Abstract
Our purpose in this study is to evaluate the clinical feasibility of deep-learning techniques for F-18 florbetaben (FBB) positron emission tomography (PET) image reconstruction using data acquired in a short time. We reconstructed raw FBB PET data of 294 patients acquired for 20 and 2 min into standard-time scanning PET (PET20m) and short-time scanning PET (PET2m) images. We generated a standard-time scanning PET-like image (sPET20m) from a PET2m image using a deep-learning network. We did qualitative and quantitative analyses to assess whether the sPET20m images were available for clinical applications. In our internal validation, sPET20m images showed substantial improvement on all quality metrics compared with the PET2m images. There was a small mean difference between the standardized uptake value ratios of sPET20m and PET20m images. A Turing test showed that the physician could not distinguish well between generated PET images and real PET images. Three nuclear medicine physicians could interpret the generated PET image and showed high accuracy and agreement. We obtained similar quantitative results by means of temporal and external validations. We can generate interpretable PET images from low-quality PET images because of the short scanning time using deep-learning techniques. Although more clinical validation is needed, we confirmed the possibility that short-scanning protocols with a deep-learning technique can be used for clinical applications.
Similar content being viewed by others
Introduction
Amyloid positron emission tomography (PET) is a nuclear medicine imaging test that shows amyloid deposits in the brain. It is currently being used in the diagnosis of Alzheimer's disease, which is known to be caused by amyloid1. Although there are some differences in the acquisition protocols that depend on the commercially available radiopharmaceuticals for amyloid PET, most of these should be taken for 10–20 min, especially for F-18 florbetaben (FBB), which needs 20 min for scanning2. Since most of the patients with memory disorder are elderly, there are complaints that it is difficult for them to lie down without movement for 20 min. Head movements due to postural discomfort during long scan acquisition can cause motion artifacts in PET images, which degrade their diagnostic value. Some elderly patients actually needed re-scanning (or additional radiation exposure) because of a poor image due to movement. Thus, the demand for shortening scan time is growing with the increasing use of PET for patients with dementia. However, PET images obtained from short scanning times can suffer from a low signal-to-noise ratio and have reduced diagnostic reliability as well.
Recently, deep-learning techniques for image restoration have been widely applied to medical images, including computed tomography (CT), magnetic resonance imaging (MRI), and PET3,4,5,6,7,8,9,10,11. Some of them have used the deep-learning techniques for low-dose PET image restoration and have shown potential for reducing noise artifacts3,4,5,6,7,8. There have been only a few studies on reducing noise and improving the quality of images taken by reducing the acquisition time of brain PET7. They have used additional MR information obtained from a PET/MR scanner to restore brain PET images. However, a PET/MR scanner is costly and is not yet widely installed. Since PET/CT scanners are used in most hospitals, a restoration technique using only PET without MRI information is needed.
In this study, we applied the deep-learning technique for short-scanning FBB PET image restoration. The proposed method uses PET images only, without additional information, such as MRI or CT. We did qualitative and quantitative analyses to evaluate the clinical applicability of the proposed method.
Materials and methods
The Institutional Review Board (IRB) of Dong-A University Hospital reviewed and approved this retrospective study protocol (DAUHIRB-17-108). The IRB waived the need for informed consent, since only anonymized data would be used for research purposes. We used all methods in accordance with the relevant guidelines and regulations.
Patients and F-18 FBB brain PET acquisition
For training and internal validation of our deep-learning algorithm, we enrolled 294 patients with clinically diagnosed cognitive impairment who had received FBB PET between December 2015 and May 2018 retrospectively in this study. We also randomly collected 30 patients who had FBB PET from January to May 2020 for temporal validation. Out of 30 patients, we excluded two patients because of insufficient clinical information, and finally 28 patients participated. In this study, we excluded patients with head movement during PET scanning. All the FBB PET examinations were done using a Biograph mCT flow scanner (Siemens Healthcare, Knoxville, TN, USA). The PET/CT imaging was done according to the routine examination protocol of our hospital, which is the same method used in the previous study published by our group12. We injected 300 MBq F-18 florbetaben intravenously into the patients and started PET/CT acquisition 90 min after the radiotracer injection. A helical CT scan was carried out with a rotation time of 0.5 s at 120 kVp and 100 mAs, without an intravenous contrast agent. A PET scan followed immediately, and the image was acquired for 20 min with the list mode. All the images were acquired from the skull vertex to the skull base. We reconstructed the list mode PET data for 20 min into a 20-min static image (PET20m) and used it as the full-time ground-truth image. We also reconstructed a short-scanning static PET image (PET2m) using the first 2-min data from the total list mode PET data. We used the same parameters to acquire both PET20m and PET2m images.
