Abstract
Immune-checkpoint inhibition provides an unmatched level of durable clinical efficacy in various malignancies. Such therapies promote the activation of antigen-specific T cells, although the precise targets of these T cells remain unknown. Exploiting these targets holds great potential to amplify responses to treatment, such as by combining immune-checkpoint inhibition with therapeutic vaccination or other antigen-directed treatments. In this scenario, the pivotal hurdle remains the definition of valid HLA-restricted tumour antigens, which requires several levels of evidence before targets can be established with sufficient confidence. Suitable antigens might include tumour-specific antigens with alternative or wild-type sequences, tumour-associated antigens and cryptic antigens that exceed exome boundaries. Comprehensive antigen classification is required to enable future clinical development and the definition of innovative treatment strategies. Furthermore, clinical development remains challenging with regard to drug manufacturing and regulation, as well as treatment feasibility. Despite these challenges, treatments based on diligently curated antigens combined with a suitable therapeutic platform have the potential to enable optimal antitumour efficacy in patients, either as monotherapies or in combination with other established immunotherapies. In this Review, we summarize the current state-of-the-art approaches for the identification of candidate tumour antigens and provide a structured terminology based on their underlying characteristics.
Key points
-
Immune-checkpoint inhibition has profoundly changed the paradigm for the care of several malignancies. Although these therapies activate antigen-specific T cells, the precise mechanisms of action and their specific targets remain largely unknown.
-
Anticancer immunotherapies encompass two fundamentally different therapeutic principles based on knowledge of their therapeutic targets, that either have been characterized (antigen-aware) or have remained elusive (antigen-unaware).
-
HLA-presented tumour antigens of potential therapeutic relevance can comprise alternative or wild-type amino acid sequences and can be subdivided into different categories based on their mechanisms of formation.
-
The available methods for the detection of HLA-presented antigens come with intrinsic challenges and limitations and, therefore, warrant multiple lines of evidence of robust tumour specificity before being considered for clinical use.
-
Knowledge obtained using various antigen-detection strategies can be combined with different therapeutic platforms to create individualized therapies that hold great promise, including when combined with already established immunotherapies.
-
Tailoring immunotherapies while taking into account the substantial heterogeneity of malignancies as well as that of HLA loci not only requires innovative science, but also demands innovative approaches to trial design and drug regulation.
Similar content being viewed by others
Introduction
In 1900, Paul Ehrlich developed the concept of magic bullets (Zauberkugeln) as agents that specifically target harmful substances while sparing the body itself, thus limiting their toxicity1. Originally applied to antimicrobial agents, the quest for such magic bullets has since been a crucial goal in other areas of medical research and has become particularly relevant in the field of cancer immunology.
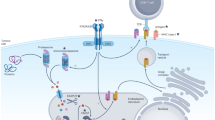
Immunotherapeutic approaches have been applied in the treatment of patients with cancer for over a century, including, among others, allogeneic haematopoietic cell transplantation (HCT), adoptive cell transfer, targeted monoclonal antibodies (which might also engage the immune system), immune-checkpoint inhibition (ICI) and other cellular therapies. Immunotherapies are directed against either known target antigens, and can thus be categorized as antigen-aware therapy (AaT), or antigens that currently remain uncharted (antigen-unaware therapy, AuT), with the latter category including therapies that induce and/or exploit basic immunological mechanisms or involve unmanipulated cell products. Established target antigens include cell-surface antigens such as proteins that are accessible and thus targetable using antibodies or human leukocyte antigen (HLA)-restricted peptide antigens (protein fragments). The latter are presented to antigen-specific T cells by molecules of the major histocompatibility complex (MHC, referred to as HLA when discussing human MHCs) and comprise either cytosolic proteins that have been processed by proteasomes (HLA class I) or peptides derived mainly from extracellular proteins (HLA class II). In this way, the protein pool accessible to the immune system is massively increased, enabling interactions with intracellular peptides that would otherwise be inaccessible.
Allogeneic HCT was the first example of a clinically effective active immunotherapy, although the mechanism of action was not originally appreciated. This treatment was initially believed to repopulate the defective bone marrow of patients with haematological malignancies using cells derived from a compatible donor, following elimination of the host’s defective haematopoietic stem cells using chemotherapy and/or radiotherapy2. Over the years, immunosurveillance of the underlying disease by the transplanted cells (the graft) was identified as the main mechanism leading to durable disease remission3,4. In this situation, alloreactivity is known to be mediated by donor T cells5, although the identity of the corresponding targeted HLA-restricted antigens remains largely unknown6,7. This lack of knowledge of the target antigen repertoire is also a feature of various other adoptive anticancer strategies, such as adoptive T cell transfer8. By contrast, monoclonal antibodies are necessarily directed against defined, specific cell-surface antigens, such as CD20 or HER2 (refs9,10,11,12).
In addition to clinically approved antibodies and antibody constructs directed against various cell-surface proteins, antibodies directed against HLA–peptide complexes (such as T cell receptor (TCR)-mimic antibodies) are also currently under active development13. The same holds true for chimeric antigen receptor (CAR) T cells, which are autologous T cells genetically engineered to express a CAR targeting a cell-surface protein, combined with activation of co-stimulatory proteins to promote cellular survival and/or differentiation upon target binding14. Although CARs require target antigens to be accessible from the cell surface15,16,17,18,19, targeting HLA-presented peptides would potentially enable access to antigens derived from the intracellular proteome20. For all of these approaches, the prior identification of suitable target antigens remains crucial21,22, albeit with an additional level of heterogeneity created by strategies directed against HLA-restricted antigens, owing to the extensive allelic variability of the genes encoding these proteins23.
With the introduction of antibody-mediated ICI, active immunotherapy has now entered standard clinical practice in the adjuvant24, advanced-stage and/or metastatic25,26,27,28 and neoadjuvant settings29 in patients with various types of solid tumours. ICI can currently be classified as a treatment with unknown targets (AuT), because disruption of PD-1–PD-L1 and/or cytotoxic T lymphocyte protein 4–CD28 signalling leads to the activation of unselected and unknown antigen-specific T cells. A supposed mechanism of action of immune-checkpoint inhibitors includes the activation of a pre-existing tumour-specific T cell repertoire. The addition of effective targeted immunotherapies with diligently curated antigens that are intended to direct immunological activity towards specific targets might, therefore, provide the highest possible level of antitumour efficacy, in line with the concept of antitumour ‘magic bullets’1.
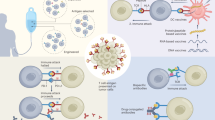
The majority of targeted therapies provide limited improvements in progression-free and/or overall survival30; however, immunotherapies could potentially lead to long-term survival, if not ultimately cure31,32. Patients receiving immunotherapies targeting specific antigens should be selected on an individual basis because most malignancies, even those of an apparently identical histological subtype, have substantial levels of variation across their mutational and antigenic repertoires33. Such approaches will, therefore, ultimately need to be adapted to provide a uniquely assembled drug product (UADP) for each individual patient (Fig. 1) or might even necessitate the selection of individual targets followed by the synthesis of a uniquely designed drug product (UDDP) based on analyses of autologous tumour samples34. Respective in-depth investigations of individual tumours could yield one, or even several, suitable individual antigens, which could be targeted using the available immunotherapeutic platforms including vaccines, TCR-mimic antibodies, CAR T cells or adoptive transfer of preselected antigen-specific T cells33,35,36,37,38,39. ICI enables pre-existing antitumour immune responses to be reawakened, although only their direction against tumour-specific antigens (TSAs) would allow these agents to be referred to as magic bullets.
Three levels of biomarker-driven adaptation of drug products for (immuno)therapies directed against given malignancies for clinical application can be achieved via different strategies. a | Stratification. The allocation of patients to specific therapies is directed by the assessment of established predefined biomarkers. In the absence of a respective selection criterion, therapy is not administered (represented by Ø in the figure). Usually, the active pharmaceutical ingredients (APIs) used and also the resulting stratified drug product (SPD) are manufactured and shelved. Such therapies are employed either as a defined pharmaceutical product or as an API used in combination with autologous or allogeneic cells for release as a drug product31. In this scenario, the principle of either antigen-aware therapy or antigen-unaware therapy may apply. b | Warehousing. A pre-characterized set of APIs (coloured squares) is manufactured and stored as separate components that may be individually assembled (active adaptation), for example, representing a selection of APIs defined by biomarkers assessed in autologous tumour tissues (coloured circles under ‘theranostics’; colouring reflects the biomarker-defined components of the warehouse represented below). The combination of individually selected API components results in a uniquely assembled drug product (UADP), which consists of a defined number of APIs from the warehouse (represented as colours in each vial under drug product reflecting the components selected from the warehouse; respective components from the warehouse are labelled with checkmarks). The resulting drug product can be considered a UADP, which can be administered either directly as a pharmacologically defined drug product or using autologous or allogeneic cellular drug products. For this strategy, the applied therapeutic principle has to be generally antigen aware. c | Individualization. Here, active ingredients are adapted according to patient-individual biomarkers (represented by colours in different shadings in accordance with the colouring of the silhouettes) and therefore per se cannot be pre-manufactured and stored. Hence, individualization requires de novo identification, characterization, selection and manufacturing of APIs for every given patient, based on analysis of autologous tumour (and non-malignant) tissues. The individual compilation of APIs and their formulation results in a uniquely designed drug product (UDDP). When administered as a pharmacologically defined product, the resulting therapy is antigen-aware (for example, for vaccines targeting individual specified mutated HLA ligands), or, if UDDPs are generated using autologous or allogeneic cell products, these therapies may be either characterized as antigen-aware (for example, when T cells are primed with known antigens) or antigen-unaware (for example, when T cells are primed with undefined tumour lysates). SDP, standardized drug product.
In this Review, we provide an overview and clarified nomenclature designed to describe the spectrum of (targetable) HLA-presented antigens. We also describe the identification and characterization of such antigens, as well as the opportunities and challenges associated with each approach. Finally, we discuss the emerging consequences for treatment stratification, (active) adaptation and/or individualization of treatments and the resulting regulatory consequences.
