Abstract
Group3 medulloblastoma (MBG3) that predominantly occur in young children are usually associated with MYC amplification and/or overexpression, frequent metastasis and a dismal prognosis. Physiologically relevant MBG3 models are currently lacking, making inferences related to their cellular origin thus far limited. Using in utero electroporation, we here report that MBG3 mouse models can be developed in situ from different multipotent embryonic cerebellar progenitor cells via conditional expression of Myc and loss of Trp53 function in several Cre driver mouse lines. The Blbp-Cre driver that targets embryonic neural progenitors induced tumors exhibiting a large-cell/anaplastic histopathology adjacent to the fourth ventricle, recapitulating human MBG3. Enforced co-expression of luciferase together with Myc and a dominant-negative form of Trp53 revealed that GABAergic neuronal progenitors as well as cerebellar granule cells give rise to MBG3 with their distinct growth kinetics. Cross-species gene expression analysis revealed that these novel MBG3 models shared molecular characteristics with human MBG3, irrespective of their cellular origin. We here developed MBG3 mouse models in their physiological environment and we show that oncogenic insults drive this MB subgroup in different cerebellar lineages rather than in a specific cell of origin.
Similar content being viewed by others
Introduction
Medulloblastoma (MB), a malignant embryonal brain tumor with a peak incidence in childhood, exhibits considerable molecular heterogeneity with the existence of at least four distinct subgroups—Wingless (WNT), Sonic Hedgehog (SHH), Group3 (G3) and Group4 (G4).1, 2 These subgroups have distinct characteristics with respect to age, gender, prognosis and response to therapy.3 The genetic and phenotypic differences of MB subgroups are in part attributable to differences in their cellular origin.4 Identification of the cellular origin of a tumor often relies on the molecular characterization of normal and tumor tissues; however, tumor-specific somatic alterations likely confound such analysis. Thus, the generation of genetically engineered animals bearing tumors arisen under physiological conditions provides a context for investigating oncogenic events underlying the transformation of a normal cell into a tumor cell in vivo.
Spatial and temporal specification of cell fate within the cerebellum is well characterized,5 and has been integral to the identification of molecular signatures associated with cerebellar neuronal subtypes along the spectrum of neurogenesis.6 These neuronal lineages provide a means to assess the effect of genetic modifications (e.g. activation of oncogenes and deletion of tumor suppressor genes) on a specific neuronal sub-population, and thus to develop genetically engineered MB mouse models.7 A number of studies have uncovered distinct cells of origin for MB subgroups: genetic ablation of Ptch1 enables descendants of cerebellar stem cells (e.g. granule neuron precursors (GNPs) and Nestin-positive (+) cells) to form SHH MBs (MBSHH),8, 9, 10 whereas expression of a constitutively active mutant form of Ctnnb1 in dorsal hindbrain progenitors, generates WNT MBs (MBWNT).11 In contrast, forced activation of WNT signaling in GNPs impairs their proliferation and induces differentiation,12, 13, 14 which highlights the differences in cancer susceptibility among neural progenitors under the influence of the same oncogenic insult. Enforced expression of MYCN under the glutamate transporter 1 (Glt1) promoter or in neonatal cells positive for the glial fibrillary acidic protein (GFAP+) induces MBs with high expression of Kcna1, a known marker of G4 MBs (MBG4),15 whereas MYCN overexpression in Trp53-null GNPs or embryonic cerebellar stem cells triggers MBSHH.15, 16, 17 These studies demonstrate that genetic insults to specific cerebellar cell types can influence the subgroup-specific characteristics of MBs.
MYC-driven MBs are mainly classified as G3 (MBG3) and represent one of the most aggressive subgroup.2, 3 They are characterized by frequent metastasis at diagnosis and are often associated with a dismal outcome.18 So far, several orthotopic models of MYC-driven MBG3 were developed in mice by Myc and Trp53 dysregulation.12, 16, 19 We and another group independently reported the first orthotopic mouse model of MBG3 by overexpressing Myc in combination with functional loss of Trp53 in postnatal cerebellar cells selected by fluorescence-activated cell sorting for the basic helix–loop–helix transcription factor Atoh1-positive (Atoh1+) and Prominin/CD133-positive (Prom1+) cells.12, 16 Nevertheless, while human MBG3 have been found in relatively young children,20 the role of Myc expression in transforming embryonic stem/progenitors into malignant cells under physiological conditions has not yet been tested. We here report the first MBG3 model from embryonic cerebellar cells by Myc activation and loss of Trp53 function using in utero electroporation (EP)-based in vivo gene transfer combined to a Cre/LoxP-mediated technology. The present study also provides the opportunity to trace tumor growth with bioluminescence and fluorescent proteins, which will help in the future to not only understand cellular and molecular mechanisms of tumorigenesis but also to undertake further preclinical trials.
Results
EP of embryonic cerebellar neuroepithelium with Myc and Trp53DN induces MBG3
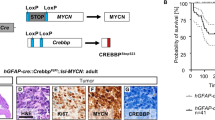
Human MBG3 is typically restricted to infants and younger children,20 prompting us to assess the predisposition of embryonic cerebellar progenitors to initiate MBG3. Previous orthotopic MBG3 models from postnatal progenitors required loss of Trp53 function to overcome cell death caused by excess Myc expression.12 Consistent with these results, the oncogenic transcriptional repressor growth factor independent 1, known to antagonize Trp53,21 was subsequently shown to cooperate with Myc to induce MBG3 in an orthotopic transplant model.22 Indeed, MYC-TP53 dysfunction has been reported in relapsed human MBG3.23 We reasoned that we could use Myc expression and a dominant-negative form of Trp53 to model MBG3 using embryonic cells. In vivo EP of plasmids into mouse embryos is an effective method to transfer genes into cerebellar progenitors under physiological conditions.11, 24 To avoid dilution of plasmid-driven gene expression as cells divide, we used the Tol2 transposon-mediated genomic integration system.25 We constructed two Tol2 cis-flanked CAG promoter-driven plasmids: Tol2-CAG-LoxP-DsRed2-LoxP (LRL)-Myc-IRES-Luciferase (Luc)-Tol2 (pT2K LRL-Myc-IRES-Luc) that strongly express both Myc and Luc after excision of the LRL cassette in the presence of the Cre recombinase protein (Figures 1a and b) and Tol2-CAG-Trp53DN-copGFP-Tol2, encoding a dominant-negative form of Trp53 (Trp53DN)26 fused to a T2A peptide with arthropod-derived green fluorescent protein (copGFP) (pT2K Trp53DN-copGFP). To target broadly cerebellar neural stem/progenitor cells, as the first step these plasmids were electroporated into brain lipid-binding protein (Blbp)-Cre-IRES-nuclear localized LacZ (nlacZ) embryos at embryonic (E) day 13.5 together with the Tol2 transposase (T2TP)-carrying plasmid (pCAGGS T2TP) that lacks Tol2 cis elements to prevent multiple gene ‘hopping’ (Figure 1c). At E13.5, Cre expression was restricted to the cerebellar ventricular zone (VZ) and the external granule layer (EGL), the two germinal zones of the cerebellum (Supplementary Figures 1a and b′). Lineage tracing analysis in [Blbp-Cre-IRES-nlacZ; R26-LSL-EYFP] mice identified broadly distributed enhanced yellow fluorescence protein-positive (EYFP+) cells at P7 (Supplementary Figures 1c and d). While Calb1+ Purkinje cells lack EYFP (Supplementary Figure 1e), the great majority of cerebellar cells, including inhibitory cerebellar interneuron progenitors (Pax2+), granule cells (Pax6+) as well as Bergmann glia and multipotent progenitors (Sox2+), expressed EYFP (Supplementary Figures 1f–h). We electroporated the LRL-EGFP gene27 into the cerebellar neuroepithelium of [Blbp-Cre-IRES-nlacZ] embryos by EP at E13.5 to confirm that Cre-mediated recombination of transfected plasmids occurred in vivo. Two days after EP (E15.5), most of the labeled cells expressed enhanced green fluorescent protein (EGFP) rather than DsRed2 (Supplementary Figures 1i and j), which is consistent with the distribution of Cre+ cells revealed by X-gal staining (Supplementary Figure 1k). We also observed a small population of EGFP+ cells in the choroid plexus (CP) (Supplementary Figure 1j). Immunohistochemistry (IHC) revealed successful recombination of the transfected genes within Pax6+ cells in the EGL, Sox2+ cells in the VZ and Pax2+ cells migrating inwards deep into the cerebellum (Supplementary Figures 1l–n).
