR22, 35 also occurred in murine hepatocytes undergoing EMT. As shown in Figure 3a, Snail immunoprecipitation was found to recruit HOTAIR and none of the other control transcripts (i.e. L34 and GAPDH pre-mRNAs and the known EZH2-interactor lncRNA named steroid receptor RNA activator (SRA36). The specificity of HOTAIR binding to Snail was also confirmed by RIP assays with another transcriptional factor, that is, HNF4α (Supplementary Figure 3). On the other hand, as expected, EZH2 was found able to recruit both HOTAIR and SRA (Figure 3a).
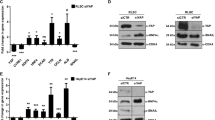
Identification of the Snail/HOTAIR/EZH2 complex. (a) RIP assays with rabbit polyclonal anti-Snail, anti-EZH2 or preimmune IgG on 24 h TGFβ-treated (TGFβ) or untreated (NT) murine hepatocyte cell extracts. RNA levels in immunoprecipitates were determined by qRT–PCR. HOTAIR lncRNA and, as controls, ribosomal L34 RNA, GAPDH pre-mRNA and SRA lncRNA were reported as percentage with respect to 1/10th of the input sample (TGFβ treated in the hotair panel or NT in the other panels) (% Input). Data are means ±s.e.m. of three independent experiments. (b) Co-immunoprecipitation of Snail and EZH2. Immunoprecipitations with rabbit polyclonal anti-Snail, anti-EZH2, preimmune IgG or no antibody (NoAb) were performed on protein extracts from murine hepatocyte cells treated with TGFβ for 24 h and silenced for HOTAIR (siHotair) or GFP (siCtr) as control. Immunoblots were performed using anti-Snail and anti-EZH2 antibodies. (c) RIP assays with rabbit polyclonal anti-Snail, anti-EZH2 or preimmune IgG on TGFβ-treated (TGFβ) or untreated (NT) human hepatoma cell extracts. RNA levels in immunoprecipitates were determined by RT–qPCR. HOTAIR lncRNA and, as controls, ribosomal L34 RNA, GAPDH pre-mRNA and SRA lncRNA were reported as percentage with respect to 1/10th of the input sample (TGFβ treated in the hotair panel or NT in the other panels) (% Input). Data are means ± s.e.m. of three independent experiments. (d) Co-immunoprecipitation of Snail and EZH2. Immunoprecipitations with rabbit polyclonal anti-Snail, anti-EZH2 or preimmune IgG were performed on protein extracts from human hepatoma cells treated with TGFβ for 24 h and silenced for HOTAIR (siHotair) or GFP (siCtr) as control. Immunoblots were performed using anti-Snail and anti-EZH2 antibodies.
Furthermore, reciprocal Snail and EZH2 co-immunoprecipitation experiments revealed the formation of a Snail/HOTAIR/EZH2 complex in TGFβ-treated cells, while in TGFβ-treated cells silenced for HOTAIR this interaction was lost (Figure 3b).
Our conclusions are not limited to the murine model. Indeed, the human HOTAIR lncRNA shares partial but significant sequence similarity (55% identity) with its mouse homolog (Supplementary Figure 4). Also, well conserved are the EZH2 and Snail proteins (92.8% and 99.3% sequence identity, respectively). This opened the need of exploiting our results in human cells; consequently, we extended our analysis to human Hep3B HCC lines. Notably, consistent with the hypothesis that their close evolutionary relationship corresponds to a conserved similar function, Snail and HOTAIR, as well as EZH2 and HOTAIR, form a complex also in TGFβ-treated human hepatoma cells (Figures 3c and d): Snail immunoprecipitation and, as expected, EZH2 immunoprecipitation specifically retrieved endogenous HOTAIR in RIP assays (Figure 3c); moreover, the formation of a Snail/HOTAIR/EZH2 complex was confirmed by reciprocal Snail and EZH2 co-immunoprecipitations (Figure 3d).
Taken together, these data suggest that Snail may (directly or indirectly) interact with HOTAIR and HOTAIR can mediate the Snail/EZH2 interaction.
Next, we aimed to prove the potential HOTAIR–Snail direct interaction. As the in vitro interaction between PCR2 components and lncRNAs has been recently reported somewhat promiscuous,23, 24 to distinguish between the direct lncRNA–protein interactions that occur in the cell from those that only associate in solution and to exclude indirect binding, we accomplished in vivo assays (according to McHugh et al.27 and Chu et al.37) in murine hepatocytes. RNA pull-down was performed either without or after UV crosslinking and subsequent purification of complexes after washes in non-denaturing or in denaturing conditions. The UV crosslink/denaturing wash protocol allows the identification of chromatin-extracted proteins only when directly interacting with specific RNAs, while indirect interactions are disrupted (see Materials and methods).