Characterizing HLA-presented antigens
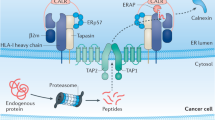
Tumour antigens can emerge during all stages of protein synthesis and degradation, including transcription and translation, and their emergence can also involve post-translational modifications as well as alterations in antigen processing and presentation (Fig. 2a; Table 1). These antigens can be divided into TSAs and tumour-associated antigens (TAAs) according to whether they are presented by HLAs expressed exclusively on tumour cells (mainly HLA class I) or to a variable extent on non-malignant tissues, respectively.
a | Schematic overview of cellular processes involved in emergence, processing and HLA class I presentation of antigens. Respective changes in cellular processes (for example, modifications during translation or post-transcriptional modifications) that are altered leading to the formation of respective antigen classes are marked. b | Overview of different classes of HLA-presented antigens. Blue bars indicate whether antigens comprise alternative and/or wild-type sequences (or both). Other specific characteristics of the different tumour-specific antigens (TSAs) and tumour-associated antigens (TAAs) described in this Review are provided as a table labelled with traffic light colours: green indicates that the antigen characteristics are in accordance with the respective criteria; red indicates that the antigen characteristics are discordant with the respective criteria; and orange indicates that the respective criteria are not a defining property of this antigen class. For cryptic antigens, question marks indicate missing experimental evidence. CA, cryptic antigen; mut., mutated; oex, overexpressed; ptm, post-translational modification; roe, reactivated ontogenetic expression; spl., spliced; TAP, transporter-associated with antigen processing; TCR, T cell receptor; wt, wild-type.
Tumour antigens possess either a wild-type (self) or altered (foreign) amino acid sequence. Among antigens with an altered sequence, TSAs emerge owing to somatic mutations that confer a non-synonymous amino acid sequence that is not present in the germline DNA and is therefore unique to the malignant tissue. These alterations can arise from several different types of mutations including single-nucleotide variants (SNVs), nucleotide insertions and/or deletions (indels) and/or frameshifts. Such antigens are referred to as mutated tumour-specific antigens throughout this article, thus avoiding the term neoantigen, which is often used inconsistently (Box 1)40.
The discovery and characterization of TSAs are feasible using a diverse range of high-throughput multi-omics analyses, including next-generation sequencing (NGS) and tandem mass spectrometry (MS/MS). Application of these methods results in two fundamentally different approaches to antigen discovery: computational prediction of putative HLA-presented antigenic peptides41,42,43 based on their HLA-binding properties44 and direct identification of naturally presented HLA-eluted ligands and their annotation to protein sequence databases45, respectively. Thus, only MS/MS-based approaches enable physical confirmation of presentation of antigenic peptides as HLA ligands. Combined use of NGS and MS/MS enables the characterization of TSAs on two crucial levels: a first level where (1) genetic information from annotated NGS data is used to infer HLA presentation in silico, which may entail a relevant fraction of false-positive findings. This information can be complemented by (2) MS/MS data confirming predicted HLA ligands in silico, thereby being a much more conservative approach.
The characterization of an existing T cell repertoire in blood and/or tumour-infiltrating lymphocytes provides an alternative indirect method for confirming the natural presentation of HLA ligands and, among other aspects, their capacity for target cell lysis46,47,48,49. This characterization can be achieved using several methods, including by screening predicted peptides50 or using cDNA libraries51.
Alternative sequence antigens
Genome-/exome-derived mutated TSAs
Alterations characterized by NGS may either be limited to the exome, a proportion of about 1% of the genome established as protein coding52, or exceed it (whole genome). All gene sequences encoded synonymously in non-malignant and malignant tissues are generally accepted as part of the individual’s wild-type genome. By contrast, non-synonymous variants might encode sequence alterations in naturally occurring proteins, which — when translated — result in novel protein sequences that could potentially give rise to HLA ligands to which the immune system has never previously been exposed and are therefore highly immunogenic (Table 1). Limiting the analysis of exon-encoded somatic variants to those with transcriptomic evidence (requiring transcript coverage53) is another method of restricting the size of the search space.
Initially, TSAs were mainly predicted based on the presence of non-synonymous SNVs located in exons, disregarding those created by frameshift and indel mutations54,55. Irrespective of this limitation, only a few TSAs among HLA class I ligands could be eluted directly from HLAs and characterized using MS/MS56,57, which also require low detection thresholds and a very sensitive detection methodology. Of note, all mutated TSAs detected using these methods are restricted to individual tumours. Shared mutated TSAs that can be detected in more than one patient are currently elusive58,59. The reasons why TSAs seem to be so difficult to detect have yet to be completely elucidated but might reflect technical limitations of the employed detection and/or discovery methodologies (Table 2), as well as immunoediting by the patient’s initial antitumour immune response, leading to elimination of the most antigenic tumour clones. Furthermore, T cell recognition of TSAs might lead to the upregulation of inhibitory ligands, such as PD-L1, and contribute to the development of an immunosuppressive tumour microenvironment, both of which might impede the expansion and response of antigen-specific T cells60,61. In summary, antigen-specific T cells against a specific mutated TSA can frequently be detected in the absence of a robust antitumour immune response35,49,50,59,62, and thus the antigen either must have previously been present, but has been removed by immunoediting59, or still prevails but remains undetectable by MS/MS.
Exonic variants potentially leading to mutated TSAs (Fig. 2b) derived from frameshift mutations, such as those created by indels, occur with increased frequency in patients with renal cell carcinoma (RCC; median eight to ten indels per tumour versus a median of four in a pan-cancer cohort)63 and in those with tumours of a microsatellite instability-high (MSI-H) phenotype (frameshift mutations in one of four genes examined have been detected in 75% of such tumours)64, both of which are characterized by enhanced responsiveness to ICI65,66,67. A potential explanation for this responsiveness is provided by the higher immunogenicity of indel-derived as compared with SNV-derived peptides, resulting in the generation of greater numbers of novel and immunologically relevant HLA ligands63. The safety and initial clinical performance of a personalized vaccine comprising a mixture of peptides derived from up to 20 tumour antigens (including predicted indel-derived peptides) have been tested in patients with previously treated high-risk melanoma in an early-phase clinical trial38. Notably, the immunogenicity of selected predicted indel-derived peptides was greater than that of their SNV-derived counterparts38.
Gene fusions are characteristic of, if not pathognomonic for, a multitude of haematological and solid cancers68,69. Many of these events have been characterized as driver mutations, and can thus be successfully targeted70, although resistance mutations in these fusion proteins can result in therapeutic failure71. Thus far, predicted TSAs derived from such fusions have not been successfully validated as being naturally presented by HLA ligands, according to MS/MS-based analyses.
The safety and efficacy of immunogenic peptide vaccines based on predicted mutated TSAs have been investigated in several clinical trials involving patients with chronic myeloid leukaemia or sarcoma. These vaccines have been proven to be safe and immunogenic, albeit without producing clinically meaningful responses in most patients72,73,74,75,76.
TSAs derived from splicing
Analysis of the tumour transcriptome can provide additional evidence that a specific mutation is translated into a protein. Furthermore, variations acquired during transcription, including cancer-specific fusions of transcripts and mRNA splicing events, might lead to the creation of alternatively spliced TSAs, which are undetectable using NGS-based exome sequencing. Thus, alternative splicing and/or editing of RNA transcripts is a potential supplementary source of HLA-presented peptides with altered sequences (spliced TSAs)77,78,79 (Fig. 2b). Indeed, mutations that influence the splicing machinery, such as by creating new splice sites80,81, by dysregulating the spliceosome82,83,84 or by enabling intron retention85,86, or mRNA splice junction mutations87, might all be additional sources of alternative ribosomal products. With advances in RNA sequencing technology, including the ability to assess splice variants and ribosome profiling (RiboSeq), previously unassigned antigens (cryptic antigens) can now be characterized88, which potentially extends the number of targets for antigen-specific immunotherapies79.
A pan-cancer analysis of data from 8,705 tumours analysed as part of The Cancer Genome Atlas (TCGA) project89 has revealed a high number of exon–exon neojunctions (mean of 930 per sample), which is potentially relevant to TSA discovery. The detection of alternatively spliced TSAs and the assessment of their potential for HLA presentation remain challenging77, although initial studies have confirmed the existence of exon–exon junction-derived proteins that are predicted to be capable of HLA binding77,79,86,87,90,91. The widespread MS/MS-based validation of alternatively spliced TSAs is still pending, although a T cell repertoire has been established for certain variants, such as D393-CD20 (ref.92).
In addition to mRNA splicing, proteasomal splicing might also be a source of TSAs93,94, although their formation and frequency of occurrence currently remains controversial95,96,97,98. Basic characterizations of antigenic peptides produced by peptide splicing in the proteasome have already been conducted using cDNA-based approaches51,99. Additional alterations at the protein level (such as phosphorylation of serine or threonine residues) are conceivable, for example, when the respective post-translational modifications are retained in peptides and subsequently presented by HLAs. These changes form the basis for modified potentially immunogenic self-peptides, such as TSAs comprising phosphorylated peptides100,101 as well as glycated peptides102. Defective ribosomal products (DRIPs) by definition do not influence the transcript-encoded amino acid sequence103, whereas changes in proteasomal processing might give rise to alternative sequence TSAs104,105. Currently, most peptides deemed to originate from proteasomal splicing can strictly be regarded as cryptic antigens.
Cryptic antigens
An additional space exists within the genome that might also be a source of HLA-presented peptides that are not synonymous with the exome-encoded proteome. This dark matter of the proteome derived from the genome outside regions assumed to be translated106, yielding cryptic peptides (Box 1), has not been extensively investigated to date but has been suggested as a relevant source of TSAs88,107 with estimates indicating that it accounts for at least 10% of all peptides presented by HLA class I molecules108. However, as the estimates provide only approximate numbers, the true quantity of such antigens is unknown. The discovery of such antigens remains challenging and might be improved in future, although MS/MS combined with the latest sequencing software, as well as RiboSeq (an NGS-based method of determining which RNAs are actively translated), provide the necessary technical foundations. The extent to which such cryptic peptides are tumour specific is also currently unknown.
