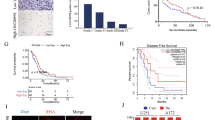
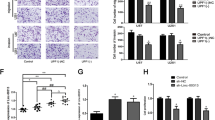
Abstract
Accumulating studies have demonstrated the importance of long noncoding RNAs (lncRNAs) during oncogenic transformation. However, because most lncRNAs are currently uncharacterized, the identification of novel oncogenic lncRNAs is difficult. Given that intergenic lncRNA have substantially less sequence conservation patterns than protein-coding genes across species, evolutionary conserved intergenic lncRNAs are likely to be functional. The current study identified a novel intergenic lncRNA, LINC00461 (ECONEXIN) using a combined approach consisting of searching lncRNAs by evolutionary conservation and validating their expression in a glioma mouse model. ECONEXIN was the most highly conserved intergenic lncRNA containing 83.0% homology with the mouse ortholog (C130071C03Rik) for a region over 2500 bp in length within its exon 3. Expressions of ECONEXIN and C130071C03Rik were significantly upregulated in both human and mouse glioma tissues. Moreover, the expression of C130071C03Rik was upregulated even in precancerous conditions and markedly increased during glioma progression. Functional analysis of ECONEXIN in glioma cell lines, U87 and U251, showed it was dominantly located in the cytoplasm and interacted with miR-411-5p via two binding sites within ECONEXIN. Inhibition of ECONEXIN upregulated miR-411-5p together with the downregulation of its target, Topoisomerase 2 alpha (TOP2A), in glioma cell lines, resulting in decreased cell proliferation. Our data demonstrated that ECONEXIN is a potential oncogene that regulates TOP2A by sponging miR-411-5p in glioma. In addition, our investigative approaches to identify conserved lncRNA and their molecular characterization by validation in mouse tumor models may be useful to functionally annotate novel lncRNAs, especially cancer-associated lncRNAs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet 2015; 47: 458–468.
Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 2016; 164: 550–563.
Wahlestedt C . Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov 2013; 12: 433–446.
Schmitt AM, Chang HY . Long noncoding RNAs in cancer pathways. Cancer Cell 2016; 29: 452–463.
Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011; 477: 295–300.
Ke J, Yao YL, Zheng J, Wang P, Liu YH, Ma J et al. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget 2015; 6: 21934–21949.
Katsushima K, Natsume A, Ohka F, Shinjo K, Hatanaka A, Ichimura N et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat Commun 2016; 7: 13616.
Kapranov P, Willingham AT, Gingeras TR . Genome-wide transcription and the implications for genomic organization. Nat Rev Genet 2007; 8: 413–423.
Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009; 458: 223–227.
Quinn JJ, Zhang QC, Georgiev P, Ilik IA, Akhtar A, Chang HY . Rapid evolutionary turnover underlies conserved lncRNA-genome interactions. Genes Dev 2016; 30: 191–207.
Washietl S, Kellis M, Garber M . Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res 2014; 24: 616–628.
Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I . Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep 2015; 11: 1110–1122.
Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP . Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 2011; 147: 1537–1550.
Kutter C, Watt S, Stefflova K, Wilson MD, Goncalves A, Ponting CP et al. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet 2012; 8: e1002841.
Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011; 25: 1915–1927.
Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell 2011; 146: 209–221.
Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell 2014; 53: 1005–1019.
Oliver PL, Chodroff RA, Gosal A, Edwards B, Cheung AF, Gomez-Rodriguez J et al. Disruption of Visc-2, a brain-expressed conserved long noncoding RNA, does not elicit an overt anatomical or behavioral phenotype. Cereb Cortex 2015; 25: 3572–3585.
Tay Y, Rinn J, Pandolfi PP . The multilayered complexity of ceRNA crosstalk and competition. Nature 2014; 505: 344–352.
Li JH, Liu S, Zhou H, Qu LH, Yang JH . starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014; 42: D92–D97.
Carthew RW, Sontheimer EJ . Origins and mechanisms of miRNAs and siRNAs. Cell 2009; 136: 642–655.
Ma JB, Ye K, Patel DJ . Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 2004; 429: 318–322.
Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2013; 2: e01749.
Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S . The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci 2007; 120: 2498–2506.