As shown in Figure 4a a pool of specific DNA oligonucleotides was found able to capture HOTAIR from chromatin. Snail was found to bind directly and specifically to HOTAIR in cells overexpressing both Snail and HOTAIR and, notably, in cells overexpressing only Snail, which induces endogenous HOTAIR expression (Figure 4b, left panel, and Figure 2b). These results are confirmed in TGFβ-treated cells (Supplementary Figure 5). This occurs in non-denaturing conditions and, notably, in denaturing condition only after UV crosslinking, thus proving a direct HOTAIR–Snail interaction (Figure 4b and Supplementary Figure 5). RNAse treatment of these samples impedes the binding between HOTAIR and both SNAIL and EZH2, further demonstrating that these interactions are RNA-mediated (Supplementary Figure 6). As a negative control, pre-ribosomal 45S RNA was also pulled down by specific complementary oligonucleotides; these did not retrieve HOTAIR in none of the experimental conditions and no interaction with Snail or EZH2 was found (Figures 4a and b and Supplementary Figure 5). Notably, in parallel experiments, the interaction between EZH2 and HOTAIR, previously described in vitro,22, 35 has been found to occur also in vivo in these experimental conditions and independently from Snail (Figure 4b, right panel and Supplementary Figure 5). Again this occurs in non-denaturing conditions and, notably, in denaturing condition only after UV crosslinking, proving a direct HOTAIR–EZH2 interaction.
Identification of the Snail/HOTAIR/EZH2 complex within the chromatin. (a) RNA pull-down for HOTAIR in the ChIRP analysis. Nuclei were prepared from cells overexpressing HOTAIR (HOTAIR), Snail (Snail) or both (HOTAIR/Snail) and, as control, transfected with the empty vectors (Ctr). Biotinylated complementary DNA probes effectively retrieved HOTAIR RNA, as compared with L34 and GAPDH. Note that in the double Snail/HOTAIR transfection the amount of each plasmid was halved. All the experiments have been performed in triplicate, after UV crosslinking (+UV, in denaturing condition) or not (−UV, in non-denaturing condition). As a negative control, specific pre-ribosomal 45S probes have been used in the pull-down assays, not retrieving HOTAIR in none of the tested experimental conditions. Data were reported as percentage with respect to the Input sample (% RNA retrieved). Means±s.e.m. are shown. (b) Western blot analysis for Snail (left panels) and EZH2 (right panels) of the chromatin fraction bound to HOTAIR, showing the direct interaction between both these proteins and HOTAIR. Proteins were obtained from total cell lysates (TOT) or from chromatin pulled down with probes recognizing HOTAIR (Hotair) or pre-ribosomal RNA 45S (45S). Cells were overexpressing HOTAIR (HOTAIR), Snail (Snail), both (HOTAIR/Snail) or the empty vectors (Ctr), as in (a). All the experiments have been performed in triplicate after UV crosslinking (+UV in denaturing condition) or not (−UV, in both denaturing and non-denaturing conditions). (c) ChIRP–qPCR analysis of the DNA in the chromatin fraction bound to HOTAIR. Crosslinked samples were as in (a). Data show the enrichment of HOTAIR on the Snail consensus binding sites on the murine promoters of HNF4α, E-cadherin and HNF1α only in the presence of Snail. Timm and Snail promoters were used as negative controls. Values derived from three independent experiments are expressed as the percentage of the Input chromatin (% Input) and reported as means ± s.e.m.
In summary, we demonstrated the in vivo direct interactions between Snail and HOTAIR and EZH2 and HOTAIR, linking a transcriptional repressor to the main writer of the chromatin-repressive marks, the catalytic member of the PCR2 complex EZH2. This evidence, together with the functional requirement of HOTAIR in the Snail-mediated repression, leads to the hypotheses that Snail conveys the PRC2 functional epigenetic activity on Snail-specific epithelial target genes through the enrollment of HOTAIR.
The Snail/HOTAIR/EZH2 complex has a functional role in epithelial gene repression
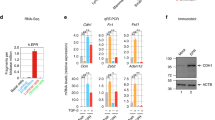
To reveal the Snail/HOTAIR/EZH2 complex genome occupancy, we analyzed the Snail binding sites on DNA of the chromatin fraction pulled down by HOTAIR from in vivo UV crosslinked cells (note that the ChIRP, whose first step was described above, allows the identification of RNA locally bound to both proteins and DNA).37 HOTAIR-bound DNA was enriched in Snail-specific epithelial target genes (i.e. E-boxes on HNF4α, HNF1α and E-cadherin promoters) in the chromatin fraction of Snail-overexpressing cells (Figure 4c). As negative controls the Snail-unresponsive Timm and Snail promoters were used.
Moreover, ChIP assays for the binding of Snail and EZH2 to the same epithelial genes were performed and H3K27me3 on the corresponding binding sites analyzed (Figure 5, Figure 6 and Supplementary Figure 7). Hepatocytes treated or not with TGFβ (inducing Snail and EMT) were analyzed in the presence or absence of small interfering RNAs (siRNAs) specifically targeting HOTAIR (expressed in cells undergoing EMT). As shown in Figure 5a, Snail was bound to its consensus binding sites (E-boxes) on HNF4α, HNF1α and E-cadherin promoters in TGFβ-treated cells independently from HOTAIR expression (Figure 5a). Interestingly, the HOTAIR-independent Snail occupancy on epithelial gene promoters did not coincide with the local H3K27me3. In fact, H3K27me3 levels increased in TGFβ-treated cells compared with untreated cells on Snail binding sequences of HNF4α, HNF1α and E-cadherin promoters (Figure 5b); however, in HOTAIR-interfered cells, H3K27 trimethylation was found at lower levels with respect to control cells (in which endogenous HOTAIR was present; Figure 5b) in spite of TGFβ treatment. Notably, EZH2 was recruited to the same Snail binding sites only in TGFβ-treated HOTAIR-expressing cells (Figure 6a). Furthermore, analysis has been extended to TGFβ-treated Snail-silenced cells in which the presence of HOTAIR did not cause the EZH2 recruitment to the same promoters (Supplementary Figure 7). As negative controls for these experiments the Snail-unresponsive Timm and Snail promoters were used.