Generation of MBG3 using in utero EP-based gene transfer approach. (a) Schematic representation of conditional transcriptional activation of Myc and Luc in the presence of the Cre recombinase. (b) Immunoblotting with antibodies against Myc and Luc of protein lysates from HEK293T cells untransfected (lane 1) or transfected with Myc-IRES-Luc (lane 2) or LRL-Myc-IRES-Luc with (lane 3) or without (lane 4) the Cre expression vector. Note: Luc expression is regulated downstream of the internal ribosomal entry site (IRES) and expressed at low levels in the absence of Cre. (c) Schematic representation of a sagittal section of embryonic hindbrain at E13.5, when in utero EP was performed. Cre was expressed in the EGL and VZ of [Blbp-Cre-IRES-nlacZ] mice. Plasmid DNAs were injected into the fourth ventricle and transfected into cells by delivering electric pulses through the ventricule/hindbrain. (d) Bioluminescence imaging of 2-week-old [Blbp-Cre-IRES-nlacZ] or wild-type (WT) mice into which LRL-Myc-IRES-Luc, Trp53DN-EGFP and T2TP were transduced by EP. (e) Serial bioluminescence imaging of a single representative animal bearing MB at a few week intervals after birth. (f) Kaplan–Meier survival curve of animals subjected to EP with Myc and Trp53DN (n=8/8, red curve), and Trp53DN alone (n=0/6, blue curve). (g) Representative H&E-stained section of Blbp-MYC MB. Inset (white rectangle) shows high magnification. Scale bar=300 and 50 μm (for insets). (h–k) H&E staining (h), IHC with antibodies against Luc and Ki67 (i) and Npr3 (j) and copGFP expression (k) in a Blbp-MYC MB. Scale bar=50μm. (l) Heat map of subgroup-specific signature genes across mouse models of MBWNT and MBSHH, GNPs and Blbp-MYC MB.
We next performed in utero EP of LRL-Myc-IRES-Luc and T2TP with or without Trp53DN-copGFP in [Blbp-Cre-IRES-nlacZ] embryos at E13.5 and subsequently monitored Luc activation in neonatal mice by in vivo bioluminescence. Strong Luc signal was detected in the head of Cre-carrying, but not wild-type (Cre-negative) mice (Figure 1d). In the absence of Trp53DN-copGFP, the bioluminescence signal disappeared by 5 weeks after birth and no MBs developed within 5 months after birth (n=6, Supplementary Figure 2a). These data indicate that overexpression of Myc alone is insufficient to induce MBs in vivo. Intriguingly, in one out of six cases, a CP carcinoma (CPC) was induced by Myc overexpression in the absence of Trp53 loss (Supplementary Figure 2b), which is consistent with the activation of EGFP expression downstream of LRL in electroporated CP cells (Supplementary Figure 1j). In contrast, triple EP of LRL-Myc-IRES-Luc, Trp53DN-copGFP and T2TP resulted in cerebellar tumors with 100% penetrance. Luc signals increased steadily (Figure 1e) and all mice were killed within 8 weeks of birth due to progressive tumor burden (Figure 1f). Six out of 14 mice developed only CPCs in the fourth ventricle (Supplementary Figures 2b–d and Table 1). All the other tumors were diagnosed as MBs with large-cell anaplastic characteristics (hereafter called Blbp-MYC MB) (Figures 1e–h). Tumors were highly proliferative (Ki67+) with MBG3 characteristics, namely Npr3 expression (Figures 1i and j). Importantly, tumor cells strongly expressed copGFP (Figure 1k), indicating that Trp53 inactivation is a prerequisite of MBG3 formation. EP-induced Blbp-MYC MBs also showed strong expression of known stem cell marker genes (Figure 1l). These tumors displayed gene expression profiles for the most part different from Myc-driven CPCs; however, some genes were shared with MBG3 (e.g. Npr3) (Supplementary Figures 2e and f). Hematoxylin and eosin (H&E)-stained sections confirmed leptomeningeal metastasis of the tumors (Supplementary Figures 3a and b). The luciferase signal expanded from the hindbrain to the cerebral cortex as tumors developed, invading the olfactory bulb (Supplementary Figures 3c–h), implying a high tendency of Blbp-MYC MBs for metastatic dissemination as reported for human MBG3.1 No tumors developed without Myc overexpression (Figure 1f), excluding the possibility that Blbp-MYC MBs were accidentally caused by transposition of Tol2-flanked transgenes, rather than due to Myc oncogenic properties. Thus, in vivo EP generated MBs from Blbp+ neural stem/progenitor cells under physiological conditions.