In addition, we carried out external validation, and obtained data used in the preparation of the external validation from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu). Among the subjects who underwent FBB PET, we randomly selected 60 patients, and excluded two patients because of inconsistency in the brain amyloid plaque load (BAPL) scoring. Finally, 58 patients were involved.
The characteristics of all subjects included in this study are summarized in Table 1.
Deep-learning method
Network architecture
We adopted a generative adversarial network that consists of two competing neural networks with an additional pixelwise loss13. The schematic diagram of the proposed network is shown in Fig. 1. The generator (\(G\)) is trained to generate a synthetic PET20m-like (sPET20m) image from the noisy PET2m image, and the discriminator (\(D\)) is trained to distinguish sPET20m images generated by the generator from real PET20m image. In the training procedure, the discriminator enables the generator to provide more realistic sPET20m images14. Pixelwise loss is defined as a mean-squared error between sPET20m images and original PET2m images, which prevents the generator from changing small anomalies or structures of PET2m images during training15.
The schematic diagram of the adversarial network used in this study (top left). In this proposed network, the discriminator (top right) and the generator (bottom row) are shown, and the generator is constructed using the deep convolutional framelets. The numbers below the rectangular boxes indicate the number of filters.
The generator is constructed using the deep convolutional framelets, which consist of encoder-decoder structures with skipped connections16. Both encoder and decoder paths contain two repeated \(3\times 3\) convolutions (conv), each followed by a batch normalization (bnorm) and a leaky rectified linear unit (LReLU)17, 18. A 2-D Haar wavelet de-composition (wave-dec) and re-composition (wave-rec) are used for down-sampling and up-sampling, respectively, of the features19. In the encoder path, three high-pass filters after wavelet de-composition skip directly to the decoder path (arrow marked by ‘skip’), and one low-pass filter (marked by ‘LF’) is concatenated with the features in the encoder path at the same step (arrow marked by ‘skip & concat’). At the end, a convolution layer with a \(1\times 1\) window is added to match the dimension of input and output images. The numbers below the rectangular boxes in Fig. 1 indicate the number of filters. The architecture of deep convolutional framelets is similar to that of the U-net20, a standard multi-scale convolutional neural network (CNN) with skipped connections. The difference is in using the wavelet de-composition and re-composition, instead of max-pooling and un-pooling, for down-sampling and up-sampling, respectively. Additional skip connections of high-frequency filters help to train the detailed relationship between PET2m and PET20m images.
For the discriminator, we adopted the standard CNN without a fully connected layer. The discriminator contains three convolution layers with a \(4\times 4\) window and strides of two in each direction of the domain, each followed by a batch normalization and a leaky ReLU with a slope of 0.2. At the end of the architecture, a \(1\times 1\) convolution is added to generate a single-channel image.
Datasets for training and internal validation
In the dataset of the 294 patients’ PET images (70 image slices/patient), we randomly divided the training and internal validation datasets into 80% and 20%, and used 236 patients’ images as the training dataset and used 58 patients’ images as the internal validation dataset. The original size of the PET images was \(400\times 400\). In order to improve training effectiveness, we cropped all \(400\times 400\) images to \(224\times 224\) pixels around the center of an image in both horizontal and vertical directions. Here, only background (i.e., zero-valued) information was removed. We used the cropped images as input and label datasets for the proposed deep-learning network. In the testing procedure, we resized the images corrected by means of the trained generator to \(400\times 400\) by adding the rows and columns of zeros at the top, bottom, left, and right sides of the images (i.e., zero padding). We did not use data augmentations such as rotation or flipping for training.