Snail binding sites chromatin trimethylation requires HOTAIR. (a) qPCR analysis of ChIP assays with anti-Snail antibody (IP Snail) and, as control, normal rabbit IgG (IgG) on chromatin from murine hepatocyte cells silenced for HOTAIR (siHotair) or GFP (siCtr), as control, and treated (+TGFβ) or not (−TGFβ) with TGFβ for 24 h. Data show the direct recruitment of endogenous Snail on the correspondent consensus binding sites on the murine promoters of HNF4α, E-cadherin and HNF1α. Timm promoter was used as a negative control. Values derived from five independent experiments are reported as means ±s.e.m. and expressed as percentage of the Input chromatin (% Input). Statistically significant differences are reported (*P<0.05, **P<0.01, ***P<0.001). (b) qPCR analysis of ChIP assays with anti-H3K27me3 antibody (IPH3K27me3) and, as controls, normal rabbit IgG (IgG) on chromatin from murine hepatocyte cells silenced for HOTAIR (siHotair) or GFP (siCtr), as control, and treated (+TGFβ) or not (−TGFβ) with TGFβ for 24 h. Data show the enrichment of H3K27 trimethylation on the Snail consensus binding sites on the murine promoters of HNF4α, E-cadherin and HNF1α, as above. Timm promoter was used as a negative control. Values derived from five independent experiments are reported as means ± s.e.m. and expressed as the percentage of the Input chromatin (% Input). Statistically significant differences are reported (*P<0.05, **P<0.01).
EZH2 is recruited by Snail to its target genes by means of HOTAIR. (a) qPCR analysis of ChIP assays with an anti-EZH2 antibody (IP EZH2) and, as controls, normal rabbit IgG (IgG) on chromatin from murine hepatocyte cells silenced for HOTAIR (siHotair) or GFP (siCtr), as control, and treated (+TGFβ) or not (−TGFβ) with TGFβ for 24 h. Data show the recruitment of EZH2 on the Snail consensus binding sites on the murine promoters of HNF4α, E-cadherin and HNF1α and, as control, its displacement from the HNF4α binding site on Snail promoter.29 Timm promoter was used as unresponsive control sequence. Values derived from five independent experiments are reported as means ± s.e.m. and expressed as the percentage of the Input chromatin (% Input). Statistically significant differences are reported (*P<0.05; **P<0.01; ***P<0.001). (b) qPCR analysis of ReChIP assays of samples TGFβ treated for 24 h and silenced for HOTAIR (siHotair) or GFP (siCtr), as control. Values, derived from three independent experiments, are reported as means ±s.e.m. and expressed as the percentage of the Input chromatin (% Input).
ReChIP experiments, as expected, confirmed that Snail recruits EZH2 on its target gene promoters in a HOTAIR-dependent manner (Figure 6b). These data, together with those previously showed in Figure 2, provided evidence for a Snail functional impairment, in the repression of epithelial genes, in the absence of HOTAIR.
Discussion
The main finding of this investigation is that the Snail-repressive activity conveys the action of EZH2, the main writer of chromatin-repressive marks, to specific sites by means of the direct interaction with the lncRNA HOTAIR. The functional Snail/HOTAIR/EZH2 complex was found instrumental for the execution of the EMT by epithelial gene repression.
The mechanism conferring local specificity to PRCs is the matter of a long-lasting open question, whose resolution requires the investigation of the various biological processes involving PRCs and, conceivably, also a number of specific molecular players. PRC2 complex was described to have a broad RNA interactome,38, 39 although its RNA-binding specificity has been recently disputed. Recent studies, in fact, provided evidence for promiscuity of PRC2 in vitro binding to long transcripts.23, 24 In vivo data to our knowledge are limited to the analysis by Davidovich et al.23 of RIP-seq and ChIP-seq available data sets, drawing a model in which PRC2 binding serves as a checkpoint to prevent escape from silencing.40 Conceivably, the PRC2 binding in vivo to RNA(s) and the recruitment of this complex to specific loci are highly regulated, cell-type and/or context-specific steps that modulate the EZH2 enzymatic activity in the local chromatin environment. In this regard, the possible role(s) of lncRNAs is/are so far largely unknown.
The human lncRNA HOTAIR was previously described to bind to PRC2 and to the lysine-specific demethylase 1, both fundamental for gene silencing by H3K27 trimethylation and H3K4 demethylation, respectively.22, 35 Notably, evidence showing that manipulating HOTAIR levels reprograms cell state by genome-wide retargeting of PRC221 highlights an active role of this lncRNA in the modulation of PRC2-specific binding to chromatin, even if the mechanism is not yet clarified.
