Abstract
The intestinal microbiota is an important contributor to the health of preterm infants, and may be destabilized by a number of environmental factors and treatment modalities. How to promote the development of a healthy microbiota in preterm infants is largely unknown. We collected fecal samples from 45 breastfed preterm very low birth weight (birth weight < 1500 g) infants from birth until 60 days postnatal age to characterize the intestinal microbiota development during the first weeks of life in preterm infants. Fecal microbiota composition was determined by 16S rRNA amplicon sequencing. The main driver of microbiota development was gestational age; antibiotic use had strong but temporary effects and birth mode had little influence. Microbiota development proceeded in four phases indicated by the dominance of Staphylococcus, Enterococcus, Enterobacter, and finally Bifidobacterium. The Enterococcus phase was only observed among the extremely premature infants and appeared to delay the microbiota succession. The results indicate that hospitalized preterm infants receiving breast milk may develop a normal microbiota resembling that of term infants.
Similar content being viewed by others
Introduction
Prematurity is a risk factor for infant morbidity and mortality1, and is associated with a high risk of bacterial and inflammatory diseases, such as sepsis and necrotizing enterocolitis (NEC). Preterm infants have an immature intestine with underdeveloped peristalsis, barrier function and immunity, which makes the intestine a potential source of infections and inflammation2,3. Several studies have shown that the intestinal microbiota in preterm infants differ from that of healthy term infants4,5,6,7,8,9,10,11. Common anomalies are the bloom of opportunistic and potentially pathogenic bacteria such as Enterobacter, Enterococcus, and Staphylococcus4,5,6,7. Many common practices, caesarean section12,13,14, antibiotic use15, and formula feeding14,15,16, may disrupt normal microbiota development. Moreover, studies with small groups of infants have indicated that both NEC and late-onset sepsis are associated with alterations in the microbiota8,9,10,11, and that the microbiota composition correlates with signs of pain and distress17. Modification of the microbiota by probiotics or breast milk may reduce the risk of NEC in preterm infants18,19. Thus, promoting a healthy microbiota development in preterm infants is important, and a key question is whether natural colonization and succession of the microbiota is possible in these infants. For rational decision-making and consideration of therapies in the future, information on the microbiota development in preterm infants is essential. Although studies on the preterm microbiota have been conducted4,5,6,7,8,9,10,11, they involve only a handful of infants and none show detailed microbiota succession.
To establish a defined and predictive picture of the preterm infant microbiota development, we analyzed 262 fecal samples from 45 preterm infants using deep 16S rRNA amplicon sequencing.
Methods
Study design and participants
The samples were collected during a randomized controlled trial conducted in 2010 at Akershus and Oslo University Hospitals, Norway. The study was approved by the Regional Committee for Medical and Health Research Ethics, Norway (Reference number: 2009/1946) and performed in accordance with the principles of the Helsinki Declaration. Written informed consent was obtained from the parents. The study was registered in Clinical Trials (registration number NCT01103219) on 12. April 2010. The trial was designed to investigate the effect of enhanced nutrient supply to very low birth weight infants (VLBW; birth weight < 1500 g) as previously described20.
All VLBW infants born between August 17th and December 21st 2010 in the study hospitals were eligible for inclusion. Exclusion criteria were congenital malformations, chromosomal abnormalities, critical illness with short life expectancy and clinical syndromes known to affect growth and development. Infants were included after written informed parental consent was obtained and randomized into two nutrition groups within 24 h after birth. Fifty-seven infants were eligible for inclusion but 7 infants were not included due to parental consent refusal (1), critically sick mother/sibling (3), congenital anomaly (1) and omissions during enrollment (2). Thus, 50 infants were included in the trial. Three infants died during the first weeks of life and two infants were excluded due to critical illness and congenital heart disease. Hence, 45 VLBW infants were included in the present study investigating the microbiota development. Twenty-four of these infants received enhanced nutrient supply, i.e. increased supply of energy, protein, fat, the long-chain polyunsaturated fatty acids (PUFA) and vitamin A20. Twenty-one infants were classified as extremely premature (EP) and 24 as moderately or very premature (MVP). Caesarean delivery was common, especially in the MVP group (Table 1).
