Abstract
Combining HLA-DQ–gluten tetramers with mass cytometry and RNA sequencing analysis, we find that gluten-specific CD4+ T cells in the blood and intestines of patients with celiac disease display a surprisingly rare phenotype. Cells with this phenotype are also elevated in patients with systemic sclerosis and systemic lupus erythematosus, suggesting a way to characterize CD4+ T cells specific for disease-driving antigens in multiple autoimmune conditions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data and code availability
The raw sequences of the RNA-seq data are deposited at the EGA European Genome Phenome Archive (https://ega-archive.org/studies/EGAS00001003017). The source code is available on https://github.com/eivindgl/natmed_gluten_tcell_mass_cytometry. All other data supporting the findings of this study are available from the corresponding authors on request.
References
Abadie, V. et al. Annu. Rev. Immunol. 29, 493–525 (2011).
Rao, D. A. et al. Nature 542, 110–114 (2017).
Sollid, L. M. et al. Gut 41, 851–852 (1997).
Di Niro, R. et al. Nat. Med. 18, 441–445 (2012).
Steinsbo, O. et al. Nat. Commun. 5, 4041 (2014).
Hoydahl, L. S., et al. Gastroenterology https://doi.org/10.1053/j.gastro.2018.12.013 (2018).
Moens, L. et al. Front. Immunol. 5, 65 (2014).
Kooy-Winkelaar, Y. M. et al. Proc. Natl Acad. Sci. USA 114, E980–E989 (2017).
Christophersen, A. et al. United European Gastroenterol. J. 2, 268–278 (2014).
Raki, M. et al. Proc. Natl Acad. Sci. USA 104, 2831–2836 (2007).
Du Pre, M. F. et al. Am. J. Gastroenterol. 106, 1147–1159 (2011).
Cook, L. et al. J. Allergy Clin. Immunol. 140, 1592–1603 (2017).
Risnes, L. F. et al. J. Clin. Invest. 128, 2642–2650 (2018).
Yu, W. et al. Immunity 42, 929–941 (2015).
Su, L. F. et al. Immunity 38, 373–383 (2013).
Ludvigsson, J. F. et al. Gut 63, 1210–1228 (2014).
Marsh, M. N. et al. Baillieres Clin. Gastroenterol. 9, 273–293 (1995).
Oberhuber, G. et al. Eur. J. Gastroenterol. Hepatol. 11, 1185–1194 (1999).
Anderson, R. P. et al. Nat. Med. 6, 337–342 (2000).
van den Hoogen, F. et al. Ann. Rheum. Dis. 72, 1747–1755 (2013).
Hochberg, M. C. Arthritis Rheum. 40, 1725 (1997).
Newell, E. W. et al. Immunity 36, 142–152 (2012).
Newell, E. W. et al. Nat. Biotechnol. 31, 623–629 (2013).
Mei, H. E. et al. J. Immunol. 194, 2022–2031 (2015).
Shan, L. et al. Science 297, 2275–2279 (2002).
Quarsten, H. et al. J. Immunol. 167, 4861–4868 (2001).
Christophersen, A. et al. J. Immunol. 196, 2819–2826 (2016).
Sollid, L. M. et al. Immunogenetics 64, 455–460 (2012).
Molberg, Ø. et al. Methods Mol. Med. 41, 105–124 (2000).
Bodd, M. et al. Eur. J. Immunol. 43, 2605–2612 (2013).
van der Maaten, L. et al. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Nowicka, M. et al. F1000Res. 6, 748 (2017).
Andy, L. et al. R News 2, 18–22 (2002).
Patro, R. et al. Nat. Methods 14, 417–419 (2017).
Love, M. I. et al. Genome Biol. 15, 550 (2014).