Here, in the frame of EMT, Snail is pinpointed to have the function of directing PRC2 to various epithelial targets through the direct enrollment of HOTAIR (see scheme in Figure 7), not ruling out the possibility that HOTAIR also contributes to the site specificity. In vivo evidence extending the scaffold role of HOTAIR to a transcription factor, namely Snail and also highlighting in vivo the direct HOTAIR–EZH2 interaction, has been gathered by means of RNA pull-down after UV crosslinking of cells and purification of complexes in denaturing conditions.27, 37 Furthermore, the bridging role of both murine and human HOTAIR, allowing Snail–EZH2 interaction, was established by RIP assays, coupled to co-immunoprecipitation experiments in both murine and human cells. Further experiments are required to address the possibility that other nucleic acid/proteins interact with the Snail/HOTAIR/EZH2 complex.
With respect to the functional role of the Snail/HOTAIR/EZH2 complex, Snail triggers EMT of the hepatocyte in a HOTAIR-dependent manner, impacting chromatin modifications causal to its repressive role through the site-specific recruitment of the complex HOTAIR/EZH2. This was observed specifically for genes having a pivotal functional role in epithelial morphogenesis and differentiation (i.e. HNF4α, HNF1α and E-cadherin). ChIRP and ChIP experiments allowed concluding that the HOTAIR/SNAIL/EZH2 complex is localized and functional at Snail binding sites, on the epithelial gene promoters, only if Snail is present. Notably, HOTAIR impairment impacted Snail/EZH2 interaction in vitro (see 'Co-immunoprecipitation') and in vivo: ChIP assays of TGFβ-treated hepatocytes, in the absence or presence of HOTAIR, demonstrated that Snail occupancy on target promoters appears independent of HOTAIR, whereas the Snail-repressive activity, and the related modifications of chromatin marks guiding the outcome of the EMT, depend on HOTAIR/EZH2 recruitment. Thus, Snail is, in its functional state on the epithelial genes we tested, part of a ribonucleoprotein complex containing HOTAIR and the chromatin modifier EZH2. In other words, our data provide this paradigm of function: a DNA-binding transcriptional factor (i.e. Snail) conveys to specific sites a general chromatin modifier (i.e EZH2) by direct interaction with an lncRNA (i.e. HOTAIR). The EZH2 functional activity in EMT reaches specific chromatin loci by means of this interaction. Although we focused on a limited number of Snail target genes, we cannot exclude that this lncRNA can act on a larger battery of genes. Interestingly, a computational prediction of transcription factor binding sites governing mRNA expression dynamics suggested Snail as a transcription factor that mediates Polycomb targeting.41 Consistently, EZH2 was found to regulate E-cadherin expression42, 43, 44, 45 in a Snail-dependent manner.18, 45 Moreover, the upregulation of both Snail and EZH2 was correlated to HCC progression and aggressiveness,46 whereas their silencing significantly reversed tumorigenicity.47
HOTAIR, in the frame of our observations, appears epistatic to Snail. Previous correlative evidence should be considered in line with this: HOTAIR overexpression was correlated with metastasis and poor prognosis in breast,21 colorectal,48 nasopharyngeal49 and in liver cancer.25, 50 Moreover, forced HOTAIR expression in epithelial cancer cells was found to cause PRC2 complex occupancy pattern retargeting in parallel with metastatic properties acquisition.21 While our investigation focuses on HOTAIR as Snail corepressor, it appears conceivable that HOTAIR informational capacity has other functions. Notably, other transcriptional factors may convey HOTAIR/EZH2 to different chromatin contexts in EMT and/or in other cellular processes.
Overall our work contributes to the understanding of how EZH2 gets to its genomic targets. The current study reveals a mechanism by which a DNA-binding transcriptional factor recruits a general chromatin modifier to specific sites by means of an lncRNA and emphasizes the role of HOTAIR as epistatic to the master repressor Snail in EMT.
Materials and methods
Cell culture conditions
Non-tumorigenic hepatocytes51, 52 were grown on collagen I-coated dishes in RPMI-1640 supplemented with 10% fetal bovine serum (Gibco Life Technology, Monza, Italy), 50 ng/ml epidermal growth factor, 30 ng/ml insulin-like growth factor II (PeproTech Inc., Rocky Hill, NJ, USA), 10 μg/ml insulin (Roche, Mannheim, Germany) and antibiotics. Where reported, cells were treated with 5 ng/ml TGFβ1 (PeproTech Inc.) for 24 h. Human Hep3B cells were grown in DMEM supplemented with 10% fetal bovine serum (Gibco) and antibiotics.