Four types of antibiotics were given to the infants: aminoglycosides (gentamycin, N = 10) usually combined with ampicillin/ekvacillin (N = 26), and vancomycin (N = 6) usually combined with cephalosporin (N = 12). Ampicillin and cephalosporin were not given alone. The antibiotics were all given intravenously. Vancomycin was not given to any of the infants during the first 10 days of life.
Fecal sample collection
Fecal samples were collected from diapers at different intervals from birth until discharge from the hospitals (up to 60 days postnatal age). The median (range) number of samples per infant was 6 (2–11). Fresh samples were collected from diapers and frozen at −80 °C until analyzed. In total 262 fecal samples were analyzed.
16S rRNA amplicon sequencing and pre-processing
The microbiota composition of the fecal samples was analyzed using 16S rRNA amplicon sequencing with Illumina MiSeq. DNA was extracted using the repeated bead beating method, and for sequencing we used the V1-V3 primers forward AGAGTTTGATCMTGGCTCAG and reverse GTATTACCGCGGCTGCTG. The reads were processed and analyzed using the R-package mare21, which utilizes USEARCH22 for quality filtering, clustering of reads into species-like operational taxonomic units (OTUs), and taxonomic annotation. The analysis procedure was validated using artificial microbial communities of known composition21. Specifically, we assessed how the observed microbiota composition depended on read length. We found that the forward reads trimmed to 100–150 nucleotides resulted in better correspondence to the actual composition in the artificial communities than using long, merged paired-end reads (Supplementary Fig. 1). Thus, we used only the forward reads, which we truncated to 150 nt, to remove low-quality bases at the end of the reads. The 150nt reads were quality-filtered. All sequences representing less than 0.001% of the total reads were eliminated, as rare reads are likely to contain errors or chimaeras. The reads were OTU-clustered for richness analysis. The OTUs were not annotated, as OTU-clustering is a potential source of taxonomic errors. Instead, we taxonomically annotated the reads. After quality filtering, we had on average 68 000 reads per sample, ranging from 23 to 282 000. The samples with <100 reads were meconium samples, in which very low abundance of bacteria is expected, and therefore we considered the low number to be biologically relevant. For the meconium samples we obtained on average 26 000 (23–98 570) reads. We included also 14 empty samples as negative controls in the sequencing run. The number of reads obtained for these samples varied between 0 and 822 (median = 37, mean = 164). We compared the composition observed in the meconium samples with <1000 reads to that in the negative control samples (Supplementary Fig. 2). Four of the meconium samples had control-like compositions high in lactobacilli, suggestive of contamination. These few meconium samples were unlikely to alter the results, and they were kept in the analysis.
Statistical analyses
The statistical analyses were conducted using the R package mare21, which implements tools from the R package vegan23. Principal coordinates analysis (PCoA), which summarizes the multivariate microbiota data into a few variables, was conducted using Bray-Curtis dissimilarities with the R-package vegan23. Associations between the most abundant bacterial groups with the host factors postmenstrual age, postnatal age, and birth mode (vaginal/cesarean) as well as antibiotic courses (aminoglycoside or vancomycin) were tested using generalized additive mixed models (GAMM), with negative binomial error distribution (function gam in the package mgcv24), subject ID as the random variable, and the number of reads as the offset.
Data availability
The data are available in Supplementary Table 1. The DNA sequences will be submitted to European Nucleotide Archive (http://www.ebi.ac.uk/ena).
Term infant samples
To compare the microbiota development in the premature infants to the microbiota in term-born healthy infants, we included previously reported 16S rRNA–based microbiota composition data from a Dutch infant cohort25 in which 9 fecal samples were collected from 187 infants during the first 15 postnatal weeks (total 665 samples).