Acknowledgements
We thank the patients participating in this study, S. Furholm, C. Hinrichs and M. H. Bakke for collecting patient material at the Endoscopy Unit (Oslo University Hospital—Rikshospitalet), the Stanford-LPCH Vaccine Program with S. Swope and S. Mackey for the study of patients with influenza virus infection, A. Nau (Davis group), M. Leipold (The Human Immune Monitoring Center, Stanford University) and Brith Bergum (Flow Cytometry Core Facility, University of Bergen) for technical assistance with the Helios mass cytometers, B. Simonsen and S. R. Lund (Sollid group) for producing the biotinylated HLA-DQ2.5:gluten molecules, M. K. Johannesen (Sollid group) for laboratory technical assistance, K. J. Rolfsen (University of Oslo) for producing the cookies for gluten challenge, G. K. Sandve (University of Oslo) for critical inputs on RNA-seq data analysis in addition to V. K. Sarna (University of Oslo) and L. Chung (Stanford Medicine) for help with the clinical assessment of patients. We also thank the Flow Cytometry Core Facility (Oslo University Hospital—Rikshospitalet and Radiumhospitalet) and the Stanford Shared FACS Facility for technical assistance. We express our gratitude to the funding bodies of this research: The University of Oslo Scientia Fellows program, co-funded by the University of Oslo World-leading research program on human immunology (WL-IMMUNOLOGY) (L.M.S.) and by the EC FP7 Maria Sklodowska-Curie COFUND Programme (GA 609020) (A.C. and L.M.S.); Stiftelsen KG Jebsen (project SKGJ-MED-017) (L.M.S. and K.E.A.L.); The Unger-Vetlesen Medical Fund (A.C.); The U.S.-Norwegian Fulbright Foundation for educational exchange (A.C.); Fondsstiftelsen (Oslo University Hospital) (A.C.); The Howard Hughes Medical Institute (M.M.D.); The Simons Foundation (M.M.D.); The National Institutes of Health; U19-AI057229 (M.M.D.), U19-AI110491 (P.J.U.), UL1 TR001085 (C.L.D.) and R01 AI125197-01 (P.J.U.); The Donald E. and Delia B. Baxter Foundation (P.J.U.); The Henry Gustav Floren Trust (P.J.U.); and a gift from Elizabeth F. Adler (P.J.U.).
Author information
Authors and Affiliations
Contributions
A.C., L.M.S. and M.M.D. conceptualized the study and drafted the manuscript with support from E.G.L. and O.S. A.C. developed the protocol for class II tetramer staining combined with mass cytometry, established the mass cytometry staining panels and performed the flow cytometry and most mass cytometry staining experiments. E.S. performed the mass cytometry staining experiments on influenza samples and some autoimmune samples. O.S. and L.M.S. designed the RNA-seq study and O.S. prepared the libraries for RNA-seq. RNA-seq data were analyzed by E.G.L. and C.K. The mass cytometry data were analyzed by E.G.L. and A.C. The CeD patient material was organized by K.E.A.L., S.D.-K. and S.Z. Material from patients with autoimmune disorders other than CeD was organized by Ø.M., P.J.U., M.R.-P. and J.F.S. Material from patients during and after influenza infection was organized by C.L.D. Critical manuscript revisions were done by E.S., C.K., S.D.-K., S.Z., Ø.M., P.J.U., M.R.-P., J.F.S, C.L.D. and K.E.A.L.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Establishing HLA class II tetramer staining with mass cytometry.
a, Gluten-specific T cell clone binding a corresponding or negative control HLA-DQ2.5:gluten tet reagent metal tagged with secondary binding to PE, APC or FITC (one T cell clone in one experiment). b, Comparison of tet staining in mass cytometry and flow cytometry with a gluten-specific T cell clone binding the corresponding or non-corresponding HLA-DQ2.5:gluten tet reagent (n = 8 distinct T cell clones in two mass cytometry and two flow cytometry experiments, respectively). c, Tet enrichment of a gluten-specific T cell clone binding the corresponding PE-Cy7-coupled HLA-DQ2.5:gluten tet reagent (one T cell clone in one experiment). The T cell clone was spiked into PBMCs, enriched with anti-Cy7 beads and metal tagged with anti-PE (one T cell clone in one experiment). d, Unspecific HLA-DQ2.5:gluten tet binding was excluded with APC-Cy7-coupled HLA-DQ2.5:CLIP2 and metal-tagged anti-APC (n = 7 patients with UCeD and 10 controls in 9 experiments. Here, two distinct T cell clones and three PBMC samples were analyzed in two pilot experiments before the tet staining approach was established).
Extended Data Fig. 2 Gating strategy for cells analyzed with mass cytometry.
From the initial plot to the plot and gate that encounters CD4+ blood or gut T cells. Anti-CD45 coupled with 89Y or 108Pd was used for sample barcoding.