Mo CF, Wu FC, Tai KY, Chang WC, Chang KW, Kuo HC et al. Loss of non-coding RNA expression from the DLK1-DIO3 imprinted locus correlates with reduced neural differentiation potential in human embryonic stem cell lines. Stem Cell Res Ther 2015; 6: 1.
Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 2010; 29: 3082–3093.
Moran I, Akerman I, van de Bunt M, Xie R, Benazra M, Nammo T et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab 2012; 16: 435–448.
Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 2016; 44: D110–D115.
Holmberg J, He X, Peredo I, Orrego A, Hesselager G, Ericsson C et al. Activation of neural and pluripotent stem cell signatures correlates with increased malignancy in human glioma. PLoS ONE 2011; 6: e18454.
Labreche K, Simeonova I, Kamoun A, Gleize V, Chubb D, Letouze E et al. TCF12 is mutated in anaplastic oligodendroglioma. Nat Commun 2015; 6: 7207.
Ulitsky I . Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat Rev Genet 2016; 17: 601–614.
Dweep H, Gretz N . miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods 2015; 12: 697.
Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M . Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell 2014; 54: 766–776.
Pommier Y, Leo E, Zhang H, Marchand C . DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol 2010; 17: 421–433.
Chen T, Sun Y, Ji P, Kopetz S, Zhang W . Topoisomerase IIalpha in chromosome instability and personalized cancer therapy. Oncogene 2015; 34: 4019–4031.
Leonard A, Wolff JE . Etoposide improves survival in high-grade glioma: a meta-analysis. Anticancer Res 2013; 33: 3307–3315.
Clark PI, Slevin ML, Joel SP, Osborne RJ, Talbot DI, Johnson PW et al. A randomized trial of two etoposide schedules in small-cell lung cancer: the influence of pharmacokinetics on efficacy and toxicity. J Clin Oncol 1994; 12: 1427–1435.
Mistry AR, Felix CA, Whitmarsh RJ, Mason A, Reiter A, Cassinat B et al. DNA topoisomerase II in therapy-related acute promyelocytic leukemia. N Engl J Med 2005; 352: 1529–1538.
Yang G, Lu X, Yuan L . LncRNA: a link between RNA and cancer. Biochim Biophys Acta 2014; 1839: 1097–1109.
Ideue T, Hino K, Kitao S, Yokoi T, Hirose T . Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. RNA 2009; 15: 1578–1587.
Kim SU . Human neural stem cells genetically modified for brain repair in neurological disorders. Neuropathology 2004; 24: 159–171.
Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L . Mosaic analysis with double markers in mice. Cell 2005; 121: 479–492.
Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012; 22: 1775–1789.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S . MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30: 2725–2729.
Acknowledgements
This study was performed as research programs of the Project for Development of Innovative Research on Cancer Therapeutics (P-Direct), Ministry of Education, Culture, Sports, Science and Technology of Japan (YK), of the Project for Cancer Research And Therapeutic Evolution (P-CREATE) from the Japan Agency for Medical Research and development, AMED (YK), of the PRESTO, JST (YK) and of the Grant-in-Aid for Scientific Research, the Japan Society for the Promotion of Science (25290048, YK).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Deguchi, S., Katsushima, K., Hatanaka, A. et al. Oncogenic effects of evolutionarily conserved noncoding RNA ECONEXIN on gliomagenesis. Oncogene 36, 4629–4640 (2017). https://doi.org/10.1038/onc.2017.88
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.88
This article is cited by
-
LncRNA INHEG promotes glioma stem cell maintenance and tumorigenicity through regulating rRNA 2’-O-methylation
Nature Communications (2023)
-
TUG1-mediated R-loop resolution at microsatellite loci as a prerequisite for cancer cell proliferation
Nature Communications (2023)
-
Involvement of the long intergenic non-coding RNA LINC00461 in schizophrenia
BMC Psychiatry (2022)
-
HDAC6 involves in regulating the lncRNA-microRNA-mRNA network to promote the proliferation of glioblastoma cells
Journal of Experimental & Clinical Cancer Research (2022)
-
HIF-1α-activated long non-coding RNA KDM4A-AS1 promotes hepatocellular carcinoma progression via the miR-411-5p/KPNA2/AKT pathway
Cell Death & Disease (2021)