Results
Development of microbiota in preterm infants
The intestinal microbiota composition in the 45 premature neonates was longitudinally followed and was characterized by strong dominance of only a few organisms. Typically one of four genera, Bifidobacterium, Enterobacter, Staphylococcus or Enterococcus, represented >50% of the reads in a given sample (Supplementary Fig. 3, Fig. 1a). The infants often switched from one pattern of microbiota (dominating organism) to another within days (Supplementary Fig. 3). The overall microbiota composition was strongly associated with postnatal age (Fig. 1b, R2 = 0.07, p = 0.001 in permutational multivariate ANOVA) and even more clearly with postmenstrual age (Fig. 1c, R2 = 0.13, p = 0.001). After adjusting for postmenstrual age, postnatal age explained only 2% of the variation (p = 0.002). With increasing postmenstrual age, the infants progressed from Staphylococcus-Enterococcus-dominated composition to Enterobacter and finally towards Bifidobacterium-dominated microbiota (Fig. 1c).
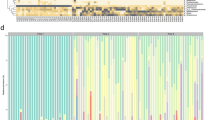
Microbiota composition in the fecal samples of preterm infants. Principal coordinates analyses (PCoA, Bray-Curtis dissimilarities) (a–c). The axes represent PCoA component scores, showing the two most important gradients differentiating the microbial communities. Each circle represents a microbial community, colored in panel (a) by the dominant organism in the community (>50% of all reads). Background color corresponds to: (b) postnatal age of the infant at the time of fecal sample collection; (c) postmenstrual age of the infant at sample collection. Blue background indicates low values, green intermediate, and orange high values. Average relative abundances of the dominant families in extremely premature (EP), moderately or very premature (MVP), compared to term infants from a Dutch cohort, at different postmenstrual ages (weeks)25 (d). Average total DNA concentration by postmenstrual age, divided to bacterial taxa based on their relative abundances in the 16S rRNA data (e).
We compared the observed microbiota composition at different postnatal ages in the EP and MVP infants to healthy term-born infants. The microbiota composition in the preterm infants progressed towards the composition observed in the term-born infants, depending on gestational age (Fig. 1d); the microbiota development in the EP group appeared to lag behind that of the MVP group (Fig. 1d), but the development was matched in both groups when considering the postmenstrual age. In fact, when comparing the MVP and the term infants at the same postmenstrual age, it appeared that the microbiota of the MVP infants progressed faster towards a strongly Bifidobacterium-dominated composition (Fig. 1d).
To assess the development in bacterial load with age, we measured the total DNA concentration in the fecal samples (Fig. 1e). For further insight, the DNA yield was subdivided to different bacterial taxa based on their relative abundances. The amount of DNA in the samples was fairly stable until 30 weeks postmenstrual age, after which it began to increase, possibly due to an increase in the absolute abundance of bifidobacteria. Simultaneously, the absolute abundance of staphylococci and enterococci appeared to decline. After 36 weeks postmenstrual age, there appeared to be a decline in DNA yield, as bifidobacteria became dominant and all other bacteria declined.
Overall, the DNA concentration increased significantly with postmenstrual age (Fig. 2a, p = 0.03, explaining 14% of the variation), and with postnatal age (Fig. 2b, p = 0.0002, 7% of the variation), indicating an increase in total bacterial abundance, and therefore active replication. The DNA concentration was not dependent on birth mode (p = 0.8, 0.1% of variation).
Development of the microbiota with postmenstrual (a,c,e) and postnatal (b,d,e) age. Association between postmenstrual and postnatal age of the infant and total DNA concentration (a,b), microbial richness (c,d), and development index (e,f, sum of the two first principal components in Fig. 1) among infants born before (extremely premature, EP), and at/after gestational week 28 (moderately or very premature, MVP). The trend lines show the best-fit (second polynomial) and the shaded areas show 95% confidence intervals.