Extended Data Fig. 3 On the CD4+ gut T cells analyzed with mass cytometry.
a, Heat map showing the fold-change expression of indicated markers in CD4+ HLA-DQ2.5:gluten tet-negative gut T cells of six patients with UCeD versus CD4+ gut T cells of seven healthy controls (five experiments in total). b, Expression level of mass cytometry panel markers (Supplementary Table 1) in gluten tet-positive and tet-negative CD4+ gut T cells in six patients with UCeD (five experiments). The y axis indicates the ArcSinh-transformed intensity. Box plots show the median frequency, the interquartile range and the whiskers show largest and smallest values below 1.5 times the interquartile range. c, t-SNE plots separately highlighting the presence of cells expressing the markers in a and b in CD4+ gut T cells merged from 1 patient with UCeD and 1 control subject (1 of n = 6 patients with UCeD and 1 of n = 7 controls, 5 experiments). For comparison, the location of HLA-DQ2.5:gluten tet-binding CD4+ gut T cells of the same patient is visualized in the upper left plot.
Extended Data Fig. 4 Mass cytometry and RNA-seq data correlation.
a,b, Correlation between RNA-seq-derived and mass cytometry-derived fold-change expression of HLA-DQ2.5:gluten tet-positive versus HLA-DQ2.5:gluten tet-negative CD4+ gut T cells in patients with UCeD (a) and versus CD4+ gut T cells in controls (corresponding data depicted as a heat map in Fig. 1e,f) (b). For RNA-seq data: n = 5 patients with UCeD, 4 control subjects in 2 experiments. For mass cytometry data: n = 6 patients with UCeD, 7 controls in 5 experiments. The shaded region indicates the 95% confidence interval around the regression line.
Extended Data Fig. 5 On the CD4+ blood T cells analyzed with mass cytometry.
a, Heat map showing the log2 fold-change expression of indicated markers in CD4+ blood T cells of seven patients with UCeD (pre-tet-enriched sample) versus ten healthy controls in nine experiments. b, Expression level of mass cytometry panel markers (Supplementary Table 1) in gluten tet-positive and pre-tet-enriched CD4+ blood T cells in seven patients with UCeD (six experiments). The y axis indicates the ArcSinh-transformed intensity values. The box plots show the median frequency, the interquartile range and the whiskers show the largest and smallest values below 1.5 times the interquartile range. c, t-SNE plots separately highlighting the presence of cells expressing the markers in a and b in CD4+ blood T cells merged from 1 healthy control and 1 patient with UCeD (1 of n = 7 patients with UCeD and 1 of n = 10 controls in 9 experiments). For comparison, the location of HLA-DQ2.5:gluten tet-binding CD4+ blood T cells of the same patient with UCeD is visualized in the upper left plot.
Extended Data Fig. 6 Flow cytometry staining confirms CXCR5/ICOS expression.
General gating strategy for flow cytometry analysis of tet-binding cells, including the expression of CXCR5 and ICOS in tet-positive and tet-negative (+/−) CD4+ blood T cells in one patient with UCeD (in one experiment).
Extended Data Fig. 7 Expression of regulatory T cell-associated markers on gluten-specific CD4+ T cells in vitro and ex vivo.
a, CD4+ blood T cells of a patient with UCeD were HLA-DQ2.5:gluten tet-sorted ex vivo and cultured in vitro with phytohemmagglutinin and irradiated PBMCs for 2 weeks before re-staining with HLA-DQ2.5:gluten tets to analyze for the expression of Foxp3 and CD25 (n = 2, 1 experiment). b, The same experiment as in a, only with tet-sorted CD4+ gut T cells from the patient in a (n = 1, 1 experiment). c, RNA-seq-derived log2 fold-change expression of glycoprotein A repetitions predominant (GARP) in tet+ (<2 GARP transcripts per million) versus tet− CD4+ gut T cells of five patients with UCeD and in tet+ of five patients with UCeD versus CD4+ gut T cells of four control subjects. GARP was differentially expressed in tet+ versus tet− cells, but not when compared to CD4+ gut T cells in controls (indicated by asterisks) (differentially expressed genes in Supplementary Table 5). d,e, Ex vivo flow cytometry staining of tet+/− CD4+ gut T cells from a patient with UCeD with anti-CD127, anti-CD25 and anti-Foxp3 (d) and summarized CD25/Foxp3 staining in gut biopsies of five patients with HLA-DQ2.5+ UCeD and one patient with UCeD HLA-DQ8+ (five experiments) (e). The median frequency and interquartile range are indicated. f,g, Tet+/− CD4+ blood T cells from a patient with UCeD with anti-CD127, anti-CD25 and anti-Foxp3 (f) and summarized CD25/FoxP3 staining in blood of five patients with UCeD and four gluten-challenged patients with UCeD (four experiments) (g). The median frequency and interquartile range are indicated. Samples in a and b were stained with a different anti-CD25 antibody than samples in d, e, f, and g.