Location of in utero EP-engineered murine MBG3 was similar to their human counterparts
In a subset of patients enrolled in the SJCRH protocol, SJMB03, we measured the location of MBG3 in magnetic resonance imaging (MRI)-acquired images and compared this localization to in utero EP-engineered murine tumors (Figure 2). All human MBG3 tumors were located near the midline in the fourth ventricle, and the median anterior–posterior location was distinct from MBWNT and MBSHH, but not MBG4 (Figures 2a–d). MBWNT tumors clustered proximal to MBG3, reflecting an origin from hindbrain progenitors in the brain stem. MBSHH tumors were dispersed distal to MBG3 in the cerebellar hemispheres, reflecting their origin from GNPs in the EGL.11 The intermediate distribution of human MBG3 tumors is consistent with an origin in the VZ or upper rhombic lip, with subsequent growth into the ventricle as observed in the Blbp-MYC mouse MBG3 (Supplementary Figures 1g and 2e–g). MRI on two of the eight tumors generated showed that the location and in vivo growth pattern of the Blbp-MYC MBG3 were remarkably similar to a typical human MBG3 (Figures 2h–m).
Localization of the Blbp-MYC mouse model and human MBG3. (a–d) Tumor-center location of human G3 (n=16, yellow) tumors in the standard atlas space, plotted along with SHH (n=22, red), WNT (n=13, blue) and G4 (n=19, green) tumors. Data are shown on atlas planes at the approximate center of the MBG3 tumor distribution: (a) coronal at y=−56, (b) sagittal at x=5 and (c) transverse at z=−35. Coordinate units are mm. The anterior–posterior location (vertical axis in c) varied with subtype (d, P<10−5) and G3 location was distinct from SHH (P=0.0005) and WNT (P=0.008), but not G4 (P=0.63). (e–g) Heat map of Blbp-MYC MB tumors (n=6). Tumors were outlined on representative H&E-stained sections (e and f) and overlayed on a parasagittal plane at x=0.12 mm (g) from the Franklin and Paxino brain atlas.28 Coordinates units are mm relative to Bregma, and color indicates cumulative number of tumors. The rostral–caudal orientation of the atlas slice is reversed for comparison with the human atlas. (h–m) Representative MRI imaging of 4-year-old human MBG3 patient (h–j) and 57-day-old mouse bearing a tumor (k–m). Colored dots highlight points for comparison of the anatomic similarity of tumor localization that was apparent in 2/6 Blbp-MYC MB studied with MRI.
Myc overexpression and functional loss of Trp53 in early neural progenitors induced hyperproliferation proximal to the cerebellar VZ
EP-based murine MB models enabled us to investigate early developmental phases of Blbp-MYC MBs and provided more insight into the cellular origin of these tumors. By P5, we observed accumulation of proliferating (Ki67+) cells electroporated with Myc (Luc+) and Trp53DN-copGFP proximal to the fourth ventricle including upper rhombic lip and cerebellar VZ of Cre-carrying animals (Figures 3a–g′). In contrast, we showed that EP with Trp53DN alone did not ectopically induce any Ki67+ clusters in copGFP+ cells (Figures 3h and i). Intriguingly, the lesions from electroporated cells were Pax6+ and negative for both Sox2 and Pax2 (Figures 3j–n′). This suggested that the hyperplasia arose from Pax6+ GNPs. Indeed, proliferating copGFP+ cells frequently accumulated in the upper rhombic lip (Supplementary Figures 4a and b) and aberrantly migrated inwardly from the EGL of the posterior cerebellum (Supplementary Figures 4c–f). The lesions were immunoreactive for Atoh1 (Figures 3o–q″), a marker of proliferating GNPs,5 but not Tbr2 (Figures 3r–t″), a marker for unipolar blush cells in the developing cerebellum.29 Deep cerebellar nuclear neurons, which are also cerebellar excitatory neurons, are born and leave from the VZ at an earlier time point (E10.5–12.5) than when EP was performed (E13.5).29 Furthermore, EP in the cerebella of Atoh1-Cre mice developed copGFP+ hyperproliferative clusters in neonatal mice (Supplementary Figures 4g and h). Consistent with the results of GNP-derived Atoh1ER-MYC orthotopic models described below, GNPs are most likely responsible for the development of hyperplastic lesions in this EP-based model.
Hyperplasia in early development of Blbp-MYC MBs. (a–h) Posterior region of P1 [Blbp-Cre-IRES-nlacZ] cerebellum electroporated with LRL-Myc-IRES-Luc, Trp53DN-copGFP and T2TP genes (a–g and j–n) and with LRL-IRES-Luc, Trp53DN-copGFP and T2TP genes (h). (a) copGFP expression. (b and c) H&E staining on the ipsilateral (electroporated, b) and contralateral (non-electroporated, c) sides. Note: b is the neighboring section of a. Arrow indicates ectopic accumulation of cells on the VZ corresponding to where copGFP-positive cells form a cluster in a. Scale bar in a is 100 μm for a–c. (d–h and j–n′) Confocal microscopy images of the hyperplasia stained with antibodies against Luc (d–e′), Ki67 (f–h), Pax2 (j), Pax6 (k–l′) and Sox2 (m-n′). GFP+ cells are devoid of Sox2 expression in the margin of the VZ (arrowheads in n′). (e′, g′, l′ and n′) High magnification view of the area outlined by white squares in (e, g′ l and n), respectively. Scale bar in j=100 μm for (e, f, h, j, k and m) and scale bar in n′=25 μm for (e′, g′, l ′ and n′). (i) Quantification of the percentage of Ki67+ and Pax6+ cells in copGFP+ cells distributed in the IX and X lobules of neonatal cerebella (n=3). The bars represent normalized mean value±s.d. (o–t‴) Confocal microscopy images the most posterior region of of P4 [Blbp-Cre-IRES-nlacZ] cerebellum electroporated with LRL-Myc-IRES-Luc, Trp53DN-copGFP and T2TP genes. (q–q‴) and (t′–t‴) are high-power view of the area q and t, respectively. (o–s) GFP expression in sagittal sections of the cerebellum counterstained with 4',6-diamidino-2-phenylindole (DAPI). IHC with Atoh1 (p) and Tbr2 (s) antibodies. Scale bar=100 μm in (p and s). Scale bar=25 μm in (q‴ and t‴). Cerebellar surface is outlined by dashed lines.
Cells committed to different types of cerebellar progenitors are susceptible to transformation by Myc overexpression with Trp53DN
[Blbp-Cre-IRES-nlacZ] mice targeted Pax2+ inhibitory interneuron progenitors as well as GNPs (Supplementary Figures 1h and n). Nevertheless, they might not allow proper evaluation of the possibility that Myc overexpression and loss of Trp53 function could transform committed inhibitory interneuron progenitors due to rapid growth of MBG3 from GNPs. We therefore used pancreas specific transcription factor 1a [(Ptf1a)-Cre] and glutamic acid decarboxylase 2 [(Gad2)-IRES-Cre] mice that specifically target committed inhibitory neuron progenitors. In parallel, [Atoh1-Cre] mice were used to control for neuronal subtype since Cre is expressed in committed excitatory neuron progenitors, including GNPs. Targeted gene expression/disruption within specific cerebellar cell populations are well characterized in [Atoh1-Cre] and [Ptf1a-Cre] mice,8, 11, 30 whereas those in [Gad2-IRES-Cre] animals are less well characterized. EP of the LRL-EGFP plasmid into E13.5 [Gad2-IRES-Cre] embryos confirmed that GFP+ cells were Pax2+ (Supplementary Figures 5a–f‴), but negative for PCNA and Pax6 (Supplementary Figures 5g–j‴) at E15.5. Furthermore, lineage-tracing analysis using the [R26-LSL-tdTomato] strain confirmed strong expression of tdTomato in Purkinje cells (Calb1+/Parv+) and cerebellar inhibitory interneurons (Parv+) (Supplementary Figures 5k–l‴), but not Bergmann glia (Sox2+) and granule cells (Pax6+) (Supplementary Figures 5m–n‴) in the adult mice. Thus, the [Gad2-IRES-Cre] and [Ptf1a-Cre] mice can be used to target cerebellar inhibitory neuron progenitors.