Simultaneous with the increase in DNA concentration, richness of the microbiota increased with postmenstrual age (Fig. 2a, p = 0.0014, explaining 20% of the variation), and with postnatal age (Fig. 2b, p = 0.02, 7% of the variation). Richness was not associated with birth mode (p = 0.19, 1% of the variation). The microbiota development with age was captured by the sum of the score on the first two principal coordinates (Fig. 1a–c). We used the PCoA score sum as the development index. This index was positively associated with postmenstrual age (Fig. 2c, p < 0.0001, 35% of the variation), but after adjusting for postmenstrual age, the association with postnatal age was not significant (Fig. 2d, p = 0.4, 1% of the variation). The development index tended to be lower in vaginally born infants (p = 0.07, 1% of the variation).
Microbiota development in phases
A detailed analysis of the most abundant genera revealed that the development of the microbiota composition was not random, but followed a pattern depending on postmenstrual age (Fig. 3). The gestational age at birth appeared to have little influence on the microbiota development, as the EP, MVP, and healthy term-born infants followed largely the same course of development. In the first 50 weeks postmenstrual age, there were four successional phases.
Phase 1 was Staphylococcus dominance, which peaked between 25 and 30 weeks postmenstrual age and ended by 35 weeks postmenstrual age, and therefore did not occur in term-born infants (Fig. 3a). The abundance of Staphylococcus was significantly associated with postmenstrual age (p < 0.0001), which explained 20% of the variation in Staphylococcus abundance, while postnatal age explained an additional 8% of the variation (p < 0.0001). Vaginally born infants had a higher abundance of Staphylococcus, although birth mode explained only 1% Staphylococcus abundance (p = 0.004). Individuality covered 8% of the variation in Staphylococcus (p = 0.016).
Phase 2 was Enterococcus dominance, peaking at 30 weeks and ending by 35 weeks postmenstrual age (Fig. 2b). Postmenstrual age explained 3% of the variation in the abundance of enterococci (p = 0.0001), and postnatal age explained 2% (p = 0.002). Birth mode was not significantly associated with the relative abundance of Enterococcus (p = 0.74). Enterococcus abundance showed high individuality, subject ID explaining 30% of the variation (p < 0.0001).
Phase 3 was Enterobacteriaceae dominance (mainly genus Enterobacter in the preterm infants), peaking on average at 35 weeks postmenstrual age (Fig. 2c). The term-born infants showed only the declining part of this successional phase. There appeared to be a modest secondary bloom of enterobacteria at 45–50 weeks postmenstrual age. Among the preterm infants, the timing of the Enterobacter peak showed extensive individual variation and consequently the postmenstrual age explained only 1% of the variation (p = 0.47), and postnatal age explained 4% (p = 0.22). Vaginal birth was associated with lower abundance of Enterobacter (p = 0.01, 1% of variation explained).
Phase 4 was characterized by a high abundance of Bifidobacterium, which began to develop gradually after 30 weeks postmenstrual age. Postmenstrual age explained 11% of the variation in Bifidobacterium abundance (p < 0.0001), and postnatal age explained 5% (p = 0.39). There was considerable individuality in Bifidobacterium abundances, ID explaining 24% (p < 0.0001), while birth mode explained 1% (p = 0.22).
Associations between microbiota and antibiotic treatments
To assess the effect of antibiotic treatments on the microbiota, we focused on the period before and after antibiotic courses, and estimated the effect of aminoglycoside and vancomycin antibiotics after adjusting for postmenstrual age, postnatal age, and birth mode. The responses to the antibiotics were generally temporary and the microbiota appeared to recover within days (Fig. 4, Supplementary Fig. 4). Furthermore, only a subset of the infants responded to the antibiotics, showing lower than expected values during the first 10 days after the beginning of the antibiotic, while others either showed no response or even the opposite, increasing response.
Effect of antibiotic treatments on the most abundant bacterial genera. Deviation from expected relative abundance of each genus, based on age of the infant and birth mode, is shown in relation to timing of the antibiotic courses. Consistently negative values after the beginning of the antibiotic course (between days 1 and 10) are interpreted as a response to the antibiotic, and the infants are categorized as responders (blue) or non-responders (red). Number of infants in each category and the p-values of the GAMM models are shown.
