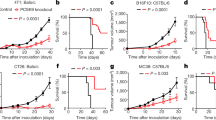
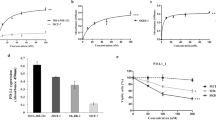
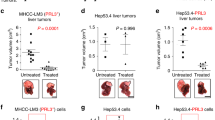
Abstract
Checkpoint blockade therapy targeting the programmed-death ligand 1 (PD-L1) and its receptor programmed cell death 1 promotes T-cell-mediated immunosurveillance against tumours, and has been associated with marked clinical benefit in cancer patients. Antibodies against PD-L1 function by blocking PD-L1 on the cell surface, but intracellular storage of PD-L1 and its active redistribution to the cell membrane can minimize the therapeutic benefits, which highlights the importance of targeting PD-L1 throughout the whole cell. Here, we show that PD-L1 is palmitoylated in its cytoplasmic domain, and that this lipid modification stabilizes PD-L1 by blocking its ubiquitination, consequently suppressing PD-L1 degradation by lysosomes. We identified palmitoyltransferase ZDHHC3 (DHHC3) as the main acyltransferase required for the palmitoylation of PD-L1, and show that the inhibition of PD-L1 palmitoylation via 2-bromopalmitate, or the silencing of DHHC3, activates antitumour immunity in vitro and in mice bearing MC38 tumour cells. We also designed a competitive inhibitor of PD-L1 palmitoylation that decreases PD-L1 expression in tumour cells to enhance T-cell immunity against the tumours. These findings suggest new strategies for overcoming PD-L1-mediated immune evasion in cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary Information files.
Change history
11 April 2019
In the version of this Article originally published, ‘palmitoyltransferase ZDHHC3 (DHHC3)’ was incorrectly referred to as an ‘acetyltransferase’ rather than an as an ‘acyltransferase’; this has now been corrected in five instances. In Fig. 3a, the label for the bottom row of the blots was mistakenly written as ‘GAPHD’; it should have read ‘GAPDH’. In the two right-most panels of Fig. 4j, the antibody labels ‘α-PD-L1’ for the reciprocal co-immunoprecipitation of DHHC3 were incorrect; they should have been ‘α-DHHC3’. These errors have been corrected in all versions of the Article.
References
Brahmer, J. R. et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 366, 2455–2465 (2012).
Sonpavde, G. PD-1 and PD-L1 inhibitors as salvage therapy for urothelial carcinoma. N. Engl. J. Med. 376, 1073–1074 (2017).
Yao, H., Wang, H., Li, C., Fang, J. Y. & Xu, J. Cancer cell-intrinsic PD-1 and implications in combinatorial immunotherapy. Front. Immunol. 9, 1774 (2018).
Rodell, C. B. et al. TLR7/8-agonist-loaded nanoparticles promote the polarization of tumour-associated macrophages to enhance cancer immunotherapy. Nat. Biomed. Eng. 2, 578–588 (2018).
Sharpe, A. H. & Pauken, K. E. The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol. 18, 153–167 (2018).
Burr, M. L. et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 549, 101–105 (2017).
Zerdes, I., Matikas, A., Bergh, J., Rassidakis, G. Z. & Foukakis, T. Genetic, transcriptional and post-translational regulation of the programmed death protein ligand 1 in cancer: biology and clinical correlations. Oncogene 37, 4639–4661 (2018).
Maj, T. et al. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nat. Immunol. 18, 1332–1341 (2017).
Snyder, A. et al. Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: an exploratory multi-omic analysis. PLoS Med. 14, e1002309 (2017).
Takeda, Y. et al. A TLR3-specific adjuvant relieves innate resistance to PD-L1 blockade without cytokine toxicity in tumor vaccine immunotherapy. Cell Rep. 19, 1874–1887 (2017).
Tang, H. et al. Facilitating T cell infiltration in tumor microenvironment overcomes resistance to PD-L1 blockade. Cancer Cell 29, 285–296 (2016).