Triple EP with LRL-Myc-IRES-Luc, Trp53DN-copGFP and T2TP plasmids was performed in the embryonic cerebellum (E13.5) of the three Cre-expressing mouse lines: [Atoh1-Cre], [Gad2-IRES-Cre] and [Ptf1a-Cre]. Although one animal developed MB 44 days after birth (Figures 4a and b; hereafter called Atoh1-MYC), the remaining two [Atoh1-Cre]-transfected mice developed CPCs by 4 weeks old, which is consistent with expression of the Cre recombinase in CP cells as well as GNPs (Supplementary Figure 5o).11 Unlike Blbp-MYC and Atoh1-MYC MBs, the bioluminescence signal intensity from transfected [Gad2-IRES-Cre] and [Ptf1a-Cre] mice gradually decayed during the first several weeks after birth, but subsequently showed a marked increase in intensity with mice succumbing to tumor burden between 9 and 14 weeks after birth (Figures 4a, c and d; hereafter called Gad2-MYC and Ptf1a-MYC MBs). Intriguingly, peak of Luc signals were detected in several brain regions in mice from both models (Figures 4c and d), implying multifocal tumor development. Histopathological analysis revealed that Ptf1a-MYC and Gad2-MYC MBs, as well as one Atoh1-MYC MB have a large-cell anaplastic MB phenotype and high expression of Npr3 (Figures 4e–j). Interestingly, hyperproliferative lesions were not found in the cerebella of [Gad2-IRES-Cre] and [Ptf1a-Cre] mice, but only in [Atoh1-Cre] neonates (Figures 4k and l). These data suggest that absolute Luc intensity was much lower in [Gad2-Cre] and [Ptf1a-Cre] mice compared with [Blbp-Cre] and [Atoh1-Cre] animals (Figures 1e and 4b–d), possibly because hyperplasia had not been initiated at neonatal stages in these mice. The delay of hyperplasia formation could explain the longer latency of Gad2-MYC and Ptf1a-MYC MBs. High RNA expression of Npr3 and stem cells markers were observed in most of Gad2-MYC and Ptf1a-MYC MBs (Figure 4m), suggesting that overexpression of Myc and Trp53DN induced MBG3s from Ptf1a+/Gad2+ inhibitory neuron progenitors as well as from GNPs targeted by Blbp-Cre animals.
MBG3 from cerebellar inhibitory interneuron progenitors. (a) Kaplan–Meier survival curve for mice developing Atoh1-MYC (n=3/3, red curve), Gad2-MYC (n=4/11, blue curve) and Ptf1a-MYC (n=3/7, green curve) MBs. Atoh1-MYC tumors developed significantly faster than Gad2-MYC MBs (P<0.0001, a two-tailed P-value). (b–d) Serial bioluminescence images of representative (b) glutamatergic (Atoh1-Cre) and (c and d) GABAergic (Gad2-IRES-Cre (c) and Ptf1a-Cre (d)) mice subjected to EP with LRL-Myc-Luc, Trp53DN-copGFP and T2TP. (e–j) Tumors stained with H&E (e, g and i) and an anti-Npr3 antibody (f, h and j). Scale bar=25 μm. (k and l) Distribution of GFP+ cells in electroporated cerebella of P2 Atoh1-Cre (k) and P3 Gad2-IRES-Cre (l) animals. (sagittal sections, scale bar=100 μm). (m) Heat maps of subgroup-specific genes differentially expressed across EP-based MBG3 models.
EP-induced Myc-driven MBs molecularly recapitulate human MBG3
Using different Cre drivers, we generated EP-based MBG3 models from different cell types (Table 1). We compared gene expression profiles of these tumors with orthotopic Myc-driven MBG3 models engineered from postnatal cerebellar progenitors. We here developed [Atoh1-CreER; Trp53Fl/−] and [Prom1-CreER; Trp53Fl/−] mice that lacked one allele of Trp53 in the germline. Mouse pups were administered tamoxifen at P0 and P1 to genetically delete the LoxP-flanked Trp53 in CreER-expressing cells, then killed at P7. Lineage tracing analysis under the same condition confirmed that Atoh1+ GNPs and Sox2+ are targeted by Atoh1-CreER and Prom1-CreER drivers, respectively (Supplementary Figure 6).
Cerebellar cells were purified by percoll density gradient centrifugation, infected in vitro with retroviruses carrying Myc and red fluorescent protein and transplanted into the cortices of naive recipient nude mice as reported previously.16 Tumors from Atoh1-CreER+ and Prom1-CreER+ cells (hereafter referred to as Atoh1ER-MYC and Prom1ER-MYC MBs, respectively) developed within 3 months after birth (Supplementary Figure 7). These tumors as well as our previous model16 exhibited large-cell anaplastic characteristics and high expression of Npr3, a faithful marker of MBG3 (Supplementary Figure 7).2
Unsupervised hierarchical clustering using the top 1000 signature genes from the 430 v.2 Affymetix microarray chip (Affymetrix, Santa Clara, CA, USA) revealed that EP-based MBG3 and orthotopic MBG3 tumors clustered separately from each other and from published murine MBWNT and MBSHH models (Figure 5a). Consistent with these results, principal component analysis using all genes measured on the arrays revealed that the MBG3 models reported here group together, were similar to our previous orthotopic models16 and were clearly separated from MBWNT and MBSHH models (Figure 5b). Of note, EP-MYC tumors did not form subclustering in a cellular origin-dependent manner (Supplementary Figure 8a). Rather, they were intermingled with each other, suggesting that dysfunction of MYC and TP53 is a strong determinant driving MBG3, overriding their origins. Consistent with this idea, cross-species analysis using orthologs between human and mouse further revealed significant similarity of EP-mediated tumors to human MBG3 (Figure 5c and Supplementary Figures 8b–d). Irrespective of the cell types examined in this study, EP-based tumors generated by aberrant activation of Myc combined with the loss of Trp53 function mimicked human MBG3.