Cell transfection
Cells were transfected with Lipofectamine 2000 transfection reagent (Invitrogen, San Diego, CA, USA), according to the manufacturer’s protocol. Equal amounts of DNA (pLPCX vector (BD Biosciences Clontech, Palo Alto, CA, USA) or pLPCX/Snail29 or pTRACER or pTRACER-HOTAIR (Gibco)) were used. Cells were analyzed 48 h post-transfection.
siRNA interference
Cells were transfected with equal amounts of siRNA against GFP (5′-GGCUACGUCCAGGAGCGCACC-3′), murine HOTAIR (5′-CUGUGUUUACAAGACCAGA-3′, 5′-CUAAGUCCUUCCAGAGAGA-3′, 5′-AAUAGAAAAACACAAAUAG-3′, 5′-CAAAUAGAAAAAACCAAUU-3′) or ON_TARGETplus Mouse Snai1 siRNA (GE Healthcare Dharmacon, Lafayette, CO, USA) by Lipofectamine 2000 (Invitrogen), according to commercial protocol and analysis was performed at 48 h.
RNA extraction, reverse transcription, reverse transcription and real-time polymerase chain reaction
RNAs were extracted by RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany) or TRIzol (Ambion, Life Technology, Monza, Italy) and reverse transcribed with iScriptTM c-DNA Synthesis Kit (Bio-Rad Laboratories Inc., Hercules, CA, USA). cDNAs were amplified by qPCR reaction using GoTaq qPCR Master Mix (Promega, Madison, WI, USA). Relative amounts, obtained with 2(−ΔCt) method, were normalized with respect to the housekeeping gene 18S rRNA.53 List of primers is reported in Table 1.
Co-immunoprecipitation
Cells were lysed in 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 5 mM EGTA (pH 8.0), 50 mm NaF (pH 8.0), 10% glycerol, 1.5 mm MgCl2, 1% Triton X-100 containing protease and phosphatase inhibitors (complete EDTA-free; Roche Applied Science, Mannheim Germany) and protein concentrations determined by Bradford method. One milligram of cell lysates, after preclearing with protein A-sepharose (GE Healthcare, Little Chalfont, Buckinghamshire, UK), was incubated with 5 μg of anti-EZH2 (sc-25383), anti-Snail (sc-28199; Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) or rabbit immunogobulin G (IgG) 12-370 (Millipore Corp., Bedford, MA, USA). The complexes were incubated for 2 h with protein A-sepharose. Immune complexes were washed, eluted and denatured in Laemmli buffer. Proteins from either cell lysates or immunoprecipitation were resolved on sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane (162-0115; Bio-Rad Laboratories). Blots were probed with primary anti-EZH2 05-1319 (Millipore Corp.) or anti-Snail L70G2 (Cell Signaling Technology Inc., Danvers, MA, USA) and immune complexes were detected with horseradish peroxidase-conjugated species-specific secondary antiserum (Bio-Rad Laboratories), followed by enhanced chemiluminescence reaction (Pierce, Rockford, IL, USA).
RNA immunoprecipitation
RIP was performed as reported in Dahm et al.54 starting from 3 mg of cleared lysate. Immunoprecipitated RNA was reverse transcribed for reverse transcription and real-time polymerase chain reaction (RT–qPCR) amplifications. List of primers is reported in Table 1.
Primary antibodies for IP were: anti-EZH2 sc-25383, anti-Snail sc-28199 (Santa Cruz Biotechnology Inc.) and anti-HNF4α sc-8987X (Santa Cruz Biotechnology Inc.).
RNA pull-down and ChIRP analysis
Complementary DNA oligonucleotides against full-length murine HOTAIR and ribosomal 45S RNA were designed (http://www.singlemoleculefish.com), compared with the mouse genome using BLAST and 3′-end biotinylated to capture DNA–RNA hybrids on streptavidin-coated magnetic beads (see Table 2).
Cells were irradiated or not in a UV crosslinker (Stratagene, Agilent Technologies, Santa Clara, CA, USA) using 0.8 J/cm2 at 254 nm, rinsed with phosphate-buffered saline, scraped and pelleted. The preparation of chromatin and all subsequent steps of ChIRP (for analysis of RNA, proteins and DNA) were differentially performed after or excluding the UV crosslinking step, according to Chu et al.39 Sonication allows to obtain a DNA size range of 100–500 bp. In RNase-treated RNA pull-down samples were digested with RNase at 37 °C for 45 min before the preclearing step. Preclearing of chromatin was performed with 50 μl of streptavidin beads/sample (Promega). Probes (100 pmol) were added to 1 mg of precleared chromatin and mixed by end-to-end rotation at 25 °C for 2 h. One hundred microliters of washed/blocked streptavidin magnetic beads (Promega) were added to the samples (30 min). Magnets allowed the capture of beads/biotin probes/RNA/chromatin complexes. Non-denaturing washes were performed according to Zhao et al.55 for the analysis of UV-untreated samples; denaturing washes were performed according to McHugh et al.27 on UV-treated and -untreated samples.
Beads were resuspended in TRIzol (Ambion, Life Technology) to extract RNA. The enrichment of specific RNA transcripts was evaluated by RT–qPCR analysis. Primers are reported in Table 1.
Beads were treated with RNAse A (Sigma-Aldrich, St Louis, MO, USA), RNAse H (New England Biolabs, Ipswich, MA, USA) and DNase I (Sigma-Aldrich) before protein elution in Laemmli. Proteins were analyzed by western blot (see section 'Co-immunoprecipitation').