In terms of total DNA, microbiota richness, and microbiota development index, 37% of the infants responded temporarily to aminoglycoside (Supplementary Fig. 4a,c,e). The rest either did not respond or showed a temporary increase in total DNA and development index (Supplementary Fig. 4a,e). Vancomycin treatment was associated with a decline in total DNA and development index in 52% of the infants, but richness was not significantly affected (Supplementary Fig. 4b,d,f).
Individual bacterial taxa did not always respond according to the total DNA response (Fig. 4). Staphylococcus and Enterobacter followed the same pattern as total DNA, showing a short-term response to aminoglycoside in 37% of the infants and to vancomycin in ca. 50%. Enterococcus showed a dramatic short-term decline in response to aminoglycoside in 56% of the infants, and increased in response to vancomycin in 74% of the infants. Bifidobacteria declined in 58% of the infants temporarily in response to aminoglycoside treatment. Vancomycin treatment was associated with either a short-term reduction of bifidobacteria, or a slower but persistent reduction.
Microbiota composition and sepsis incidence
Fourteen cases of sepsis in 10 infants were documented during the study period with fecal samples collected within five days of diagnosis. Based on blood cultures, the causative organisms of sepsis in the analyzed cases were always staphylococci. Staphylococcus was more abundant before sepsis than after (Fig. 5), and in 7/14 cases, Staphylococcus was dominant in the fecal samples either before or during sepsis. During sepsis, the dominant organism in the fecal samples was Enterobacter in five cases, Enterococcus in five cases, and Staphylococcus in four cases (Supplementary Fig. 2). Sepsis did not occur in infants with Bifidobacterium as the dominant organism; the abundance of Bifidobacterium was significantly reduced during sepsis (Fig. 5).
Discussion
We followed the development of the intestinal microbiota longitudinally in a cohort of 45 prematurely born infants from birth to discharge from hospital up to 60 days after birth. Frequent sampling enabled the detailed construction of individual developmental patterns, and characterisation of the microbiota succession. Our results show that the promotion of normal microbiota development in prematurely born infants is possible in a hospital setting, because many infants progressed to a Bifidobacterium-dominated composition which is characteristic of healthy term-born non-hospitalized infants. A major obstacle for the microbiota development among the extremely premature infants was the overgrowth of Enterococcus spp, which seemed to inhibit the normal succession by several weeks in some cases.
We found maturity, indicated by postmenstrual age, to be a major determinant of the microbiota development, and particularly of the ability of bifidobacteria to reach dominance. This has previously been observed in term and premature infants26,27. Regardless of the gestational age at birth, infants began to proceed towards a Bifidobacterium-dominated composition, an indicator of healthy microbiota in infants, after postmenstrual week 30. The results show that although the extremely premature infants had significantly more co-morbidities and required more medical interventions than the moderately premature group, both premature groups followed the same microbiota development pattern. This suggests that physical maturity is a very strong determinant of microbiota development in neonates.
In our cohort, infants with a disrupted microbial development, showing a dominance of aerobic cocci and reduced abundance of bifidobacteria, were at increased risk of sepsis. A dominance of Enterobacter spp. or other Proteobacteria, and the absence of bifidobacteria have previously been associated with risk of necrotizing enterocolitis and sepsis in preterm infants8,9,10,11; in our cohort Enterobacter was abundant in some sepsis cases, but also in many cases not developing sepsis.
As the microbiota development is dependent on infant maturity, particularly the extremely premature infants are at risk of delayed microbiota development and may require therapeutic interventions. Supplementation with bifidobacteria or bifidogenic prebiotics has been shown to increase the abundance of bifidobacteria in preterm infants, to reduce the abundance of clostridia and enterobacteria, and to decrease signs of distress17,28,29. Antibiotics may have a harmful effect on the microbiota development30. In line with this, we observed temporary changes in microbiota composition following antibiotic courses. The microbiota appeared to recover within 2–3 weeks. Importantly, the response to antibiotics depend on the susceptibility of the bacteria to the drug. For example, vancomycin-resistant enterococci prevail in hospital environments, and may prevent a beneficial response.