Generally, in silico translation of the consensus genomic sequence or even a genome individually mapped into putative proteins is feasible using methods such as six-frame translation and can be used for the discovery of cryptic TSAs, assuming that additional protein coding sequences arise under certain conditions (such as after malignant transformation)108. Furthermore, the exonic translation space might be extended upstream and downstream of coding genes to include sequences of untranslated regions, intronic sequences and intergenic regions, as well as transposable elements, as additional sources of cryptic peptides and even TSAs.
Additional potential sources of HLA-presented cryptic antigens include altered mRNA splicing events, long non-coding RNAs, small nucleolar RNAs and proteins encoded in ribosomal DNA88. The categorization of cryptic antigens needs to be viewed as dynamic, given that the ability to annotate an HLA ligand as originating from an intron, an open reading frame, a splice variant or other source is rapidly evolving owing to improvements in detection and/or analytical methods77,107. Hence, increasing numbers of cryptic antigens will be annotated, thereby defining their origins and functions in the near future.
Wild-type sequence TSAs
The majority of studies involving alternative sequence antigens have thus far focused predominantly on predicted target antigens using computational workflows, while discovery and direct verification of such peptides using MS/MS remains an exception56,57,109. Even in these studies, in which a subset of predicted TSAs was directly verified (including comparisons with the spectra of synthetic peptide mimetics), the definitive confirmation of most predicted antigens as naturally presented HLA ligands remained elusive. By contrast, MS/MS-based analyses of HLA-bound peptides eluted from native tumour tissue samples can provide comprehensive and in-depth information on their naturally presented HLA ligandome. Thus, differential comparisons with HLA ligands from benign tissues might also enable the discovery of TSAs with wild-type amino acid sequences to which the immune system has never previously been exposed. Such wild-type tumour-specific antigens may therefore also prove valuable, similar to tumour-specific antigens with reactivated early ontogenic expression56,58,110 (Fig. 2b).
The HLA ligandome is influenced by cancer-specific alterations affecting, among others, translational regulation101, protein metabolism, proteasomal processing, cytosolic proteolysis, but also alterations in antigen processing, such as those involving the transporter for antigen processing (TAP)111, which ultimately affect HLA ligand presentation112. These changes might result in the emergence of TSAs with a wild-type sequence (Table 1). Considering the poor correlation between the transcriptome and the HLA class I-presented ligandome113,114,115, a better understanding of the underlying rules that apply to antigen processing and presentation by HLAs might facilitate further target discovery105. Such peptides include antigens derived from proteins that are exclusively expressed in the early stages of ontogenesis, which can then be reactivated during oncogenesis, such as cancer–germline antigens116,117. Moreover, colorectal cancer-specific oncogenic signalling pathways118 have been suggested to leave an imprint within the tumour-exclusive HLA ligandome, possibly contributing wild-type TSAs58.
The HLA ligandome of several cancer types has been comprehensively characterized using MS/MS119. Nevertheless, the reliable identification of wild-type TSAs requires extensive experimental effort in order to ensure that such TSAs are indeed tumour exclusive and to exclude HLA presentation on non-malignant tissues58,114. This approach should include comparisons of HLA ligandome data with growing databases of HLA ligands eluted from a diverse range of non-malignant tissues120. Importantly, when comparing matched malignant and non-malignant tissue samples, fundamental differences in physiological processes might introduce a relevant source of bias affecting the HLA ligand repertoire, such as substantial differences in the expression of proliferation-associated proteins. HLA ligands arising from such physiological processes might therefore appear to be different but also lack tumour exclusivity. A scarcity of data exists on HLA-presented ligands with rare allotypes, resulting in insufficiently characterized binding motifs, that might further impair the reliable identification of wild-type TSAs. A possible strategy that could mitigate such bias is to generate large datasets and restrict the comparisons of HLA-presented peptides between tumours and non-malignant tissues to well-defined matched HLA allotypes (such as HLA-A*0201), as well as including RNA expression data113.
Tumour-associated antigens
TAAs feature wild-type amino acid sequences but — unlike TSAs — are also presented by HLAs on the surface of other non-malignant cells, making these antigens more challenging targets for therapeutic use owing to immunological tolerance and a lack of specificity. The identification of TAAs is mainly based on comparisons of the transcriptomes of malignant and non-malignant tissues to identify overexpressed gene products and MS/MS to confirm natural presentation as HLA-restricted ligands45,113,121. These analyses are often complemented by establishing a natural T cell repertoire for the putative TAA. Occasionally TAAs can even induce T cell tolerance122,123,124.
TAAs can also be targeted in the clinical setting provided that they are overexpressed on tumour cells and are otherwise only presented on non-malignant tissues that are not considered essential for survival, such as reproductive tissues114,125 or B cells11,126. Hence, large databases containing HLA ligands from non-malignant tissues as controls120,127,128 or strategies that confine ligands to single HLA alleles113 have been created or are currently under development and will provide a key resource supporting the selection of candidate TAAs that are as tumour specific as possible.
In the absence of shared TSAs, clinical interventions involving the administration of TAA sequences as vaccines have been evaluated in both early-stage and late-stage clinical trials. Many approaches involving several different delivery platforms (DNA, RNA, peptides or viral vectors) and adjuvants (inter alia Montanide ISA 51 VG, granulocyte–monocyte colony-stimulating factor or RNA) have led to the induction or re-activation of TAA-specific T cells, although no conclusive evidence of clinical efficacy has so far been established129,130,131.
Several vaccine-based approaches involving the administration of standardized peptide cocktails (as formulated drug products) containing TAAs and possibly including wild-type TSAs have advanced to the later stages of clinical development, although the efficacy of these treatments has so far been disappointing. After the emergence of promising phase II data131, a peptide vaccine (IMA901) consisting of short peptides (mostly TAAs) in combination with the tyrosine kinase inhibitor sunitinib was evaluated in a phase III trial. However, the combination failed to induce any relevant vaccine-specific T cell responses, most likely owing to inhibition of T cell priming by sunitinib132, and failed to improve outcomes compared with sunitinib monotherapy133. In addition, the anchor-modified gp100:209-217(210M) peptide vaccine failed to show clinical efficacy in patients with melanoma134.
In February 2019, the data from a two-stage phase I/II trial involving patients with glioblastoma were published. In the first stage, a warehouse-based TAA vaccine (UADP) was administered, and in the second stage, selected patients received a UDDP vaccine consisting of elongated peptides containing predicted mutated TSAs and TAAs determined using MS/MS-based tumour characterization. This study demonstrated successful induction of vaccine-specific T cells, but was not sufficiently powered to provide reliable clinical data36.
Experimental evidence for different TSAs
A central challenge in the discovery of mutated TSAs and other patient-specific drug targets is the theoretically extensive range of targets and the heuristics of selecting the most appropriate targets for therapy within an acceptable time frame. In our view, such an effort only seems feasible by triangulation135, which requires several lines of evidence (Fig. 3). Hence, genomic and/or transcriptomic and proteomic evidence needs to be combined as a basis for the selection of suitable targets. Accordingly, despite a huge number of HLA class I ligands predicted from NGS in silico data, so far only a small number of such mutated TSAs, or even more complexly generated TSAs, have been detected and verified by MS/MS (for example, using spectrum matches with sequence-identical synthetic peptides)53,56,109.
The confidence in experimental evidence for alternative sequence antigens as well as tumour-specific antigens (TSAs) with wild-type sequence, tumour-associated antigens (TAAs) and cryptic antigens is represented by the lines of different colours. These represent light of different wavelengths combined to yield white light when transitioning through the prisms (here representing triangulation integrating different lines of evidence135). The lengths of the coloured lines represent the experimental evidence graded into experimental multi-omic evidence (long lines; multiple congruent lines of evidence available), experimental evidence (medium-length lines; some merging lines of evidence available) and experimental evidence with allocation (short lines; one single line of evidence available). The colours of the lines represent exome sequencing (red), transcriptome sequencing (yellow) and MS/MS techniques, including proteomics (green) and HLA ligandomics (blue). The current situation, where the majority of tumour antigens cannot be characterized with multiple congruent lines of evidence (that is, triangulation), using both multi-omics approaches as well as indirect evidence, is symbolized by the dark side of the Moon, which exists but remains invisible to the eye. For cryptic antigens the prism and respective rays of light are inverted, as observations from MS/MS made at the HLA ligand level are attributed to a space beyond the exome, excluding exome-templated sequences. In contrast to most other antigen classes, cryptic antigens are identified using a bottom-up approach, thereby allocating HLA ligand level evidence to an ample search space comprising sequences deemed non-coding in the genome. Credit for moon image: Historical/Contributor/Getty. Adapted with permission from NASA.
Methods for antigen identification
The identification and selection of TSAs or TAAs for clinical application requires more thorough validation than simple confirmation as a tumour-specific target136. An ideal therapeutic target should have the utmost level of tumour specificity, HLA presentation and immunogenicity; antigens lacking any one of these prerequisites (including those that induce T cell anergy or tolerance122,123,124) would be therapeutically irrelevant, or even potentially harmful.
Different levels of evidence have been established for all defined categories of HLA-presented antigens (Fig. 2). The level of evidence is influenced by several factors, including the characteristics and idiosyncrasies of the various detection methods, such as the coverage of regions of interest and the as-yet-unknown probabilities of HLA presentation of specific peptides.
The coverage of the regions of interest for whole-exome sequencing (WES) and transcriptome sequencing is usually comprehensive137 (Table 2). However, because mutated regions are often either incompletely translated or remain synonymous, only a subset of these regions has the potential to encode altered protein sequences. In an analysis of patient-derived organoids, only a small fraction of WES-detected mutations (<1%) could be validated as being presented by HLAs using MS/MS109. Similar observations have been found using animal models138, cell lines and tumour tissue samples primarily from patients with melanoma56,57,139. Mutations in highly expressed transcripts have been suggested to be more likely to be detectable as HLA ligands, with a reported probability as high as ~10%113.