Gene expression analysis of EP-based novel mouse MBG3 models. (a) Unsupervised hierarchical heat map using top 1000 signature genes of murine medulloblastoma. (b) Principal component analysis across murine models and GNPs. (c) Probabilities of subgroup predictions for mouse MB models. Bars represent aggregated prediction probabilities that each mouse model belongs to each MB subgroup, as determined by a random forest classifier that was trained on expression barcodes of human and mouse MB samples. The probabilities that Ptf1a-MYC tumors belong to MBG4 are not significantly higher than Blbp-MYC MBs (P=0.90, n=9, Wilcoxon's rank-sum test). Similarly, Gad2-MYC tumors do not have significantly higher probabilities of belonging to MBG4 compared to Blbp-MYC MBs (P-value=0.48, n=10).
Discussion
Identifying the cellular origin of solid tumors is of utmost importance to better understand the transformation of a normal cell into a cancer cell, especially regarding the lineage dependency driving tumor subtype diversity. The in utero EP-based gene transfer system enabled us to examine the susceptibility of distinct cerebellar progenitors to transformation by Myc overexpression and Trp53 loss into malignant MBG3. Hyperplasia formation in [Blbp-Cre] and [Atoh1-Cre] mice at neonatal stages clearly indicated that cerebellar cellular transformation began during embryogenesis. The expression of Atoh1 but not Sox2 within the hyperplastic cells suggests a commitment of neural stem cells to GNPs before transformation ensues. In addition, the lack of hyperplasia in electroporated [Ptf1a-Cre] and [Gad2-Cre] neonatal cerebella, as well as incomplete penetrance of MBG3 from these animals, implies resistance of inhibitory interneuron progenitors to Myc-induced transformation. Thus, our study suggests that there are critical periods during which distinct neuronal progenitors are susceptible to hyperproliferation and/or transformation in response to the same oncogenic insults.
Tumor localization within the caudal cerebellum of Blbp-Cre animals was consistent with our measurements of human MBG3 tumors around the caudal midline of the fourth ventricle in tight clusters as well as with subgroup-specific categorical descriptions of human MBs based on diagnostic MRI studies.31, 32 We previously showed that quantitative tumor localization within the posterior fossa reflects the molecular and cellular context of tumorigenesis for MBWNT and MBSHH in both mice and human.11 The distribution of human MBG3 tumors suggests that tumors arise from the caudal vermis, which develops from the medial cerebellar primordium in human33 and mouse,34 and postnatal lateral-to-medial migration of postmitotic EGL granule cells that populates the expanding caudal vermis, at least in the mouse.34 Convergence toward the midline of granule cells from an extended segment of the upper rhombic lip might afford opportunities for transformation of MYC-induced cells and help to explain the low variability of MBG3 tumors location in humans.
Our results show that Myc overexpression with functional inactivation of Trp53 developed tumors from multiple cellular origins. However, Myc and Trp53DN expression also induced CPCs from [Blbp-Cre] and [Atoh1-Cre] mice that target CP cells as well as neural cells. Although CP cells partly share the same origin (e.g. Gdf7+ cells) as GNPs,35 most of the CP cells are committed to this lineage at E13.5,36 the time at which the EP was performed. [Ptf1a-Cre] and [Gad2-IRES-Cre] animals that do not target CP cells developed no CPCs in our study. It is most likely that CPCs are generated from the cells committed to CP cells. Collectively, alteration of Myc and Trp53 can induce different tumor types, MB and CPC, in a cell lineage-dependent manner (Figure 6).
Last, this study validated that our approach would be a feasible and versatile method to evaluate roles of oncogene in different cell lineages. We also recently developed EP-based SHH-driven MBs and GBMs by induction of CRISPR/Cas9-mediated somatic mutations.37 Furthermore, we also developed in vivo chronic labeling of CRISPR/Cas9-targeted cells with GFP using conditional Cas9-GFP knock-in animals.38 These EP technologies will further foster the generation of new brain tumor models for preclinical studies and will increase our understanding of the cellular and molecular mechanisms of brain tumor development.
Materials and methods
Animal husbandry
Genetically engineered mice used here were [Blbp-Cre-IRES-nLacZ] (01XM9, NCI), [Atoh1-CreER] (007684, Jax), [Prom1-CreER] (017743, Jax), [Trp53Fl/Fl] (008462, Jax), [R26-LSL-EYFP] (006148, Jax), [R26-LSL-tdTomato] (007908, Jax), [Atoh1-Cre] (011104, Jax), [Gad2-IRES-Cre] (010802, Jax), [Ptf1a-Cre] (RES184, BCBC), Trp53-null (008652, Jax) and [Trp53−/−; Cdkn2c−/−].39 Cre-carrying male were bred with CD-1 female mice (Charles River, Wilmington, MA, USA) for in utero EP. Day E0.5 was defined by the observation of a vaginal plug and the day of birth as P0. Conditional deletion of the Trp53 gene was performed by administration of tamoxifen (Sigma, St Louis, MO, USA) at P0 and P1 (4 mg/40 g body weight). All experiments were conducted in strict accordance with the National Institute of Health guidelines for the Care and Use of Laboratory Animals and according to the guidelines established by the SJCRH Institutional Animal Care and Use Committee. All procedures in the protocol were approved by the Animal Care and Use Committee (ACUC) of SJCRH (Animal Assurance Number: A3077-01) and were approved by the responsible authorities in Germany (G176/13 and G48/14).
Plasmids and virus production
For Tol2 transposon-mediated stable expression, we used pCAGGS T2TP and pT2K LRL-EGFP.25 EGFP of pT2K LRL-EGFP was replaced with either IRES-luc or Myc-IRES-luc, yielding pT2K LRL-IRES-luc and pT2K LRL-Myc-IRES-luc, respectively. The gene encoding Trp53DN26 was fused to the T2A peptide with copGFP (Trp53DN-copGFP) and was inserted into the pT2K-CAGGS plasmid, yielding pT2K Trp53DN-copGFP. Retroviruses were produced as described previously.16
Generation of mouse MBG3 models
To develop EP-based MBG3 models, in utero EP was performed.37 Only the animals showing luciferase signal by P7 were selected for further analysis. Tumor growth was measured every 1–2 week by bioluminescence imaging of luciferase activity using a Xenogen IVIS system (PerkinElmer, Waltham, MA, USA).41 Orthotopic MBG3 mouse models were generated by cranial implants of purified GNPs from [Trp53−/−; Cdkn2c−/−], [Trp53Fl/-; Atoh1-CreER], [Trp53Fl/-; Prom1-CreER] and [Trp53Fl/-] P6–7 pups, infected with retroviruses carrying Myc and red fluorescent protein according to the previous study.16 No specific randomization or blinding was performed. Tumor cell purification and genomic DNA and total RNA extraction for genotyping and Affymetrix microarray analysis were described previously.16 For detection of non-recombined and recombined alleles of Trp53 in Atoh1ER-MYC and Prom1ER-MYC MBs, 1F/1R (370 bp) and 1F/10R (612 bp) primer sets were used, respectively.42 For internal control, endogenous Prom1 (563 bp)43 and Ptch1 (220 bp)17 were detected. Comparison of survival curves was performed by calculation of two-tailed P-value using the GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA, USA).