Bellucci, R. et al. Interferon-gamma-induced activation of JAK1 and JAK2 suppresses tumor cell susceptibility to NK cells through upregulation of PD-L1 expression. OncoImmunology 4, e1008824 (2015).
Wolfle, S. J. et al. PD-L1 expression on tolerogenic APCs is controlled by STAT-3. Eur. J. Immunol. 41, 413–424 (2011).
Casey, S. C. et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 352, 227–231 (2016).
Bi, X. W. et al. PD-L1 is upregulated by EBV-driven LMP1 through NF-κB pathway and correlates with poor prognosis in natural killer/T-cell lymphoma. J. Hematol. Oncol. 9, 109 (2016).
Lim, S. O. et al. Deubiquitination and stabilization of PD-L1 by CSN5. Cancer Cell 30, 925–939 (2016).
Mognol, G. P. et al. Exhaustion-associated regulatory regions in CD8+ tumor-infiltrating T cells. Proc. Natl Acad. Sci. USA 114, E2776–E2785 (2017).
Wang, Y. et al. Regulation of PD-L1: emerging routes for targeting tumor immune evasion. Front. Pharmacol. 9, 536 (2018).
Zhang, J. et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 553, 91–95 (2018).
Li, C. W. et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat. Commun. 7, 12632 (2016).
Wang, H. et al. PD-L2 expression in colorectal cancer: independent prognostic effect and targetability by deglycosylation. OncoImmunology 6, e1327494 (2017).
Li, C. W. et al. Eradication of triple-negative breast cancer cells by targeting glycosylated PD-L1. Cancer Cell 33, 187–201.e10 (2018).
Chen, G. et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 560, 382–386 (2018).
Wang, H. et al. HIP1R targets PD-L1 to lysosomal degradation to alter T cell-mediated cytotoxicity. Nat. Chem. Biol. 15, 42–50 (2019).
Chen, M. et al. Development and validation of a novel clinical fluorescence in situ hybridization assay to detect JAK2 and PD-L1 amplification: a fluorescence in situ hybridization assay for JAK2 and PD-L1 amplification. Mod. Pathol. 30, 1516–1526 (2017).
Taguchi, T. & Misaki, R. Palmitoylation pilots Ras to recycling endosomes. Small GTPases 2, 82–84 (2011).
Runkle, K. B. et al. Inhibition of DHHC20-mediated EGFR palmitoylation creates a dependence on EGFR signaling. Mol. Cell 62, 385–396 (2016).
Gao, X. & Hannoush, R. N. Single-cell imaging of Wnt palmitoylation by the acyltransferase porcupine. Nat. Chem. Biol. 10, 61–68 (2014).
Tukachinsky, H., Petrov, K., Watanabe, M. & Salic, A. Mechanism of inhibition of the tumor suppressor Patched by Sonic Hedgehog. Proc. Natl Acad. Sci. USA 113, E5866–E5875 (2016).
Ren, J. et al. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 21, 639–644 (2008).
Weng, S. L., Kao, H. J., Huang, C. H. & Lee, T. Y. MDD-Palm: identification of protein S-palmitoylation sites with substrate motifs based on maximal dependence decomposition. PLoS ONE 12, e0179529 (2017).
Thul, P. J. & Lindskog, C. The human protein atlas: a spatial map of the human proteome. Protein Sci. 27, 233–244 (2018).
Garcia-Diaz, A. et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 19, 1189–1201 (2017).
Riaz, N. et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 171, 934–949 e915 (2017).
Linder, M. E. & Deschenes, R. J. Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84 (2007).
Horita, H., Law, A., Hong, S. & Middleton, K. Identifying regulatory posttranslational modifications of PD-L1: a focus on monoubiquitinaton. Neoplasia 19, 346–353 (2017).
Stringer, D. K. & Piper, R. C. A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. J. Cell Biol. 192, 229–242 (2011).
Takahashi, H., Mayers, J. R., Wang, L., Edwardson, J. M. & Audhya, A. Hrs and STAM function synergistically to bind ubiquitin-modified cargoes in vitro. Biophys. J. 108, 76–84 (2015).