Shotgun LC–MS/MS should be considered the best available method for detection of naturally occurring HLA ligands, albeit with several limitations (Table 2). Although progress has been made, and both techniques and devices have improved substantially, technical idiosyncrasies remain. Intrinsic methodological disadvantages include the fact that shotgun LC–MS/MS is currently not a broadly applicable quantitative technique, and the sensitivity of detection is limited by the inability to amplify sample materials, in contrast to NGS-based approaches (Table 2). These limitations can result in coverage issues and bias for certain HLA ligands, preventing their detection by MS/MS. Detection of peptides with properties that are incompatible with this methodology, such as those that are either strongly hydrophobic or hydrophilic, or insufficiently ionizable, might, therefore, fail. The main technical challenges of shotgun MS/MS consist of missing values owing to (semi-random) sampling based on precursor abundance, thereby impairing technical reproducibility. Further issues include the detection limits of the instrumentation, as well as the unknown temporal dynamics of the HLA ligandome and pre-analytical factors capable of potentially influencing the detectable HLA ligand repertoire140. An additional level of bias might be introduced by the choice of HLA alleles through the selected algorithms used and the pool of data incorporated for peptide binding prediction141,142. Owing to these various limitations, shotgun MS/MS-based proteomics is currently only likely to achieve coverage of up to ~5% of the proteome. The precise size of the HLA ligandome remains undetermined, and this uncertainty also applies to the size of the part that is ultimately missed. MS/MS-based confirmation of mutated TSAs has proven to be difficult, even with unrestricted sample access and extensively characterized ligandomes. Regardless of these challenges, available evidence supports the notion that exome-encoded mutated TSAs are rare53,56,109,139.
Setting the differences between detection methods into perspective, NGS-based WES can, with some technical limitations, reproducibly yield near-comprehensive coverage of the targeted regions (Table 2), and the same also applies to transcriptome sequencing. However, this level of coverage does not apply to proteomic methods, for which only ~50% coverage of the proteome can be achieved, even with considerable effort and under optimal circumstances143. Thus, unsurprisingly, only a small fraction of exome-encoded variants are detectable using proteomic methods (2.4% of exon-encoded and 6.7% of transcriptomic single-amino acid variants)144.
Screening for patient-specific targets
Mutated or alternatively-spliced TSAs have the potential to be entirely tumour-specific targets, when experimentally confirmed with a high level of confidence. Nevertheless, these antigens have frequently been found to lack clinical relevance, particularly those in which the tumour specificity has been predicted in silico only. Possible causes include technical aspects such as high false-positive rates145,146, but also biological reasons including reduced transcription or immunoediting might preclude presentation of the predicted TSAs on HLAs.
Post-translationally modified HLA ligands are also plausible tumour-specific targets that also require confirmation at the HLA ligand level. Post-translationally modified tumour-associated antigens should have a wild-type amino acid sequence101,102, and therefore tumour specificity is dependent upon subsequently introduced modifications.
In contrast to TAAs with reactivated early ontogenetic expression, for overexpressed tumour-associated antigens and shared TAAs, tumour specificity may be challenging to confirm (Fig. 2b), although MS/MS provides convincing evidence for presentation on HLAs53,114. While reactivated ontogenetically expressed TAAs are mostly tumour specific, owing to restricted expression in early ontogenesis and/or reproductive tissues, overexpressed TAAs and shared TAAs are also presented on non-malignant tissues and thus, by definition, do not show absolute tumour specificity. For such TAAs to be considered as targets for immunotherapy, any non-malignant tissues expressing them need to be deemed dispensable (such as reproductive organs or B cells) and they may not be expressed on any vital tissues.
Therapeutic application
Considering the potential of different antigen types and individual antigens for clinical use also requires reflection on the intended therapeutic platform. Administration of antigens as vaccines is expected to result in fewer toxicities owing to self-tolerance compared with various other approaches, including TCR-mimic antibodies, bispecific TCR molecules or even, in future, MHC-restricted antigen-specific CAR T cells147. CAR T cell-based approaches in particular require the utmost caution in terms of ascertaining both the specificity and clinical relevance of the target antigen. The importance of antigen specificity is underlined by the occurrence of fatal cardiac toxicities owing to tissue cross-reactivity in patients receiving anti-MAGE-3 CAR T cells148,149.
Overall, investigators attempting to identify and select specific tumour antigens as targets for the treatment of patients with cancer should consider the various methodological limitations, the probability that the antigens are presented and also the available experimental evidence for both their existence and clinical relevance (Table 2).
Tumour antigens of different types have specific limitations regarding their detectability using the available experimental methods (in analogy to the dark side of the Moon, which physically exists but remains invisible from Earth). Multi-omics approaches53 incorporating genomics, transcriptomics and proteomics (including HLA ligandomics) can increase the level of evidence over that obtainable with only one of these approaches and thus improve relevance and shed more light on selected antigens (Table 2).
Cryptic antigens detected using MS/MS can be attributed to defined HLA allotypes, but attempts to do so have to account for an enormous search space of potential antigens (Table 2). Large numbers of mutated TSAs can be predicted with confidence from WES and/or transcriptome sequencing data, although TSAs identified in this way frequently lack proteomic and/or ligandomic evidence (Table 2), which explains why the selection of useful candidates remains a major challenge146.
In principle, a high level of experimental evidence supports the clinical validity of both wild-type TSAs and TAAs (Table 2), although the tumour specificity of such antigens might create safety concerns if they are targeted for therapeutic purposes. The implementation of such personalized therapies also poses unique and fundamental regulatory challenges.
Regulatory aspects
Taking into account the aforementioned considerations and the high level of variability among potential tumour antigens, it becomes increasingly clear that the definition of targets for magic bullets can have far-reaching consequences for the definition of active pharmaceutical ingredients (APIs) and the resulting drug products as well as their administration, especially when the targets are applied to different therapeutic delivery platforms. This notion could also affect the definition of safety criteria and pose unprecedented challenges for clinical trials, given that a drug product could be designed and manufactured for use in only one patient (UDDP).
In 2013, Britten and co-workers defined the first set of criteria for the classification of personalized immunotherapies, introducing different approaches to precision cancer immunotherapy34. Respective levels of personalization were defined in order to guide regulatory decision-making, including a specific definition for therapies designed for individual patients only. In light of current developments, we suggest that this proposed classification as previously defined should be updated (Fig. 1).
Stratification
The first level of adaptation for drug products (Fig. 1a) remains biomarker-based stratification, whereby patients are identified according to predefined tumour features, such as the presence of certain antigens, and treated with a defined drug product according to positivity for these biomarkers34 and individual characteristics (such as HLA allotype). Examples include pharmaceutically defined storable drug products, such as antibody-based therapies including the anti-HER2 antibodies trastuzumab or pertuzumab9,12,150. Furthermore, the allocation of targeted therapies to patients with advanced-stage disease according to biomarker results from focused gene-sequencing panels enabling the identification of potentially targetable mutations is to be included in this category151. Modern cellular therapies, including anti-CD19 CAR T cells (such as axicabtagene ciloleucel and tisagenlecleucel17,19) can be regarded as stratified, representing an autologous product with a standardized vector manufactured on demand.
Another example of therapies included in this category is the discontinued dendritic cell-based immunotherapy sipuleucel-T152. Furthermore, the allocation of ICI based on a high tumour mutational load or high tumour mutational burden, PD-L1 expression, or the presence or absence of other biomarkers can be regarded as a stratified therapy. Hence, the principles of both AaT, when precise knowledge of the targeted structures is available and used as a biomarker, and AuT, when such information is unavailable, may apply, as explained for ICI above.
Warehousing
Warehousing (Fig. 1b) refers to the individual composition of a drug product assembled from a set of pre-characterized and pre-manufactured peptides as active ingredients, guided by analyses of autologous tumour tissues (theranostics). Such APIs are individually assembled and released as a uniquely composed drug product (UADP) and represent a customized medicinal product for administration in only a single specified patient. Of note, specific APIs composed of several peptides can be fully characterized (AaT) before composition, but can also include, for example, cell therapies including autologous and/or allogeneic dendritic cells stimulated with a set of defined peptides or with RNA and/or DNA transfection, as well as antigen-preselected adoptive cell therapies47. Warehousing-based concepts for peptide vaccination have already been evaluated in clinical trials36,153,154. In the future, such concepts might even include additional platforms such as antibodies or CAR T cells, should financial and regulatory circumstances allow.
Individualization
According to our terminology, individualization involves the selection of one or more APIs and the on-demand manufacture of a drug product based on patient-specific and/or tumour-specific features36,37,38,39 (Fig. 1c). This concept is particularly important for regulatory considerations. This definition of individualization denotes a fully de novo designed and manufactured product, comprising one or more individual APIs, formulated and released with the intention to use it in only one patient, resulting in a UDDP. Individualization also applies to adoptive cell products, which are also specifically produced for individual patients155 and cannot be pre-characterized (AuTs). Hence, both pharmaceutically defined APIs and cellular products might be viable options.
The highly individualized approach to therapy implicit in the use of antigen-specific immunotherapies could have far-reaching consequences for the design and approval of future clinical trials. The development of UADPs and most notably UDDPs within clinical trials does not conform with established approaches to pharmaceutical manufacturing and drug development, because the exact make-up of the drug product cannot be prespecified in the usual way (Fig. 1). In these scenarios, defining and prespecifying the process of antigen selection for inclusion in the drug product (Fig. 4), and not the UDDP itself, might be a reasonable requirement. A further consideration is that the population at risk when releasing a specific UADP or UDDP is, by definition, confined to a single named patient. Finally, the challenges created by the need for full good manufacturing practice (GMP) production of UADPs or UDDPs (as required for clinical testing) are not only limited to substantial financial and/or operational barriers. Further relevant challenges exist regarding drug manufacturing and availability in an acceptable time frame. A possible solution could be to waive in-process controls and focus on final-release controls (an approach deemed optional even under the current European Medicines Agency GMP guidelines156) in order to expedite patient access. To our knowledge, no data are available thus far on patient-specific approaches to drug design using platforms such as TCR-transfected or CAR T cells157,158, TCR-mimic antibodies13 or autologous and/or allogeneic cell products directed against individually identified targets159.