Beads were treated with RNAse A (Sigma-Aldrich) and RNAse H (New England Biolabs) and then chromatin was treated with Proteinase K (Sigma-Aldrich) at 65 °C for 45 min. DNA was extracted by phenol:chloroform:isoamyl alcohol (Life Technologies) protocol and subjected to qPCR analysis. Primers are reported in Table 3.
ChIP analysis
ChIP analysis was performed as reported previously56 by using rabbit α-Snail (H-130, sc-28199; Santa Cruz Biotechnology Inc.), rabbit α-EZH2 (H-80, sc-25383; Santa Cruz Biotechnology) or the negative control rabbit IgG (Millipore Corp., Bedford, MA, USA). Equal amounts of immunoprecipitated DNA and relative controls were used for qPCR analysis. Primers are reported in Table 3. qPCR analysis of immunoprecipitated samples (IP) and negative control (IgG) were normalized to total chromatin input and expressed as (2(−ΔCt)) x100 (% Input).
ReChIP
After anti-Snail or anti-EZH2 immunoprecipitation, ReChIP experiments were performed as reported previously,57 using the same DNA quantity for each sample. qPCR of the immunoprecipitated fragments was performed using equal amounts of DNA (primer pairs in Table 3). The reaction was performed in the CFX Connect real-time PCR detection system (Bio-Rad Laboratories Inc.). Primer efficiency, relative quantity of each immunoprecipitated fragment with respect to input and standard deviations were determined with CFX Manager software (Bio-Rad Laboratories). Relative quantities of immunoprecipitated samples were normalized with respect to each input and expressed as (2(−ΔCt)) x100 (% Input).
Histones ChIP
Histones ChIP analysis was performed by using 5 μg of H3K27me3 antibody (07-449; Millipore Corp.) or rabbit IgG (Millipore Corp.) with magnetic beads (Magna ChIP protein A magnetic beads; 16661; Millipore). After washes, samples were eluted with the elution buffer (TE 1x, sodium dodecyl sulfate 0.5%), treated with RNAse A and with proteinase K. The extracted DNA was used in the qPCR analyses (primers are listed in Table 3). Data were expressed as (2(−ΔCt)) x100 (% Input).
Immunofluorescence analysis
For indirect immunofluorescence analyses, cells were methanol-fixed, permeabilized with 0.2% Triton, incubated with donkey serum (017-000-121; Jackson ImmunoResearch Laboratories, West Grove, PA, USA), treated with mouse α-E-cadherin (610182; BD Biosciences), rabbit α-Snail (38795; Cell Signaling Technology Inc.). Secondary antibodies were: Cy3 donkey anti-mouse IgG (H+L) (715.166.150; Jackson ImmunoResearch Laboratories), Alexa Fluor 488-conjugated (A21206; Invitrogen Molecular Probes, Eugene, OR, USA). Nuclei were stained with DAPI (4',6-diamidino-2-phenylindole; 268298; Calbiochem Merck, Darmstadt, Germany). Images were examined with Nikon Microphot-FXA microscope (Nikon Corporation, Tokyo, Japan) equipped with a CCD camera. Digital images were acquired by Nikon NIS-elements software (Nikon Corporation).
Production of recombinant retroviruses and retroviral infections
BOSC 23 packaging cells were transiently transfected with pLPCX/Snail or with the pLPCX vector (used as a control).29 For retroviral infections, cells were plated 24 h before the infection and then incubated with retroviral supernatants supplemented with 4 μg/ml polybrene for 7 h and re-fed with fresh medium. Infected cells were transfected 24 h after infection with specific siRNAs and analyzed after 48 h.
Statistical analysis
Paired t-test and GraphPad Prism version 5.00 (GraphPad software, San Diego, CA, USA; http://www.graphpad.com) were used. A P-value (P) <0.05 was considered statistically significant (*P<0.05; **P<0.01 and ***P<0.001). Data were obtained from independent experiments (n=5) expressed as means ±s.e.m.
Computational analysis
Human and murine HOTAIR sequences were from RefSeq NCBI i.d.'s NR047517.1 and NR047528.1. Snail and EZH2 sequences were from UniProtKB (human EZH2 i.d.: Q15910; mouse EZH2 i.d.: Q61188; human Snail i.d.: O43623; mouse Snail i.d. Q02085).
Sequence alignment was obtained using EMBOSS Water with default parameters. Data shown in Supplementary Figure 4 were produced using an in-house developed script.
Abbreviations
- EMT:
-
epithelial-to-mesenchymal transition
- TGFβ:
-
transforming growth factor-β
- lncRNAs:
-
long non-coding RNAs
- EZH2:
-
enhancer of zeste homolog 2
- HCC:
-
hepatocellular carcinoma
- PRC2:
-
polycomb-repressive complex 2
- H3K27me3:
-
trimethylation of histone H3 lysine 27
- SRA:
-
steroid receptor RNA activator
- RT–qPCR:
-
reverse transcription and real-time polymerase chain reaction
- qPCR:
-
real-time polymerase chain reaction
- ChIP:
-
chromatin immunoprecipitation
- ChIRP:
-
chromatin isolation by RNA purification
- RIP:
-
RNA immunoprecipitation.