Wang, H. et al. HIP1R targets PD-L1 to lysosomal degradation to alter T cell-mediated cytotoxicity. Nat. Chem. Biol. 15, 42–50 (2019).
Bolhassani, A., Jafarzade, B. S. & Mardani, G. In vitro and in vivo delivery of therapeutic proteins using cell penetrating peptides. Peptides 87, 50–63 (2017).
Soragni, A. et al. A designed inhibitor of p53 aggregation rescues p53 tumor suppression in ovarian carcinomas. Cancer Cell 29, 90–103 (2016).
Liang, L. et al. A designed peptide targets two types of modifications of p53 with anti-cancer activity. Cell Chem. Biol. 25, 761–774.e5 (2018).
Tian, X. et al. Organ-specific metastases obtained by culturing colorectal cancer cells on tissue-specific decellularized scaffolds. Nat. Biomed. Eng. 2, 443–452 (2018).
Fang, C. et al. Identification of palmitoylated transitional endoplasmic reticulum ATPase by proteomic technique and pan antipalmitoylation antibody. J. Proteome Res. 15, 956–962 (2016).
Yousefi-Salakdeh, E., Johansson, J. & Stromberg, R. A method for S- and O-palmitoylation of peptides: synthesis of pulmonary surfactant protein-C models. Biochem. J. 343, 557–562 (1999).
Acknowledgements
This project was supported by grants from the National Key Research and Development Plan (2016YFC0906000, 2016YFC0906003, 2016YFC0906002 and 2017YFC0906600), National Natural Science Foundation of China (81572326, 81322036, 81421001, 81773752, 81702969, 81874050, 21335002 and 31671360), Key Program of the Science and Technology Bureau of Sichuan (number 2017SZ00005), Top-Notch Young Talents Program of China (ZTZ2015-48), ‘Tang Scholar’ programme (JX-2017), Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20152514), ‘ShuGuang’ project supported by the Shanghai Municipal Education Commission and Shanghai Education Development Foundation (15SG16), National Key Technology Support Program (2015BAI13B07 to J.X.), and Natural Science Foundation of Shanghai (18ZR1402800 to C.F.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
H.Y. performed the in vitro and in vivo experiments, analysed the data and wrote the manuscript. J.L. performed the in vivo experiments and analysed the data. C.L. performed the in vitro experiments. H.S. designed the in vivo experiments and analysed the data. J.-P.B. contributed to the experimental design, analysed the data and wrote the manuscript. H.W. performed the in vitro experiments. H.L. and C.F. contributed the anti-palmitoylation antibody and analysed the relevant data. Y.Z., L.L., X.Z. and C.W. analysed the peptide toxicity data in different organs/samples, as well as the tissue microarray data. Y.X. contributed analysis tools and designed the mutagenesis experiment. Y.C. analysed the peptide toxicity data. J.X. supervised the project, conceived and designed the experiments and peptides, analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary methods, figures and tables
Rights and permissions
About this article
Cite this article
Yao, H., Lan, J., Li, C. et al. Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat Biomed Eng 3, 306–317 (2019). https://doi.org/10.1038/s41551-019-0375-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-019-0375-6
This article is cited by
-
PITPNC1 Suppress CD8+ T cell immune function and promote radioresistance in rectal cancer by modulating FASN/CD155
Journal of Translational Medicine (2024)
-
ZDHHC5-mediated S-palmitoylation of FAK promotes its membrane localization and epithelial-mesenchymal transition in glioma
Cell Communication and Signaling (2024)
-
zDHHC20-driven S-palmitoylation of CD80 is required for its costimulatory function
Acta Pharmacologica Sinica (2024)
-
Cyclical palmitoylation regulates TLR9 signalling and systemic autoimmunity in mice
Nature Communications (2024)
-
Targeting ZDHHC21/FASN axis for the treatment of diffuse large B-cell lymphoma
Leukemia (2024)