Autologous tumour and/or non-malignant tissues are analysed using several methods complementing each other (whole-exome sequencing (WES), transcriptome sequencing and MS/MS). Suitable tumour-specific antigens (TSAs) and tumour-associated antigens (TAAs) are prioritized using (automated) bioinformatics approaches, but results may additionally be manually curated. This is performed by an interdisciplinary team of professionals, assessing limitations and potential issues of sample acquisition and preparation, antigen detection and bioinformatics, as well as active pharmaceutical ingredient (API) and drug product manufacturing, thereby potentially introducing bias but also improving safety owing to thorough target review. The identified antigens could be used therapeutically utilizing different platforms, comprising chimeric antigen receptor (CAR) T cells, TCR-mimic antibodies, bispecific TCR molecules and different vaccination approaches. The resulting drug product can either be standardized or individually assembled as an uniquely assembled drug product (UADP) or uniquely designed drug product (UDDP). αCD3, anti-CD3; scTCR, single-chain TCR; SDP, stratified drug product; TCR, T cell receptor.
Future directions
In our opinion, the future development of active antigen-directed immunotherapeutic approaches faces four major challenges (forming the acronym SAFE) that need to be addressed before therapeutic efficacy can be achieved, while limiting potential toxicities to an acceptable level.
Selection
The selection and identification of targetable HLA-presented antigens should combine several levels of evidence (triangulation) and not merely use a single method, in order to ensure a high probability of their existence135. Combined selection strategies should aim for confirmation of both HLA presentation and tumour specificity in order to ensure therapeutic relevance and to limit the potential for toxicities. For vaccine development purposes, antigens presented by both HLA class I and HLA class II should ideally be included in a drug product to ensure the activation of both CD8+ T cells and CD4+ T cells, which should synergize to promote antitumour immunity. Furthermore, the robustness of HLA presentation under physiological conditions and during treatment with different therapies should be evaluated, although this is likely to pose a major challenge for future research and has thus far not been conclusively addressed160,161. The available data currently indicate that tumour-specific HLA-presented antigens have a highly patient-specific profile, suggesting that the use of patient-adapted therapies is likely to be unavoidable36,37,38,39. The definition of therapeutic targets from individual tumours and the selection of the ideal target antigens has therefore evolved as a major demand to realize such therapies.
Adjuvantation
The clinical efficacy of immunotherapeutic approaches involving single or predefined sets of antigens has remained disappointing in late-stage development133, despite promising data from early-phase studies131. Insufficient strength of the vaccine-induced immune responses provides a plausible reason for these differences. Other promising approaches, therefore, include formulated mRNA-based vaccines39,162 or effective new adjuvants, such as those targeting STING163. We are currently working on the clinical development of a new synthetic Toll-like receptor 1/2 agonist, termed XS15, that thus far seems to be a strong adjuvant, at least in one human volunteer164. Eventually, immunogenic formulations of antigens and/or combinations with other immunomodulatory substances will be essential and these might also comprise novel therapeutic platforms (but should be reviewed elsewhere).
Feasibility
Individual assembly and manufacturing, particularly of UDDPs, requires considerable effort concerning timing, technique, logistics, production capacity and financing, which is emphasized by the experience with current therapeutic approaches such as CAR T cells15,16,17,19. With increasing levels of individualization, these efforts will need to be expanded. Vaccination-based approaches involving peptides, RNA, DNA or even dendritic cells and adoptive cell transfer might still be achievable36,37,38,39, although the generation of individualized therapies using other carrier systems such as TCR-mimic antibodies, bispecific TCR molecules, TCR gene transfer or CAR T cells featuring patient-specific CARs appears extremely challenging under the current regulations, if not impossible.
Enforcement
To deal with the substantial heterogeneity of the HLA ligandome, the regulatory landscape should be adapted to acknowledge the demands of increasing individualization of immunotherapies. This environment could otherwise become extremely challenging because extensive individualization also results in a lack of experimental and clinical evidence for the efficacy of the respective drug products in selected patient populations. This challenge is illustrated by the tissue-agnostic FDA approval of ICI in MSI-high tumours. Such drug approvals could result in patients receiving a treatment with very little evidence for efficacy in tumours of their specific histology. Treatment allocation for such a diverse range of indications can therefore have substantial limitations, and consideration of additional biomarkers might allow therapy responsiveness to be better estimated beforehand165,166.
Conclusions
Besides all the challenges and unanswered questions, current immunotherapeutic platforms comprising a diverse range of vaccination strategies and novel adjuvants, CAR T cells, adoptive cell transfer, TCR-mimic antibodies and TCR constructs, as well as ICI and HCT, already provide established methods of antigen delivery. Some of these methods have been shown to potently induce or mediate effective immune responses. Further exploiting defined tumour-specific and individually selected antigens based on the analysis of autologous tumours could focus and direct these therapeutic approaches against individual tumours. Nonetheless, suitable combinations of therapeutic platforms that consistently enable effective immunological rejection of established tumours have yet to be established. The immunotherapeutic approaches themselves might already constitute magic bullets according to Paul Ehrlich’s criteria. Although the additional focus on the most valid (magic) targets has the potential to further improve clinical efficacy, ultimately rendering them guided missiles that are able to seek and destroy their targets.
References
Schwartz, R. S. Paul Ehrlich’s magic bullets. N. Engl. J. Med. 350, 1079–1080 (2004).
Thomas, E. D., Lochte, H. L. Jr., Lu, W. C. & Ferrebee, J. W. Intravenous infusion of bone marrow in patients receiving radiation and chemotherapy. N. Engl. J. Med. 257, 491–496 (1957).
Christopher, M. J. et al. Immune escape of relapsed AML cells after allogeneic transplantation. N. Engl. J. Med. 379, 2330–2341 (2018).
Kolb, H. J. et al. Donor leukocyte transfusions for treatment of recurrent chronic myelogenous leukemia in marrow transplant patients. Blood 76, 2462–2465 (1990).
Edinger, M. et al. CD4+CD25+ regulatory T cells preserve graft-versus-tumor activity while inhibiting graft-versus-host disease after bone marrow transplantation. Nat. Med. 9, 1144–1150 (2003).
Mirza, N. et al. Graft versus self (GvS) against T-cell autoantigens is a mechanism of graft-host interaction. Proc. Natl Acad. Sci. USA 113, 13827–13832 (2016).
Wölfel, C. et al. Dissection and molecular analysis of alloreactive CD8+ T cell responses in allogeneic haematopoietic stem cell transplantation. Cancer Immunol. Immunother. 57, 849–857 (2008).
Rosenberg, S. A., Restifo, N. P., Yang, J. C., Morgan, R. A. & Dudley, M. E. Adoptive cell transfer: a clinical path to effective cancer immunotherapy. Nat. Rev. Cancer 8, 299–308 (2008).
Cameron, D. et al. 11 years’ follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive early breast cancer: final analysis of the HERceptin Adjuvant (HERA) trial. Lancet 389, 1195–1205 (2017).
McLaughlin, P. et al. Rituximab chimeric anti-CD20 monoclonal antibody therapy for relapsed indolent lymphoma: half of patients respond to a four-dose treatment program. J. Clin. Oncol. 16, 2825–2833 (1998).
Salles, G. et al. Rituximab in B-cell hematologic malignancies: a review of 20 years of clinical experience. Adv. Ther. 34, 2232–2273 (2017).
Slamon, D. J. et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 344, 783–792 (2001).
Chang, A. Y. et al. Opportunities and challenges for TCR mimic antibodies in cancer therapy. Expert. Opin. Biol. Ther. 16, 979–987 (2016).
Eshhar, Z., Waks, T., Gross, G. & Schindler, D. G. Specific activation and targeting of cytotoxic lymphocytes through chimeric single chains consisting of antibody-binding domains and the gamma or zeta subunits of the immunoglobulin and T-cell receptors. Proc. Natl Acad. Sci. USA 90, 720–724 (1993).
Kiesgen, S., Chicaybam, L., Chintala, N. K. & Adusumilli, P. S. Chimeric antigen receptor (CAR) T-cell therapy for thoracic malignancies. J. Thorac. Oncol. 13, 16–26 (2018).
Maude, S. L. et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 378, 439–448 (2018).
Neelapu, S. S. et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 377, 2531–2544 (2017).
Reinhard, K. et al. An RNA vaccine drives expansion and efficacy of claudin-CAR-T cells against solid tumors. Science 367, 446–453 (2020).
Schuster, S. J. et al. Tisagenlecleucel in adult relapsed or refractory diffuse large B-cell lymphoma. N. Engl. J. Med. 380, 45–56 (2019).
Akahori, Y. et al. Antitumor activity of CAR-T cells targeting the intracellular oncoprotein WT1 can be enhanced by vaccination. Blood 132, 1134–1145 (2018).
Metzinger, M. N. et al. Chimeric antigen receptor T-cell therapy: reach to solid tumor experience. Oncology 97, 59–74 (2019).
Wang, Z., Chen, W., Zhang, X., Cai, Z. & Huang, W. A long way to the battlefront: CAR T cell therapy against solid cancers. J. Cancer 10, 3112–3123 (2019).
Garrido, F., Aptsiauri, N., Doorduijn, E. M., Garcia Lora, A. M. & van Hall, T. The urgent need to recover MHC class I in cancers for effective immunotherapy. Curr. Opin. Immunol. 39, 44–51 (2016).
Eggermont, A. M. M. et al. Adjuvant pembrolizumab versus placebo in resected stage III melanoma. N. Engl. J. Med. 378, 1789–1801 (2018).
Cohen, E. E. W. et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): a randomised, open-label, phase 3 study. Lancet 393, 156–167 (2019).
Larkin, J. et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med. 381, 1535–1546 (2019).
Mok, T. S. K. et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet 393, 1819–1830 (2019).
Rini, B. I. et al. Atezolizumab plus bevacizumab versus sunitinib in patients with previously untreated metastatic renal cell carcinoma (IMmotion151): a multicentre, open-label, phase 3, randomised controlled trial. Lancet 393, 2404–2415 (2019).
Bott, M. J. et al. Initial results of pulmonary resection after neoadjuvant nivolumab in patients with resectable non-small cell lung cancer. J. Thorac. Cardiovasc. Surg. 158, 269–276 (2019).