References
Thiery JP, Acloque H, Huang RY, Nieto MA . Epithelial–mesenchymal transitions in development and disease. Cell 2009; 139: 871–890.
Nieto MA . The snail superfamily of zinc-finger transcription factors. Nat Rev 2002; 3: 155–166.
Peinado H, Olmeda D, Cano A . Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 2007; 7: 415–428.
Perez-Moreno MA, Locascio A, Rodrigo I, Dhondt G, Portillo F, Nieto MA et al. A new role for E12/E47 in the repression of E-cadherin expression and epithelial–mesenchymal transitions. J Biol Chem 2001; 276: 27424–27431.
Rodriguez M, Aladowicz E, Lanfrancone L, Goding CR . Tbx3 represses E-cadherin expression and enhances melanoma invasiveness. Cancer Res 2008; 68: 7872–7881.
Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004; 117: 927–939.
Javaid S, Zhang J, Anderssen E, Black JC, Wittner BS, Tajima K et al. Dynamic chromatin modification sustains epithelial-mesenchymal transition following inducible expression of Snail-1. Cell Rep 2013; 5: 1679–1689.
McDonald OG, Wu H, Timp W, Doi A, Feinberg AP . Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struct Mol Biol 2011; 18: 867–874.
Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J 2010; 29: 1803–1816.
Hou Z, Peng H, Ayyanathan K, Yan KP, Langer EM, Longmore GD et al. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol 2008; 28: 3198–3207.
Peinado H, Ballestar E, Esteller M, Cano A . Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol 2004; 24: 306–319.
Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002; 298: 1039–1043.
Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D . Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of zeste protein. Genes Dev 2002; 16: 2893–2905.
Deb G, Singh AK, Gupta S . EZH2: not EZHY (easy) to deal. Mol Cancer Res 2014; 12: 639–653.
Simon JA, Kingston RE . Occupying chromatin: polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 2013; 49: 808–824.
Courel M, Friesenhahn L, Lees JA . E2f6 and Bmi1 cooperate in axial skeletal development. Dev Dyn 2008; 237: 1232–1242.
Yu M, Mazor T, Huang H, Huang HT, Kathrein KL, Woo AJ et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Cell 2012; 45: 330–343.
Chen J, Xu H, Zou X, Wang J, Zhu Y, Chen H et al. Snail recruits Ring1B to mediate transcriptional repression and cell migration in pancreatic cancer cells. Cancer Res 2014; 74: 4353–4363.
Tien CL, Jones A, Wang H, Gerigk M, Nozell S, Chang C . Snail2/Slug cooperates with Polycomb repressive complex 2 (PRC2) to regulate neural crest development. Development 2015; 142: 722–731.
Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007; 129: 1311–1323.
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071–1076.
Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329: 689–693.
Davidovich C, Zheng L, Goodrich KJ, Cech TR . Promiscuous RNA binding by polycomb repressive complex 2. Nat Struct Mol Biol 2013; 20: 1250–1257.
Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT . Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell 2014; 55: 171–185.
Geng YJ, Xie SL, Li Q, Ma J, Wang GY . Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res 2011; 39: 2119–2128.
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol 2011; 18: 1243–1250.
McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015; 521: 232–236.
Santangelo L, Marchetti A, Cicchini C, Conigliaro A, Conti B, Mancone C et al. The stable repression of mesenchymal program is required for hepatocyte identity: a novel role for hepatocyte nuclear factor 4alpha. Hepatology 2011; 53: 2063–2074.
Cicchini C, Filippini D, Coen S, Marchetti A, Cavallari C, Laudadio I et al. Snail controls differentiation of hepatocytes by repressing HNF4alpha expression. J Cell Physiol 2006; 209: 230–238.
Cicchini C, Laudadio I, Citarella F, Corazzari M, Steindler C, Conigliaro A et al. TGFbeta-induced EMT requires focal adhesion kinase (FAK) signaling. Exp Cell Res 2008; 314: 143–152.
Cicchini C, de Nonno V, Battistelli C, Cozzolino AM, De Santis Puzzonia M, Ciafrè SA et al. Epigenetic control of EMT/MET dynamics: HNF4a impacts DNMT3s through miRs-29. Biochim Biophys Acta 2015; 1849: 919–929.
Garibaldi F, Cicchini C, Conigliaro A, Santangelo L, Cozzolino AM, Grassi G et al. An epistatic mini-circuitry between the transcription factors Snail and HNF4alpha controls liver stem cell and hepatocyte features exhorting opposite regulation on stemness-inhibiting microRNAs. Cell Death Differ 2012; 19: 937–946.
Lamouille S, Xu J, Derynck R . Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol 2014; 15: 178–196.
Huang RY, Guilford P, Thiery JP . Early events in cell adhesion and polarity during epithelial–mesenchymal transition. J Cell Sci 2012; 125: 4417–4422.
Li L, Liu B, Wapinski OL, Tsai MC, Qu K, Zhang J et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep 2013; 5: 3–12.
Wongtrakoongate P, Riddick G, Fucharoen S, Felsenfeld G . Association of the long non-coding RNA steroid receptor RNA activator (SRA) with TrxG and PRC2 complexes. PLoS Genet 2015; 11: e1005615.