Extended Data Fig. 8 Different pattern of activated CD4+ T cells in patients with autoimmune diseases versus influenza infection.
a, In Fig. 2h, t-SNE visualization and unsupervised clustering of activated (CD38+) memory (CD45RA−) CD4+ blood T cells in indicated participant groups and gluten tet-positive (tet+) cells of patients with UCeD are shown. One cluster containing 75% of tet+ cells (cluster 1) from seven patients with UCeD and one cluster dramatically upregulated in subjects with influenza infection (cluster 2) are color coded in Fig. 2h. The prevalence of activated CD4+ memory T cells belonging to cluster 1 and cluster 2, respectively, for each indicated participant group is shown. Here, we randomly selected n = 5 distinct samples per participant group, except for the gluten challenge group for which we only had n = 4 distinct samples with sufficient cells. The median frequency and the interquartile range are shown, as well as the whiskers, showing the largest and smallest values below 1.5 times the interquartile range. The single data points depict outliers. b,c, Heat map of the indicated proteins in cluster 1 and cluster 2 with absolute expression (staining intensity) (b) and versus CD4+ blood T cells depicted as the log2 fold change of the grand mean of donor marker intensity (c).
Extended Data Fig. 9 Supervised clustering model predicting the gluten-specific T cell profile.
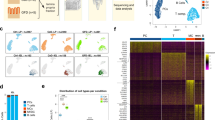
a, Diagram illustrating the workflow for model training and prediction. PBMC samples from donors with UCeD are split into two parts as indicated. One part (right) is not tet enriched and is later used for estimation of gluten-specific T cell profile cell prevalence within the sample. The tet-enriched part (left) is used to train a random forest classification model using repeated K-fold cross-validation on the phenotype of the tet-positive cells. b, Scatter plot of the mean decrease in the Gini score for each predictor provides information on how important the predictor variables are to the final model.
Extended Data Fig. 10 Cells with profile of gluten-specific CD4+ T cells in celiac, autoimmune and viral disease identified with manual gating.
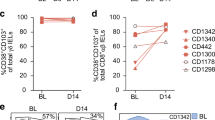
a, Manual gating strategy with markers giving a well-defined shift in staining intensity that define gluten-specific T cells, encompassing 41% and 48% of HLA-DQ2.5:gluten tet-binding CD4+ T cells in the gut and CD4+ effector memory T cells in the blood, respectively, in patients with UCeD (although the gluten-specific cells were phenotypically similar, not all of the cells had a staining intensity for all ten markers above or below the manually set threshold, as also visualized in Figs. 1c and 2c). Here, visualized in the peripheral blood of a patient with UCeD. b–d, Frequency of cells gated as in a in the gut (b) and in the blood (c) (pre-tet-enriched sample) of patients with UCeD, TCeD (gluten-free diet) and healthy controls, and in patients with TCeD before and following gluten challenge (differing from gating encountering gluten-specific cells in patients with UCeD chiefly by lower CD39 expression, as also visualized in Fig. 2f) (d). Blood and gut samples analyzed in 12 and six experiments, respectively. Gluten challenge samples were analyzed in two experiments. e,f, Frequency of cells gated as in a within patients with the indicated autoimmune disorders and different set as in b of control subjects (seven experiments in total) (e), and within a cohort during and after influenza infection (f) (three experiments in total). Statistics: an unpaired, two-tailed t-test was used and the median frequency and interquartile range are indicated in b, c and e. *A paired, two-tailed t-test was used and the lines indicate paired samples in d and f.
Supplementary information
Rights and permissions
About this article
Cite this article
Christophersen, A., Lund, E.G., Snir, O. et al. Distinct phenotype of CD4+ T cells driving celiac disease identified in multiple autoimmune conditions. Nat Med 25, 734–737 (2019). https://doi.org/10.1038/s41591-019-0403-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0403-9
This article is cited by
-
Tolerance-inducing therapies in coeliac disease — mechanisms, progress and future directions
Nature Reviews Gastroenterology & Hepatology (2024)
-
Heterogeneity of circulating CXCR5-PD-1hiTph cells in patients of type 2 and type 1 diabetes in Chinese population
Acta Diabetologica (2023)
-
HLA autoimmune risk alleles restrict the hypervariable region of T cell receptors
Nature Genetics (2022)
-
Single cell transcriptomic analysis of the immune cell compartment in the human small intestine and in Celiac disease
Nature Communications (2022)
-
Maladaptive consequences of inflammatory events shape individual immune identity
Nature Immunology (2022)