Davis, W. et al. Next-generation sequencing in 305 consecutive patients: clinical outcomes and management changes. J. Oncol. Pract. 15, e1028–e1034 (2019).
Ascierto, P. A. et al. Survival outcomes in patients with previously untreated BRAF wild-type advanced melanoma treated with nivolumab therapy: three-year follow-up of a randomized phase 3 trial. JAMA Oncol. 5, 187–194 (2019).
Herbst, R. S. et al. Long-term outcomes and retreatment among patients with previously treated, programmed death-ligand 1-positive, advanced non-small-cell lung cancer in the KEYNOTE-010 study. J. Clin. Oncol. 38, 1580–1590 (2020).
Vignard, V. et al. Adoptive transfer of tumor-reactive melan-A-specific CTL clones in melanoma patients is followed by increased frequencies of additional melan-A-specific T cells. J. Immunol. 175, 4797–4805 (2005).
Britten, C. M. et al. The regulatory landscape for actively personalized cancer immunotherapies. Nat. Biotechnol. 31, 880–882 (2013).
Parkhurst, M. R. et al. Unique neoantigens arise from somatic mutations in patients with gastrointestinal cancers. Cancer Discov. 9, 1022–1035 (2019).
Hilf, N. et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature 565, 240–245 (2019).
Keskin, D. B. et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 565, 234–239 (2019).
Ott, P. A. et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017).
Sahin, U. et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017).
Finn, O. J. & Rammensee, H. G. Is it possible to develop cancer vaccines to neoantigens, what are the major challenges, and how can these be overcome? Neoantigens: nothing new in spite of the name. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a028829 (2018).
Backert, L. & Kohlbacher, O. Immunoinformatics and epitope prediction in the age of genomic medicine. Genome Med. 7, 119 (2015).
Boegel, S., Castle, J. C., Kodysh, J., O’Donnell, T. & Rubinsteyn, A. Bioinformatic methods for cancer neoantigen prediction. Prog. Mol. Biol. Transl. Sci. 164, 25–60 (2019).
Richters, M. M. et al. Best practices for bioinformatic characterization of neoantigens for clinical utility. Genome Med. 11, 56 (2019).
Falk, K., Rotzschke, O., Stevanovic, S., Jung, G. & Rammensee, H. G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351, 290–296 (1991).
Kowalewski, D. J. & Stevanovic, S. Biochemical large-scale identification of MHC class I ligands. Methods Mol. Biol. 960, 145–157 (2013).
Dudley, M. E., Ngo, L. T., Westwood, J., Wunderlich, J. R. & Rosenberg, S. A. T-cell clones from melanoma patients immunized against an anchor-modified gp100 peptide display discordant effector phenotypes. Cancer J. 6, 69–77 (2000).
Salgaller, M. L., Weber, J. S., Koenig, S., Yannelli, J. R. & Rosenberg, S. A. Generation of specific anti-melanoma reactivity by stimulation of human tumor-infiltrating lymphocytes with MAGE-1 synthetic peptide. Cancer Immunol. Immunother. 39, 105–116 (1994).
Topalian, S. L., Kasid, A. & Rosenberg, S. A. Immunoselection of a human melanoma resistant to specific lysis by autologous tumor-infiltrating lymphocytes. Possible mechanisms for immunotherapeutic failures. J. Immunol. 144, 4487–4495 (1990).
Tran, E. et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 350, 1387–1390 (2015).
Löffler, M. W. et al. A non-interventional clinical trial assessing immune responses after radiofrequency ablation of liver metastases from colorectal cancer. Front. Immunol. 10, 2526 (2019).
Malarkannan, S., Afkarian, M. & Shastri, N. A rare cryptic translation product is presented by Kb major histocompatibility complex class I molecule to alloreactive T cells. J. Exp. Med. 182, 1739–1750 (1995).
Ng, S. B. et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461, 272–276 (2009).
Löffler, M. W. et al. Multi-omics discovery of exome-derived neoantigens in hepatocellular carcinoma. Genome Med. 11, 28 (2019).
Hacohen, N., Fritsch, E. F., Carter, T. A., Lander, E. S. & Wu, C. J. Getting personal with neoantigen-based therapeutic cancer vaccines. Cancer Immunol. Res. 1, 11–15 (2013).
Schumacher, T. N. & Schreiber, R. D. Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015).
Bassani-Sternberg, M. et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun. 7, 13404 (2016).
Kalaora, S. et al. Combined analysis of antigen presentation and T-cell recognition reveals restricted immune responses in melanoma. Cancer Discov. 8, 1366–1375 (2018).
Löffler, M. W. et al. Mapping the HLA ligandome of colorectal cancer reveals an imprint of malignant cell transformation. Cancer Res. 78, 4627–4641 (2018).
Peng, S. et al. Sensitive detection and analysis of neoantigen-specific T cell populations from tumors and blood. Cell Rep. 28, 2728–2738.e7 (2019).
Bassani-Sternberg, M. & Coukos, G. Mass spectrometry-based antigen discovery for cancer immunotherapy. Curr. Opin. Immunol. 41, 9–17 (2016).
Chen, G. et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 560, 382–386 (2018).
Bjerregaard, A. M. et al. An analysis of natural T cell responses to predicted tumor neoepitopes. Front. Immunol. 8, 1566 (2017).
Turajlic, S. et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 18, 1009–1021 (2017).
Maby, P. et al. Correlation between density of CD8+ T-cell infiltrate in microsatellite unstable colorectal cancers and frameshift mutations: a rationale for personalized immunotherapy. Cancer Res. 75, 3446–3455 (2015).
Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015).
Llosa, N. J. et al. Immunopathologic stratification of colorectal cancer for checkpoint blockade immunotherapy. Cancer Immunol. Res. 7, 1574–1579 (2019).
Mlecnik, B. et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity 44, 698–711 (2016).
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 (2013).
Rosas, G., Ruiz, R., Araujo, J. M., Pinto, J. A. & Mas, L. ALK rearrangements: biology, detection and opportunities of therapy in non-small cell lung cancer. Crit. Rev. Oncol. Hematol. 136, 48–55 (2019).
Druker, B. J. et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N. Engl. J. Med. 344, 1038–1042 (2001).
Woo, C. G. et al. Differential protein stability and clinical responses of EML4-ALK fusion variants to various ALK inhibitors in advanced ALK-rearranged non-small cell lung cancer. Ann. Oncol. 28, 791–797 (2017).
Bocchia, M. et al. Effect of a p210 multipeptide vaccine associated with imatinib or interferon in patients with chronic myeloid leukaemia and persistent residual disease: a multicentre observational trial. Lancet 365, 657–662 (2005).
Cathcart, K. et al. A multivalent bcr-abl fusion peptide vaccination trial in patients with chronic myeloid leukemia. Blood 103, 1037–1042 (2004).
Mackall, C. L. et al. A pilot study of consolidative immunotherapy in patients with high-risk pediatric sarcomas. Clin. Cancer Res. 14, 4850–4858 (2008).
Pinilla-Ibarz, J. et al. Vaccination of patients with chronic myelogenous leukemia with bcr-abl oncogene breakpoint fusion peptides generates specific immune responses. Blood 95, 1781–1787 (2000).
Rojas, J. M., Knight, K., Wang, L. & Clark, R. E. Clinical evaluation of BCR-ABL peptide immunisation in chronic myeloid leukaemia: results of the EPIC study. Leukemia 21, 2287–2295 (2007).
Frankiw, L., Baltimore, D. & Li, G. Alternative mRNA splicing in cancer immunotherapy. Nat. Rev. Immunol. 19, 675–687 (2019).
Park, J. & Chung, Y. J. Identification of neoantigens derived from alternative splicing and RNA modification. Genomics Inform. 17, e23 (2019).
Smith, C. C. et al. Alternative tumour-specific antigens. Nat. Rev. Cancer 19, 465–478 (2019).
Jaganathan, K. et al. Predicting splicing from primary sequence with deep learning. Cell 176, 535–548.e24 (2019).
Slansky, J. E. & Spellman, P. T. Alternative splicing in tumors – a path to immunogenicity? N. Engl. J. Med. 380, 877–880 (2019).
Boultwood, J., Dolatshad, H., Varanasi, S. S., Yip, B. H. & Pellagatti, A. The role of splicing factor mutations in the pathogenesis of the myelodysplastic syndromes. Adv. Biol. Regul. 54, 153–161 (2014).
Pellagatti, A. et al. Impact of spliceosome mutations on RNA splicing in myelodysplasia: dysregulated genes/pathways and clinical associations. Blood 132, 1225–1240 (2018).
Yip, B. H., Dolatshad, H., Roy, S., Pellagatti, A. & Boultwood, J. Impact of splicing factor mutations on pre-mRNA splicing in the myelodysplastic syndromes. Curr. Pharm. Des. 22, 2333–2344 (2016).
Olthof, A. M., Hyatt, K. C. & Kanadia, R. N. Minor intron splicing revisited: identification of new minor intron-containing genes and tissue-dependent retention and alternative splicing of minor introns. BMC Genomics 20, 686 (2019).
Smart, A. C. et al. Intron retention is a source of neoepitopes in cancer. Nat. Biotechnol. 36, 1056–1058 (2018).
Jayasinghe, R. G. et al. Systematic analysis of splice-site-creating mutations in cancer. Cell Rep. 23, 270–281.e3 (2018).
Chong, C. et al. Integrated proteogenomic deep sequencing and analytics accurately identify non-canonical peptides in tumor immunopeptidomes. Nat. Commun. 11, 1293 (2020).
Kahles, A. et al. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell 34, 211–224.e6 (2018).
Climente-Gonzalez, H., Porta-Pardo, E., Godzik, A. & Eyras, E. The functional impact of alternative splicing in cancer. Cell Rep. 20, 2215–2226 (2017).
Seiler, M. et al. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 23, 282–296.e4 (2018).
Vauchy, C. et al. CD20 alternative splicing isoform generates immunogenic CD4 helper T epitopes. Int. J. Cancer 137, 116–126 (2015).
Hanada, K., Yewdell, J. W. & Yang, J. C. Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature 427, 252–256 (2004).
Vigneron, N. et al. An antigenic peptide produced by peptide splicing in the proteasome. Science 304, 587–590 (2004).