Unlike most previous studies, our cohort included many infants with a fairly normal microbiota development, achieving high abundance of bifidobacteria. All infants in our study received human milk from the first days of life. Breast milk feeding is known to increase the abundance of bifidobacteria in infants31,32. The increase in bifidobacteria was achieved also in caesarean-delivered infants, demonstrating that normal microbiota development is possible even in these prematurely born, hospitalized, caesarean-delivered infants. This suggests that the recently suggested practice of vaginal seeding33, i.e. swabbing the neonate with the mother’s vaginal fluids, which has been criticized as potentially risky34, is unnecessary if breast milk (from mother or donor) is given.
Our results suggest that normal-like microbiota development in preterm infants in the hospital setting is possible, particularly among moderately premature infants receiving breast milk. In the extremely premature infants, development appeared to be disrupted by persistently increased abundance of Enterococcus. Future research on interventions to improve the microbial colonization in extremely premature infants and infants with patterns indicating abnormal developmental patterns, could improve short- and long-term outcomes.
References
Kramer, M. et al. The contribution of mild and moderate preterm birth to infant mortality. JAMA 284, 843–849 (2000).
Neu, J., Chen, M. & Beierle, E. Intestinal innate immunity: how does it relate to the pathogenesis of necrotizing enterocolitis. In: Seminars in pediatric surgerysurgery, pp.137–144: Elsevier (2005).
Malaeb, S. & Dammann, O. Fetal inflammatory response and brain injury in the preterm newborn. J. Child Neurol. 24, 1119–1126 (2009).
Magne, F. et al. Low species diversity and high interindividual variability in faeces of preterm infants as revealed by sequences of 16S rRNA genes and PCR-temporal temperature gradient gel electrophoresis profiles. FEMS Microbiol Ecol 57, 128–138 (2006).
Arboleya, S. et al. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol Ecol 79, 763–772 (2012).
Barrett, E. et al. The individual-specific and diverse nature of the preterm infant microbiota. Arch Dis Child Fetal Neonatal Ed 98, F334–40 (2013).
Moles, L. et al. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS One 8, e66986 (2013).
Wang, Y. et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. The ISME journal 3, 944–954 (2009).
Mai, V. et al. Fecal microbiota in premature infants prior to necrotizing enterocolitis. PloS One 6, e20647 (2011).
Stewart, C. et al. The preterm gut microbiota: changes associated with necrotizing enterocolitis and infection. Acta Ped 101, 1121–1127 (2012).
Mai, V. et al. Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS One 8, e52876 (2013).
Biasucci, G. et al. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev 86, S13–S15 (2010).
van Nimwegen, F. A. et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol 128, 948–55, e1-3 (2011).
Bäckhed, F. et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host & Microbe 17, 690–703 (2015).
Persaud, R. et al. Impact of perinatal antibiotic exposure on the infant gut microbiota at one year of age. Allergy, Asthma & Clin Immunol 10, A31 (2015).
Penders, J. et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118, 511–521 (2006).
Pärtty, A. et al. Effects of Early Prebiotic and Probiotic Supplementation on Development of Gut Microbiota and Fussing and Crying in Preterm Infants: A Randomized, Double-Blind, Placebo-Controlled Trial. J Pediatr 163, 1272–1277 (2013).
Boyd, C. A., Quigley, M. A. & Brocklehurst, P. Donor breast milk versus infant formula for preterm infants: systematic review and meta-analysis. Arch Dis Child Fetal Neonatal Ed 92, F169–75 (2007).
Deshpande, G. et al. Updated meta-analysis of probiotics for preventing necrotizing enterocolitis in preterm neonates. Pediatrics 125, 921–930 (2010).
Moltu, S. et al. Enhanced feeding and diminished postnatal growth failure in very-low-birth-weight infants. J Pediatr Gastroenterol Nutr 58, 344–351 (2014).
Korpela K. mare: Microbiota Analysis in R Easily. R package version 1.0, https://github.com/katrikorpela/mare (2016).