Gene expression analysis
Human Affymetrix U133 plus 2.0 expression array data for genes of interest were extracted from publicly available data sets (GSE10327, GSE12992, GSE37418 and GSE49243)40, 44, 45, 46 from the R2 software tool for analysis and visualization of genomic data (http://r2.amc.nl) (Amsterdam, The Netherlands). Additional cases came from an unpublished data set generated at the DKFZ (Heidelberg, Germany) (M Kool et al., unpublished data). Total RNAs from murine tumors were extracted and analyzed with the Affymetrix 430 v.2 chip. The microarray data of murine SHH and WNT spontaneous medulloblastoma,11, 39 as well as some of the orthotopic MBG3 models were retrieved from our previous study.16
RNA from four CPC and one MBG3 mouse tumors were extracted and analyzed by mogene 2.0 ST arrays where the MBG3 RNA was a repeated measure of a pre-existing 430 v.2 array. Data were RMA normalized in Partek Genomics Suite 6.6 (St Louis, MO, USA) and imported in STATA/MP 14.2 (StataCorp LLC, College Station, TX, USA) in which unannotated and duplicated probe sets were removed such that only the highest measured probe sets was retained for any gene. The mouse 430 v.2 array data was also deduplicated in the same manner for MBSHH and MBG3 data. These two data sets were then joined by gene symbol and a chip-based correction for each gene was calculated by finding the simple difference in RMA signal in the 430 v.2 sample that matched the mogene 2.0 ST Grp3 sample. Finally, the mogene 2.0 samples data were corrected using this factor and the resulting matrix was principal component analysis visualized. Further selected matched genes were clustered and displayed in a heat map using Partek Genomics Suite 6.6.
Antibodies
All antibodies used in this study are shown in Supplementary Table 1.
Immunohistochemistry
IHC was performed with the antibodies shown in Supplementary Table 2 according to the previous study.16 For quantification of the stained sections (five sections per brain), the number of positive (+) markers (for example, Ki67 and GFP+ cells) in the most posterior lobules (IX and X) were counted and used to calculate the ratio of positive marker (for example, Ki67+) to green fluorescence positive (GFP+) cells. Cell counting was carried out in a double-blind manner whenever applicable.
Immunoblotting
Mycoplasma-free HEK293T cells from American Type Culture Collection (Manassas, VA, USA) were transfected with pT2K-empty (negative control), pT2K Myc-IRES-Luc (positive control), and pT2K LRL-Myc-IRES-Luc with or without MSCV-Cre plasmids and harvested 2 days after transfection. Protein lysates were prepared from the transfected HEK293T cells using standard RIPA buffer (Sigma) and used 10 μg of the lysates for immunoblotting.
Neurosphere culture and differentiation assay
Cerebellar cells from postnatal (P) day P7 [Prom1-CreER; R26-LSL-EYFP] mice treated with tamoxifen at P0 and P1 were purified as published previously,16 followed by fluorescence-activated cell sorting sorting of EYFP expressing cells. Sorted cells were plated in serum-free Neurobasal media supplemented with B27 and N2 (Invitrogen, Carlsbad, CA, USA) (passage 0). Basic fibroblast growth factor (50 ng/ml; Peprotech, Rocky Hill, NJ, USA) and epidermal growth factor (EGF) (50 ng/ml; Peprotech) were added every 3 days and neurospheres were passaged every 7–10 days. After the second passage, cells were dissociated with trypsin and subsequently plated on Matrigel-coated coverslips. For differentiation assays, basic fibroblast growth factor and EGF were removed and platelet-derived growth factor-AA (10 ng/ml; Sigma) was added for 2 weeks. Cultured cells were fixed with 4% paraformaldehyde/phosphate-buffered saline, followed by immunofluorescence.
MRI imaging
In patients, preoperative imaging was acquired at the local hospital where each patient had tumor resection before referral for protocol-based therapy at SJCRH, and analyzed, as reported previously.11 The research protocol was approved by the SJCRH Institutional Review Board and all patients gave written informed consent to participate. The preoperative imaging examination varied among patients, but always included T1-weighted scans before and after administration of a gadolinium-based contrast agent. The slice thickness was 4 or 5 mm and the in-plane resolution was 0.8 to 1 mm. T1-weighted images from all patients were wrapped, or spatially normalized,47 to the standard brain space defined by the Montreal Neurological Institute (MNI) brain template.48, 49 The MNI space approximates the proportional stereotaxic brain space of Talairach and Tournoux,50 which was extended to the cerebellum in the atlas by Schmahmann et al.51 Spatial normalization was performed with Statistical Parametric Mapping software (SPM5; Wellcome Institute of Neurology, London, UK) using default parameters, except that the brain bounding box was extended from 50 to 80 mm in the negative z-direction for sagittal images and all output images were resliced to 1 mm x 1 mm x 1 mm resolution. The T1-weighted images without contrast agent were used for spatial normalization because the tumor contrast was low, which would minimize adverse effects of the tumor on the accuracy of spatial normalization. To characterize the accuracy of spatial normalization we defined a set of landmarks from the MNI space that were identified in the normalized images from each patient. The landmarks and their MNI coordinates (x, y, z) included the center of the anterior commissure (0, 5, −4), the cerebral aqueduct (0, −30, −10), a point on the posterior aspect of the brain stem in the fourth ventricle (0, −34, −26), the apex of the cerebellum in the midline (0, −54, 4) and lateral extremes of the cerebellum on the right (59, −58, −39) and left (−59, −58, −39).51 The locations of landmarks in the normalized images of each patient were measured with the 3D Slicer software (version 3.4, http://www.slicer.org).52
Cross-species analysis
To classify mouse models molecularly, all analyses were performed in the R environment (v.3.1.2) with CRAN and Bioconductor (v.3.0) packages, including GEOquery (v.2.32), frma (v.1.18), sva (v.3.12), randomForest (v.4.6) and ggplot2 (v.1.0). Public expression data of human MBs were collated from previous studies.40, 45, 46, 53, 54 Raw expression data of human MBs on the Affymetrix Human Genome U133 Plus 2.0 array platform (Affymetrix) were retrieved from GEO accessions GSE10327, GSE12992, GSE19404, GSE35493 and GSE37418. This data set was integrated with an expression data set (on the Affymetrix Mouse 430 2.0 array platform) of previously published mouse models of MBWNT, MBSHH and MBG3 (accessions GSE33199 and GSE24628)11, 16 in addition to expression data of mouse models generated in the current study. The raw data was normalized using fRMA,55 transformed into z-scores using Barcode,56 adjusted for center-specific biases and cross-species differences using ComBat, and converted into expression barcodes (non-expressed vs expressed) with a logarithm (base 10) of odds score threshold of 12. The human samples and the mouse samples of previously published models were used to train a random forest classifier.56 The expression barcodes of the data sets were visualized by principal component analysis. The integrated data set (expression barcodes) of human MBs and previously published mouse MB models was used to train a random forest classifier.57 For computational efficiency, genes determined to be non-expressed across all samples were removed before training. The trained classifier was subsequently used to predict the molecular subgroups (WNT, SHH, G3, G4) of each mouse tumor sample in the present study by its expression barcode. For each sample, the distribution of votes across the classes was interpreted as the posterior probability distribution of the sample belonging to each class. The posterior probability of a mouse model belonging to class, given the expression data of samples of the model, was calculated as

which follows from Bayes’ rule and the assumption that samples of the model are conditionally independent.