Chu C, Qu K, Zhong FL, Artandi SE, Chang HY . Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 2011; 44: 667–678.
Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 2009; 106: 11667–11672.
Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R . PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol 2013; 20: 1258–1264.
Kretz M, Meister G . RNA binding of PRC2: promiscuous or well ordered? Mol Cell 2014; 55: 157–158.
Arnold P, Scholer A, Pachkov M, Balwierz PJ, Jorgensen H, Stadler MB et al. Modeling of epigenome dynamics identifies transcription factors that mediate Polycomb targeting. Genome Res 2013; 23: 60–73.
Fujii S, Ochiai A . Enhancer of zeste homolog 2 downregulates E-cadherin by mediating histone H3 methylation in gastric cancer cells. Cancer Sci 2008; 99: 738–746.
Cao Q, Yu J, Dhanasekaran SM, Kim JH, Mani RS, Tomlins SA et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 2008; 27: 7274–7284.
Nolan KD, Franco OE, Hance MW, Hayward SW, Isaacs JS . Tumor secreted Hsp90 subverts polycomb function to drive prostate tumor growth and invasion. J Biol Chem 2015; 290: 8271–8282.
Tong ZT, Cai MY, Wang XG, Kong LL, Mai SJ, Liu YH et al. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene 2012; 31: 583–594.
Yonemitsu Y, Imazeki F, Chiba T, Fukai K, Nagai Y, Miyagi S et al. Distinct expression of polycomb group proteins EZH2 and BMI1 in hepatocellular carcinoma. Hum Pathol 2009; 40: 1304–1311.
Chen Y, Lin MC, Yao H, Wang H, Zhang AQ, Yu J et al. Lentivirus-mediated RNA interference targeting enhancer of zeste homolog 2 inhibits hepatocellular carcinoma growth through down-regulation of stathmin. Hepatology 2007; 46: 200–208.
Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011; 71: 6320–6326.
Nie Y, Liu X, Qu S, Song E, Zou H, Gong C . Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci 2013; 104: 458–464.
Bolotin E, Liao H, Ta TC, Yang C, Hwang-Verslues W, Evans JR et al. Integrated approach for the identification of human hepatocyte nuclear factor 4alpha target genes using protein binding microarrays. Hepatology 2010; 51: 642–653.
Cicchini C, Amicone L, Alonzi T, Marchetti A, Mancone C, Tripodi M . Molecular mechanisms controlling the phenotype and the EMT/MET dynamics of hepatocyte. Liver Int 2015; 35: 302–310.
Amicone L, Spagnoli FM, Späth G, Giordano S, Tommasini C, Bernardini S et al. Transgenic expression in the liver of truncated Met blocks apoptosis and permits immortalization of hepatocytes. EMBO J 1997; 16: 495–503.
Schmittgen TD, Zakrajsek BA . Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT–PCR. J Biochem Biophys Methods 2000; 46: 69–81.
Dahm GM, Gubin MM, Magee JD, Techasintana P, Calaluce R, Atasoy U . Method for the Isolation and Identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts using RIPChip. J Vis Exp 2012; 67: 3851.
Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT . Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008; 322: 750–756.
Colletti M, Cicchini C, Conigliaro A, Santangelo L, Alonzi T, Pasquini E et al. Convergence of Wnt signaling on the HNF4alpha-driven transcription in controlling liver zonation. Gastroenterology 2009; 137: 660–672.
Battistelli C, Busanello A, Maione R . Functional interplay between MyoD and CTCF in regulating long-range chromatin interactions during differentiation. J Cell Sci 2014; 127: 3757–3767.
Acknowledgements
We thank I Bozzoni and SA Ciafrè for suggestions and critical revision of the manuscript. This was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC) IG 14114; Ministero della Salute; Ministero Università e Ricerca Scientifica PRIN 20108XYHJS; Epigenomics Flagship Project-EPIGEN.
Author contributions
CB participated to the experimental design and the interpretation of results, performed cell cultures and molecular analysis experiments; CC designed the research plan, coordinated the experimental work, interpreted the results and wrote the manuscript; LS performed cell cultures and molecular analysis experiments; AT and LG coordinated and performed bioinformatics analysis; VdN performed molecular analysis; GG performed immunofluorescence analysis; LA and FJG contributed to the critical revision; MT designed the research plan, coordinated the experimental work, interpreted the results and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Battistelli, C., Cicchini, C., Santangelo, L. et al. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene 36, 942–955 (2017). https://doi.org/10.1038/onc.2016.260
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.260
This article is cited by
-
Generation of glioblastoma in mice engrafted with human cytomegalovirus-infected astrocytes
Cancer Gene Therapy (2024)
-
The lncRNA HOTAIR: a pleiotropic regulator of epithelial cell plasticity
Journal of Experimental & Clinical Cancer Research (2023)
-
Extracellular signal-Regulated Kinase 5 (ERK5) is required for the Yes-associated protein (YAP) co-transcriptional activity
Cell Death & Disease (2023)
-
EZH2-Myc driven glioblastoma elicited by cytomegalovirus infection of human astrocytes
Oncogene (2023)
-
Epigenetic regulation of hybrid epithelial-mesenchymal cell states in cancer
Oncogene (2023)