Edgar, R. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Oksanen, J. et al. vegan: Community Ecology Package. R package version 2.0-6 (2013).
Wood, S. N. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. Journal of the Royal Statistical Society (B) 73(1), 3–36 (2011).
Zijlmans, M. A., Korpela, K., Riksen-Walraven, J. M., de Vos, W. M. & de Weerth, C. Maternal prenatal stress is associated with the infant intestinal microbiota. Psychoneuroendocrinology 53, 233–245 (2015).
Butel, M. J. et al. Conditions of bifidobacterial colonization in preterm infants: a prospective analysis. J Pediatr Gastroenterol Nutr 44, 577–582 (2007).
Dogra, S. et al. Dynamics of Infant Gut Microbiota Are Influenced by Delivery Mode and Gestational Duration and Are Associated with Subsequent Adiposity. mBio 6, e02419–14 (2015).
Mohan, R. et al. Effects of Bifidobacterium lactis Bb12 supplementation on intestinal microbiota of preterm infants: a double-blind, placebo-controlled, randomized study. J Clin Microbiol 44, 4025–4031 (2006).
Underwood, M. A. et al. A randomized placebo-controlled comparison of 2 prebiotic/probiotic combinations in preterm infants: impact on weight gain, intestinal microbiota, and fecal short-chain fatty acids. J Pediatr Gastroenterol Nutr 48, 216–225 (2009).
Kuppala, V. S. et al. Prolonged initial empirical antibiotic treatment is associated with adverse outcomes in premature infants. J Pediatr 159, 720–725 (2011).
Grönlund, M. et al. Maternal breast‐milk and intestinal bifidobacteria guide the compositional development of the Bifidobacterium microbiota in infants at risk of allergic disease. Clin & Exp Allergy 37, 1764–1772 (2007).
Garrido, D., Barile, D. & Mills, D. A. A Molecular Basis for Bifidobacterial Enrichment in the Infant Gastrointestinal Tract. Adv Nutr 3, 415S–421S (2012).
Dominguez-Bello, M. G. et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med 22, 250–253 (2016).
Cunnington, A. J. et al. “Vaginal seeding” of infants born by caesarean section. BMJ 352, i227, https://doi.org/10.1136/bmj.i227 (2016).
Acknowledgements
The study was funded by the Research Council of Norway and Throne Holst Foundation as well as Health Region South-Eastern Trust of Norway, and by the Finnish Academy of Sciences and the Netherlands Organization for Scientific Research. We would like to thank the hospital staff at Akershus and Oslo university hospitals for their support in the recruitment of families.
Author information
Authors and Affiliations
Contributions
K.K. analyzed the data and wrote the manuscript. W.M.d.V. supervised the work and contributed to the writing of the manuscript. E.W.B. conceptualized and designed the randomized controlled trial, included participants and collected the samples, contributed to the clinical interpretation the data and writing of the manuscript. S.J.M., K.S., and B.N. conceptualized and designed the randomized controlled trial, included participants and collected the samples. A.G., A.E.R. K.B., P.O.I., and C.A.D. conceptualized and designed the randomized controlled trial. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Korpela, K., Blakstad, E.W., Moltu, S.J. et al. Intestinal microbiota development and gestational age in preterm neonates. Sci Rep 8, 2453 (2018). https://doi.org/10.1038/s41598-018-20827-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-20827-x
This article is cited by
-
The role of gut microbiota in human metabolism and inflammatory diseases: a focus on elderly individuals
Annals of Microbiology (2024)
-
Neonatal Necrotizing Enterocolitis: An Update on Pathophysiology, Treatment, and Prevention
Pediatric Drugs (2024)
-
Determinants of microbial colonization in the premature gut
Molecular Medicine (2023)
-
Cystic Fibrosis-Related Gut Dysbiosis: A Systematic Review
Digestive Diseases and Sciences (2023)
-
Impact of early antibiotic exposure on the risk of colonization with potential pathogens in very preterm infants: a retrospective cohort analysis
Antimicrobial Resistance & Infection Control (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.