Accession number
All microarray data has been submitted to the US National Institute of Health GEO database under the accession numbers GSE65888 and GSE33199.
References
Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC et al. The genetic landscape of the childhood cancer medulloblastoma. Science 2011; 331: 435–439.
Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M et al. Dissecting the genomic complexity underlying medulloblastoma. Nature 2012; 488: 100–105.
Northcott PA, Korshunov A, Pfister SM, Taylor MD . The clinical implications of medulloblastoma subgroups. Nat Rev Neurol 2012; 8: 340–351.
Gilbertson RJ, Ellison DW . The origins of medulloblastoma subtypes. Annu Rev Pathol 2008; 3: 341–365.
Roussel MF, Hatten ME . Cerebellum development and medulloblastoma. Curr Top Dev Biol 2011; 94: 235–282.
Dastjerdi FV, Consalez GG, Hawkes R . Pattern formation during development of the embryonic cerebellum. Front Neuroanat 2012; 6: 10.
Wu X, Northcott PA, Croul S, Taylor MD . Mouse models of medulloblastoma. Chin J Cancer 2011; 30: 442–449.
Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell 2008; 14: 123–134.
Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell 2008; 14: 135–145.
Li P, Du F, Yuelling LW, Lin T, Muradimova RE, Tricarico R et al. A population of Nestin-expressing progenitors in the cerebellum exhibits increased tumorigenicity. Nat Neurosci 2013; 16: 1737–1744.
Gibson P, Tong Y, Robinson G, Thompson MC, Currle DS, Eden C et al. Subtypes of medulloblastoma have distinct developmental origins. Nature 2010; 468: 1095–1099.
Pei Y, Moore CE, Wang J, Tewari AK, Eroshkin A, Cho YJ et al. An animal model of MYC-driven medulloblastoma. Cancer Cell 2012; 21: 155–167.
Lorenz A, Deutschmann M, Ahlfeld J, Prix C, Koch A, Smits R et al. Severe alterations of cerebellar cortical development after constitutive activation of Wnt signaling in granule neuron precursors. Mol Cell Biol 2011; 31: 3326–3338.
Poschl J, Grammel D, Dorostkar MM, Kretzschmar HA, Schuller U . Constitutive activation of beta-catenin in neural progenitors results in disrupted proliferation and migration of neurons within the central nervous system. Dev Biol 2013; 374: 319–332.
Swartling FJ, Savov V, Persson AI, Chen J, Hackett CS, Northcott PA et al. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell 2012; 21: 601–613.
Kawauchi D, Robinson G, Uziel T, Gibson P, Rehg J, Gao C et al. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer Cell 2012; 21: 168–180.
Zindy F, Uziel T, Ayrault O, Calabrese C, Valentine M, Rehg JE et al. Genetic alterations in mouse medulloblastomas and generation of tumors de novo from primary cerebellar granule neuron precursors. Cancer Res 2007; 67: 2676–2684.
Roussel MF, Robinson GW . Role of MYC in medulloblastoma. Cold Spring Harb Perspect Med 2013; 3: a014308.
Hanaford AR, Archer TC, Price A, Kahlert UD, Maciaczyk J, Nikkhah G et al. DiSCoVERing Innovative Therapies for rare tumors: combining genetically accurate disease models with in silico analysis to identify novel therapeutic targets. Clin Cancer Res 2016; 22: 3903–3914.
Gajjar AJ, Robinson GW . Medulloblastoma-translating discoveries from the bench to the bedside. Nat Rev Clin Oncol 2014; 11: 714–722.
Khandanpour C, Phelan JD, Vassen L, Schutte J, Chen R, Horman SR et al. Growth factor independence 1 antagonizes a p53-induced DNA damage response pathway in lymphoblastic leukemia. Cancer Cell 2013; 23: 200–214.
Northcott PA, Lee C, Zichner T, Stutz AM, Erkek S, Kawauchi D et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 2014; 511: 428–434.
Hill RM, Kuijper S, Lindsey JC, Petrie K, Schwalbe EC, Barker K et al. Combined MYC and P53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease. Cancer Cell 2015; 27: 72–84.
Kawauchi D, Saito T . Transcriptional cascade from Math1 to Mbh1 and Mbh2 is required for cerebellar granule cell differentiation. Dev Biol 2008; 322: 345–354.
Sato Y, Kasai T, Nakagawa S, Tanabe K, Watanabe T, Kawakami K et al. Stable integration and conditional expression of electroporated transgenes in chicken embryos. Dev Biol 2007; 305: 616–624.
Bowman T, Symonds H, Gu L, Yin C, Oren M, Van Dyke T . Tissue-specific inactivation of p53 tumor suppression in the mouse. Genes Dev 1996; 10: 826–835.
Yokota Y, Saito D, Tadokoro R, Takahashi Y . Genomically integrated transgenes are stably and conditionally expressed in neural crest cell-specific lineages. Dev Biol 2011; 353: 382–395.
Paxinos G, Franklin KB . The Mouse Brain in Stereotaxic Coordinates, 3rd edn. Academic Press: New York, NY, USA, 2008.
Hevner RF, Hodge RD, Daza RA, Englund C . Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci Res 2006; 55: 223–233.
Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV et al. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron 2005; 47: 201–213.
Wefers AK, Warmuth-Metz M, Poschl J, von Bueren AO, Monoranu CM, Seelos K et al. Subgroup-specific localization of human medulloblastoma based on pre-operative MRI. Acta Neuropathol 2014; 127: 931–933.
Perreault S, Ramaswamy V, Achrol AS, Chao K, Liu TT, Shih D et al. MRI surrogates for molecular subgroups of medulloblastoma. Am J Neuroradiol 2014; 35: 1263–1269.