Faridi, P. et al. Response to comment on “A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands”. Sci. Immunol. 4, eaaw8457 (2019).
Faridi, P. et al. A subset of HLA-I peptides are not genomically templated: evidence for cis- and trans-spliced peptide ligands. Sci. Immunol. 3, eaar3947 (2018).
Liepe, J. et al. A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 354, 354–358 (2016).
Rolfs, Z., Muller, M., Shortreed, M. R., Smith, L. M. & Bassani-Sternberg, M. Comment on “A subset of HLA-I peptides are not genomically templated: evidence for cis- and trans-spliced peptide ligands”. Sci. Immunol. 4, eaaw1622 (2019).
Boon, T. & Van Pel, A. T cell-recognized antigenic peptides derived from the cellular genome are not protein degradation products but can be generated directly by transcription and translation of short subgenic regions. A hypothesis. Immunogenetics 29, 75–79 (1989).
Abelin, J. G. et al. Complementary IMAC enrichment methods for HLA-associated phosphopeptide identification by mass spectrometry. Nat. Protoc. 10, 1308–1318 (2015).
Cobbold, M. et al. MHC class I-associated phosphopeptides are the targets of memory-like immunity in leukemia. Sci. Transl. Med. 5, 203ra125 (2013).
Malaker, S. A. et al. Identification of glycopeptides as posttranslationally modified neoantigens in leukemia. Cancer Immunol. Res. 5, 376–384 (2017).
Yewdell, J. W., Anton, L. C. & Bennink, J. R. Defective ribosomal products (DRiPs): a major source of antigenic peptides for MHC class I molecules? J. Immunol. 157, 1823–1826 (1996).
Rock, K. L., Farfan-Arribas, D. J., Colbert, J. D. & Goldberg, A. L. Re-examining class-I presentation and the DRiP hypothesis. Trends Immunol. 35, 144–152 (2014).
Yewdell, J. W., Dersh, D. & Fahraeus, R. Peptide channeling: the key to MHC class I immunosurveillance? Trends Cell Biol. 29, 929–939 (2019).
Diederichs, S. et al. The dark matter of the cancer genome: aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol. Med. 8, 442–457 (2016).
Andersen, R. S. et al. High frequency of T cells specific for cryptic epitopes in melanoma patients. Oncoimmunology 2, e25374 (2013).
Laumont, C. M. et al. Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames. Nat. Commun. 7, 10238 (2016).
Newey, A. et al. Immunopeptidomics of colorectal cancer organoids reveals a sparse HLA class I neoantigen landscape and no increase in neoantigens with interferon or MEK-inhibitor treatment. J. Immunother. Cancer 7, 309 (2019).
Singh-Jasuja, H., Emmerich, N. P. & Rammensee, H. G. The Tubingen approach: identification, selection, and validation of tumor-associated HLA peptides for cancer therapy. Cancer Immunol. Immunother. 53, 187–195 (2004).
Marijt, K. A. et al. Identification of non-mutated neoantigens presented by TAP-deficient tumors. J. Exp. Med. 215, 2325–2337 (2018).
Leone, P. et al. MHC class I antigen processing and presenting machinery: organization, function, and defects in tumor cells. J. Natl Cancer Inst. 105, 1172–1187 (2013).
Fritsche, J. et al. Translating immunopeptidomics to immunotherapy-decision-making for patient and personalized target selection. Proteomics 18, e1700284 (2018).
Schuster, H. et al. The immunopeptidomic landscape of ovarian carcinomas. Proc. Natl Acad. Sci. USA 114, E9942–E9951 (2017).
Weinzierl, A. O. et al. Distorted relation between mRNA copy number and corresponding major histocompatibility complex ligand density on the cell surface. Mol. Cell Proteom. 6, 102–113 (2007).
Chaux, P. et al. Identification of five MAGE-A1 epitopes recognized by cytolytic T lymphocytes obtained by in vitro stimulation with dendritic cells transduced with MAGE-A1. J. Immunol. 163, 2928–2936 (1999).
Chen, Q. et al. Characterization of antigen-specific CD8+ T lymphocyte responses in skin and peripheral blood following intradermal peptide vaccination. Cancer Immun. 5, 5 (2005).
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Freudenmann, L. K., Marcu, A. & Stevanovic, S. Mapping the tumour human leukocyte antigen (HLA) ligandome by mass spectrometry. Immunology 154, 331–345 (2018).
Marcu, A. et al. The HLA ligand atlas. A resource of natural HLA ligands presented on benign tissues. Preprint at bioRxiv https://doi.org/10.1101/778944 (2019).
Weinschenk, T. et al. Integrated functional genomics approach for the design of patient-individual antitumor vaccines. Cancer Res. 62, 5818–5827 (2002).
Atanackovic, D. et al. Booster vaccination of cancer patients with MAGE-A3 protein reveals long-term immunological memory or tolerance depending on priming. Proc. Natl Acad. Sci. USA 105, 1650–1655 (2008).
Gnjatic, S. et al. NY-CO-58/KIF2C is overexpressed in a variety of solid tumors and induces frequent T cell responses in patients with colorectal cancer. Int. J. Cancer 127, 381–393 (2010).
Wolchinsky, R. et al. Antigen-dependent integration of opposing proximal TCR-signaling cascades determines the functional fate of T lymphocytes. J. Immunol. 192, 2109–2119 (2014).
Chakraborty, N. G. et al. Recognition of PSA-derived peptide antigens by T cells from prostate cancer patients without any prior stimulation. Cancer Immunol. Immunother. 52, 497–505 (2003).
Kowalewski, D. J. et al. HLA ligandome analysis identifies the underlying specificities of spontaneous antileukemia immune responses in chronic lymphocytic leukemia (CLL). Proc. Natl Acad. Sci. USA 112, E166–E175 (2015).
Caron, E. et al. An open-source computational and data resource to analyze digital maps of immunopeptidomes. eLife 4, e07661 (2015).
Shao, W. et al. The systeMHC atlas project. Nucleic Acids Res. 46, D1237–D1247 (2018).
Rittig, S. M. et al. Long-term survival correlates with immunological responses in renal cell carcinoma patients treated with mRNA-based immunotherapy. Oncoimmunology 5, e1108511 (2016).
Shima, H. et al. Randomized phase II trial of survivin 2B peptide vaccination for patients with HLA-A24-positive pancreatic adenocarcinoma. Cancer Sci. 110, 2378–2385 (2019).
Walter, S. et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat. Med. 18, 1254–1261 (2012).
Jaini, R., Rayman, P., Cohen, P. A., Finke, J. H. & Tuohy, V. K. Combination of sunitinib with anti-tumor vaccination inhibits T cell priming and requires careful scheduling to achieve productive immunotherapy. Int. J. Cancer 134, 1695–1705 (2014).
Rini, B. I. et al. IMA901, a multipeptide cancer vaccine, plus sunitinib versus sunitinib alone, as first-line therapy for advanced or metastatic renal cell carcinoma (IMPRINT): a multicentre, open-label, randomised, controlled, phase 3 trial. Lancet Oncol. 17, 1599–1611 (2016).
Schwartzentruber, D. J. et al. gp100 peptide vaccine and interleukin-2 in patients with advanced melanoma. N. Engl. J. Med. 364, 2119–2127 (2011).
Munafo, M. R. & Davey Smith, G. Robust research needs many lines of evidence. Nature 553, 399–401 (2018).
Ghosh, M. et al. Guidance document: validation of a high-performance liquid chromatography-tandem mass spectrometry immunopeptidomics assay for the identification of HLA class I ligands suitable for pharmaceutical therapies. Mol. Cell Proteom. 19, 432–443 (2020).
Wang, Q., Shashikant, C. S., Jensen, M., Altman, N. S. & Girirajan, S. Novel metrics to measure coverage in whole exome sequencing datasets reveal local and global non-uniformity. Sci. Rep. 7, 885 (2017).
Gubin, M. M. et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577–581 (2014).
Yadav, M. et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515, 572–576 (2014).
Gfeller, D. & Bassani-Sternberg, M. Predicting antigen presentation–what could we learn from a million peptides? Front. Immunol. 9, 1716 (2018).
Andreatta, M. & Nielsen, M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 32, 511–517 (2016).
Rammensee, H., Bachmann, J., Emmerich, N. P., Bachor, O. A. & Stevanovic, S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50, 213–219 (1999).
Bekker-Jensen, D. B. et al. An optimized shotgun strategy for the rapid generation of comprehensive human proteomes. Cell Syst. 4, 587–599.e4 (2017).
Wang, D. et al. A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol. Syst. Biol. 15, e8503 (2019).
Beaubier, N. et al. Integrated genomic profiling expands clinical options for patients with cancer. Nat. Biotechnol. 37, 1351–1360 (2019).
Wilson, E. A. & Anderson, K. S. Lost in the crowd: identifying targetable MHC class I neoepitopes for cancer immunotherapy. Expert. Rev. Proteom. 15, 1065–1077 (2018).
Maus, M. V. et al. An MHC-restricted antibody-based chimeric antigen receptor requires TCR-like affinity to maintain antigen specificity. Mol. Ther. Oncolytics 3, 1–9 (2016).
Cameron, B. J. et al. Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Sci. Transl. Med. 5, 197ra103 (2013).
Linette, G. P. et al. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood 122, 863–871 (2013).
Swain, S. M. et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N. Engl. J. Med. 372, 724–734 (2015).
Seol, Y. M. et al. A pilot prospective study of refractory solid tumor patients for NGS-based targeted anticancer therapy. Transl. Oncol. 12, 301–307 (2019).
Kantoff, P. W. et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N. Engl. J. Med. 363, 411–422 (2010).
Noguchi, M. et al. Phase II study of personalized peptide vaccination for castration-resistant prostate cancer patients who failed in docetaxel-based chemotherapy. Prostate 72, 834–845 (2012).
Terasaki, M. et al. Phase I trial of a personalized peptide vaccine for patients positive for human leukocyte antigen–A24 with recurrent or progressive glioblastoma multiforme. J. Clin. Oncol. 29, 337–344 (2011).