Cho KH, Rodriguez-Vazquez JF, Kim JH, Abe H, Murakami G, Cho BH . Early fetal development of the human cerebellum. Surg Radiol Anat 2011; 33: 523–530.
Sgaier SK, Millet S, Villanueva MP, Berenshteyn F, Song C, Joyner AL . Morphogenetic and cellular movements that shape the mouse cerebellum; insights from genetic fate mapping. Neuron 2005; 45: 27–40.
Cheng FY, Huang X, Sarangi A, Ketova T, Cooper MK, Litingtung Y et al. Widespread contribution of Gdf7 lineage to cerebellar cell types and implications for hedgehog-driven medulloblastoma formation. PLoS One 2012; 7: e35541.
Hunter NL, Dymecki SM . Molecularly and temporally separable lineages form the hindbrain roof plate and contribute differentially to the choroid plexus. Development 2007; 134: 3449–3460.
Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, Zapatka M, Northcott PA, Schramm K et al. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun 2015; 6: 7391.
Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014; 159: 440–455.
Uziel T, Zindy F, Xie S, Lee Y, Forget A, Magdaleno S et al. The tumor suppressors Ink4c and p53 collaborate independently with Patched to suppress medulloblastoma formation. Genes Dev 2005; 19: 2656–2667.
Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One 2008; 3: e3088.
Murphy BL, Obad S, Bihannic L, Ayrault O, Zindy F, Kauppinen S et al. Silencing of the miR-17–92 cluster family inhibits medulloblastoma progression. Cancer Res 2013; 73: 7068–7078.
Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A . Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet 2001; 29: 418–425.
Zhu L, Gibson P, Currle DS, Tong Y, Richardson RJ, Bayazitov IT et al. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature 2009; 457: 603–607.
Kool M, Jones DT, Jager N, Northcott PA, Pugh TJ, Hovestadt V et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 2014; 25: 393–405.
Fattet S, Haberler C, Legoix P, Varlet P, Lellouch-Tubiana A, Lair S et al. Beta-catenin status in paediatric medulloblastomas: correlation of immunohistochemical expression with mutational status, genetic profiles, and clinical characteristics. J Pathol 2009; 218: 86–94.
Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L et al. Novel mutations target distinct subgroups of medulloblastoma. Nature 2012; 488: 43–48.
Ashburner J, Friston KJ . Nonlinear spatial normalization using basis functions. Hum Brain Mapp 1999; 7: 254–266.
Brett M . The MNI brain and the Talairach Atlas Neuroimaging Laboratory: Washington University, St Louis, CA, USA 2005.
Evans AC, Collins DL, Mills SR, Brown ED, Kelly RL, Peters TM 3D Statistical Neuroanatomical Models from 305 MRI Volumes. Proceedings of the IEEE-Nuclear Science Symposium and Medical Imaging Conference. 1993, pp 1813–1817.
Talairach J, Tournoux P . Co-Planar Stereotaxic Atlas of the Human Brain. Thieme: New York, NY, USA, 1988.
Schmahmann JD, Doyon J, Toga AW, Petrides M, Evans AC . MRI Atlas of the Human Cerebellum. Academic Press: San Diego, CA, USA, 2000.
Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S et al. 3D Slicer as an Image Computing Platform for the Quantitative Imaging Network. Magnetic Resonance Imaging 2012; 30: 1323–1341.
Rogers HA, Ward JH, Miller S, Lowe J, Coyle B, Grundy RG . The role of the WNT/beta-catenin pathway in central nervous system primitive neuroectodermal tumours (CNS PNETs). Br J Cancer 2013; 108: 2130–2141.
Birks DK, Donson AM, Patel PR, Sufit A, Algar EM, Dunham C et al. Pediatric rhabdoid tumors of kidney and brain show many differences in gene expression but share dysregulation of cell cycle and epigenetic effector genes. Pediatr Blood Cancer 2013; 60: 1095–1102.
McCall MN, Bolstad BM, Irizarry RA . Frozen robust multiarray analysis (fRMA). Biostatistics 2010; 11: 242–253.
McCall MN, Jaffee HA, Zelisko SJ, Sinha N, Hooiveld G, Irizarry RA et al. The Gene Expression Barcode 3.0: improved data processing and mining tools. Nucleic Acids Res 2014; 42: D938–D943.
Liaw A, Wiener M . Classification and regression by randomForest. R News 2002; 2: 18–22.
Wiener ALaM . Classification and regression by randomForest. R News 2002; 2: 18–2.
Acknowledgements
We thank Drs Y Takahashi (Kyoto University) and K Kawakami (National Institute of Genetics, Japan) for generously providing Tol2 transposon related vectors; L Linke and N Mack (DKFZ), J Grenet, D Farmer, S Wilkerson, S Smith and S Robinson for excellent technical assistance, J Morris and M Loyd for Affymetrix microarray analysis; Drs R Ashmun and A-M Hamilton Easton for fluorescence-activated cell sorting analysis; M Payton and C Winter for in vivo imaging of tumor growth; M Johnson and S Brown for orthotopic transplants; Dr MA Scoggins, L Stokes and A Owens for processing and analysis of human images; Drs O Witt, K Reifenberg, N Denk and K Dell for helpful assistance with animal experiments at DKFZ; Dr C Wright (Vanderbilt University), Y Nakano and M Horiguchi (Kyoto University) for mice; Drs H Liu and W Feng (DKFZ) for sharing R26-LSL-tdTomato mice and helpful discussions; Drs Y Ouyang (SJCRH) and F Bestvater (DKFZ) for Light Microscopy. This work was funded in part by the National Institute of Health Grants CA-096832, CA-21765 (to MFR), HD-049888, RR-029005 (to RJO), the Anderson fellowship and the Deutsche Forschungsgemeinschaft, KA 4472/1-1 (to DK), the Helmholtz Alliance ‘Preclinical Comprehensive Cancer Center’ Grant HA-305 (to PL, JG and SMP) and the American Lebanese Syrian Associated Charities of SJCRH.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Kawauchi, D., Ogg, R., Liu, L. et al. Novel MYC-driven medulloblastoma models from multiple embryonic cerebellar cells. Oncogene 36, 5231–5242 (2017). https://doi.org/10.1038/onc.2017.110
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.110
This article is cited by
-
Heterogeneity and tumoral origin of medulloblastoma in the single-cell era
Oncogene (2024)
-
MYC overexpression and SMARCA4 loss cooperate to drive medulloblastoma formation in mice
Acta Neuropathologica Communications (2023)
-
ARF suppression by MYC but not MYCN confers increased malignancy of aggressive pediatric brain tumors
Nature Communications (2023)
-
Unified rhombic lip origins of group 3 and group 4 medulloblastoma
Nature (2022)
-
Cytotoxic effects and tolerability of gemcitabine and axitinib in a xenograft model for c-myc amplified medulloblastoma
Scientific Reports (2021)