Nguyen, L. T. et al. Phase II clinical trial of adoptive cell therapy for patients with metastatic melanoma with autologous tumor-infiltrating lymphocytes and low-dose interleukin-2. Cancer Immunol. Immunother. 68, 773–785 (2019).
European Medicines Agency. Guideline on the requirements for the chemical and pharmaceutical quality documentation concerning investigational medicinal products in clinical trials. https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-requirements-chemical-pharmaceutical-quality-documentation-concerning-investigational_en.pdf (2017).
Chen, L., Qiao, D., Wang, J., Tian, G. & Wang, M. Cancer immunotherapy with lymphocytes genetically engineered with T cell receptors for solid cancers. Immunol. Lett. 216, 51–62 (2019).
Salter, A. I., Pont, M. J. & Riddell, S. R. Chimeric antigen receptor-modified T cells: CD19 and the road beyond. Blood 131, 2621–2629 (2018).
Ali, M. et al. Induction of neoantigen-reactive T cells from healthy donors. Nat. Protoc. 14, 1926–1943 (2019).
Caron, E. et al. The MHC I immunopeptidome conveys to the cell surface an integrative view of cellular regulation. Mol. Syst. Biol. 7, 533 (2011).
Reits, E. A. et al. Radiation modulates the peptide repertoire, enhances MHC class I expression, and induces successful antitumor immunotherapy. J. Exp. Med. 203, 1259–1271 (2006).
Armbruster, N., Jasny, E. & Petsch, B. Advances in RNA vaccines for preventive indications: a case study of a vaccine against rabies. Vaccines 7, 132 (2019).
Gutjahr, A. et al. The STING ligand cGAMP potentiates the efficacy of vaccine-induced CD8+ T cells. JCI Insight 4, e125107 (2019).
Rammensee, H. G. et al. A new synthetic toll-like receptor 1/2 ligand is an efficient adjuvant for peptide vaccination in a human volunteer. J. Immunother. Cancer 7, 307 (2019).
Eso, Y., Shimizu, T., Takeda, H., Takai, A. & Marusawa, H. Microsatellite instability and immune checkpoint inhibitors: toward precision medicine against gastrointestinal and hepatobiliary cancers. J. Gastroenterol. 55, 15–26 (2020).
Lee, J. S. & Ruppin, E. Multiomics prediction of response rates to therapies to inhibit programmed cell death 1 and programmed cell death 1 ligand 1. JAMA Oncol. 5, 1614–1618 (2019).
Brossart, P. et al. Identification of HLA-A2-restricted T-cell epitopes derived from the MUC1 tumor antigen for broadly applicable vaccine therapies. Blood 93, 4309–4317 (1999).
Pittet, M. J. et al. Melan-A/MART-1-specific CD8 T cells: from thymus to tumor. Trends Immunol. 23, 325–328 (2002).
Acknowledgements
The authors thank L. Yakes from the Department of Immunology at the University of Tübingen for language editing and editorial assistance, and A. Marcu from the Department of Immunology at the University of Tübingen for helpful discussions.
Author information
Authors and Affiliations
Contributions
All authors made a substantial contribution to all aspects of the preparation of the manuscript.
Corresponding author
Ethics declarations
Competing interests
S.P.H. has acted as an advisory board member for Abbvie, Bristol–Myers Squibb, MSD and Roche, and is a co-inventor of several patents owned by Immatics Biotechnologies. M.W.L. is a co-inventor of several patents owned by Immatics Biotechnologies, and has acted as a consultant and/or advisory board member for Boehringer Ingelheim. H.-G.R. has ownership interest (including shares) in CureVac, Immatics Biotechnologies and Synimmune, is a co-inventor and/or has shared interests in several patents held by CureVac, Immatics Biotechnologies and Synimmune, and is a co-inventor and shares the patent for the adjuvant candidate XS15. P.B. has acted as a consultant or advisory board member for Amgen, AstraZeneca, Bristol–Myers Squibb, MSD and Roche, has received speaker’s fees from Abbvie, AstraZeneca, Bristol-Myers Squibb, and MSD, and has ownership interests in Immatics Biotechnologies.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
HLA Alleles: http://hla.alleles.org/alleles
Glossary
- Antigens
-
Immunologically recognizable cell-surface structures.
- Antigen-aware therapy
-
(AaT). An active immunotherapy leading to the activation of and/or constituting antigen-specific cells designed to target known and/or well-defined antigens.
- Antigen-unaware therapy
-
(AuT). An active immunotherapy leading to the activation of and/or constituting antigen-specific cells designed to targeting uncharacterized antigens.
- Cell-surface antigens
-
Proteins or parts thereof located on the outer side of the cell membrane that are therefore accessible from outside the cell.
- T cell repertoire
-
The fraction of T cells capable of reacting against a defined HLA-restricted antigen, as detected by assays designed to measure immune cell function, without confirmation of lytic activity.
- Uniquely assembled drug product
-
(UADP). Drug product, or set of drug products assembled and (pre-)manufactured individually that can then be assembled and administered to a selected patient or patients based on the presence of biomarkers indicating responsiveness to a predesigned set of active ingredients (for example, a peptide warehouse).
- Uniquely designed drug product
-
(UDDP). A drug product specifically defined and manufactured for a specific patient according to individual clinical features that is not intended to be administered to any other individual.
- Tumour-specific antigens
-
(TSAs). HLA ligands exclusively presented on tumours that are not presented on any other tissues.
- Stratification
-
Allocation of a therapy with a defined or invariant drug product based on the presence of one or more specific biomarkers.
- Individualization
-
The fully individualized design of a drug product for a specific patient based on biomarker analyses of an individual tumour or tumours (a variant drug product).
- Tumour-associated antigens
-
(TAAs). HLA-presented peptides present on a tumour that can also be presented on other non-malignant tissues.
- Mutated tumour-specific antigens
-
HLA-binding peptides with tumour-specific alternative sequences (compared with the germline sequence) derived from one of several types of germline mutations including single-nucleotide variants, indels, fusions and others. Synonymously referred to as mutated neoantigens.
- Neoantigen
-
A term commonly used to describe tumour-specific antigens (TSAs). This term has mostly been used to describe mutated TSAs and can cause confusion when also used to describe wild-type sequence TSAs.
- Next-generation sequencing
-
(NGS). Massively parallel sequencing technology enabling the rapid, high-throughput analysis of nucleic acid sequences including those from the whole genome, exome or transcriptome.
- Tandem mass spectrometry
-
(MS/MS). Liquid chromatography followed by two-stage mass spectrometry including ionization of samples followed by detection of the parental mass and the subsequent fragmentation and assessment of respective fragment masses.
- Predicted HLA ligands
-
Amino acid sequences with defined HLA-binding motifs that can be inferred in silico using computational algorithms.
- Alternatively spliced TSAs
-
An HLA-binding peptide with a tumour-specific alternative amino acid sequence (such as that containing a neojunction) derived from alternative mRNA splicing events.
- Cryptic antigens
-
HLA-eluted peptides detected by mass spectrometry that lack clear correlates in the consensus coding sequence (exome).
- Proteasomal splicing
-
Proteasome-catalysed peptide sequence modification through ligation of other liberated protein fragments.
- Defective ribosomal products
-
(DRIPs). Prematurely terminated and misfolded peptides produced from translation of bona fide mRNA in the appropriate reading frames.
- Dark matter of the proteome
-
The proteomic correlate of genomic dark matter, located outside the coding region and assumed not to be translated.
- Consensus genomic sequence
-
The calculated order of most frequent residues, either nucleotides or amino acids, found at each position in a sequence alignment within the genome.
- Naturally presented HLA ligands
-
Peptides presented by MHC that can be detected using MS/MS following MHC immunoprecipitation or by T cell based detection assays.
- HLA ligandome
-
A naturally occurring, non-immunologically validated HLA-presented peptide repertoire, characterized by MS/MS after HLA immunoprecipitation and peptide elution.
- Wild-type tumour-specific antigens
-
HLA-eluted peptides with wild-type sequences (compared with the relevant germline sequence) that nonetheless have tumour-specific presentation, and to which the immune system has not been previously exposed and not represented on benign tissues.
- Tumour-specific antigens with reactivated early ontogenic expression
-
Tumour-specific HLA ligands (compared with the germline sequence) derived from proteins that are usually physiologically restricted to the early stages of ontogenetic expression, including cancer–testis antigens.
- Post-translationally modified tumour-associated antigens
-
HLA ligands with wild-type (or in rare cases also alternative) sequences derived from tumour-specific post-translational modifications.
- Overexpressed tumour-associated antigens
-
An HLA ligand that is typically overexpressed on tumour cells, albeit with lower levels of expression on non-malignant tissues.
- Tumour mutational load
-
The sum total of somatic mutations located in the exomes of cancer cells. Tumour mutational load should not be viewed as equivalent to tumour mutational burden.
- Tumour mutational burden
-
A biomarker used for predicting therapy response in clinical trials that usually reflects the tumour mutational load of selected genes, as assessed primarily using the Foundation Medicine gene panel, but also potentially other assays (such as the MSKCC-IMPACT panel). This term only applies directly to this reduced proportion of mutated genes and does not indicate the entire tumour mutational load.
Rights and permissions
About this article
Cite this article
Haen, S.P., Löffler, M.W., Rammensee, HG. et al. Towards new horizons: characterization, classification and implications of the tumour antigenic repertoire. Nat Rev Clin Oncol 17, 595–610 (2020). https://doi.org/10.1038/s41571-020-0387-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41571-020-0387-x
This article is cited by
-
mRNA-based precision targeting of neoantigens and tumor-associated antigens in malignant brain tumors
Genome Medicine (2024)
-
Challenges in the discovery of tumor-specific alternative splicing-derived cell-surface antigens in glioma
Scientific Reports (2024)
-
Neoantigen-targeted TCR-engineered T cell immunotherapy: current advances and challenges
Biomarker Research (2023)
-
Immunogenic cell death in cancer: concept and therapeutic implications
Journal of Translational Medicine (2023)
-
Antigen presentation in cancer — mechanisms and clinical implications for immunotherapy
Nature Reviews Clinical Oncology (2023)