Network training
In our study, we ran training for 200 epochs using Adam solver with a learning rate of 0.0002, and a mini-batch size of 1021. It was implemented using TensorFlow on a CPU (Intel Core i9-7900X, 3.30 GHz) and a GPU (NVIDIA, Titan Xp. 12 GB) system22. It took about 68 h to train the network. The network weights followed a Gaussian distribution, with a mean of 0 and a standard deviation of 0.01.
Assessment of image quality
We compared the image quality of PET2m and the synthesized sPET20m images with the original PET20m images using the peak signal-to-noise ratio (PSNR), structural similarity (SSIM), and normalized root mean-square error (NRMSE). The SSIM index depends on the parameters \({C}_{1}= {({K}_{1}L)}^{2}\) and \({C}_{2 }= {({K}_{2}L)}^{2}\), where \(L\) is the dynamic range of pixel values and \(K\) is some constant8. In our study, we chose \({C}_{1} = {\left(0.0002\times 65535\right)}^{2}\mathrm{ and }{C}_{2} = {\left(0.0007\times 65535\right)}^{2}\). The proposed method was also compared with the standard U-net method.
For further analysis, we calculated the Standardized Uptake Value Ratio (SUVR) using PMOD 3.6 software (PMOD Technologies, Zurich, Switzerland)23. We obtained the transformation matrix of each participant by fusing the CT template of the PMOD and the CT image of the participant. PET images were then spatially normalized by using the transformation matrix of each participant and were applied to an automated anatomical labeling template of PMOD (Hammers atlas). We spatially normalized all pairs of sPET20m and PET20m images to the Montreal Neurological Institute (MNI) spatial templates and applied the Hammers atlas. By reconstructing the volume-of-interests of the atlas, the representative areas were set up as the striatum, frontal, parietal, temporal and occipital lobes, and global brain. We calculated the SUVRs of the representative areas and used the cerebellar cortex as the reference region. We compared the difference of SUVRs of the identical area between sPET20m and PET20m images.
Clinical interpretations
For visual interpretation, three nuclear medicine physicians with certification and experience in amyloid PET readings participated (YJ and DY have over 15 years and JE has 4 years of experience in nuclear medicine; all of them also have 4 years of experience in amyloid PET assessment). They were blinded to the clinical data and independently read all PET images of the internal validation dataset.
Turing test
We did two Turing tests and evaluated all PET images of the internal validation dataset. First, of all the sPET20m and PET20m images, we randomly selected 58 images and presented them to the physicians one by one for them to decide whether the PET image was real or synthetic (Test 1). Second, we presented a pair of sPET20m and PET20m images of the same patient to the physicians to find the original PET20m image (Test 2). We anonymized all PET images and randomized the order of PET images.
BAPL score
We gave all anonymized sPET20m images of the internal validation dataset to the physicians to interpret and score according to the conventional interpretation protocol. All the sPET20m images were classified into three groups according to the BAPL scoring system. BAPL score is a specialized, predefined three-grade scoring system for F-18 FBB PET wherein measurements are made by the physician based on the visual assessment of the subject’s amyloid deposits in the brain24. BAPL scores of 1 (BAPL 1), 2 (BAPL 2), and 3 (BAPL 3) indicate no amyloid load, minor amyloid load, and significant amyloid load, respectively. Therefore, BAPL 1 indicates a negative amyloid deposit, whereas BAPL 2 and BAPL 3 represent positive amyloid deposits. In this study, we treated the BAPL score read from the PET20m images as the ground-truth score set by consensus among the three physicians. We measured the accuracy of the BAPL score for each physician. We also analyzed the agreements between the BAPL score of sPET20m and PET20m images for each physician.
Temporal and external validations
We additionally verified our model by measuring PSNR, SSIM, NRMSE, and SUVR by means of temporal and external validation. The patient characteristics of our temporal and external validation datasets are illustrated in Table 1. We performed all analyses of temporal validation in the same manner as used in internal validation. We did external validation using a public FBB dataset from ADNI. ADNI datasets contain a series of 4 \(\times\) 5 min of FBB PET images. The proposed model trained on our institute dataset (i.e., pair of 2-min and 20-min images) was tested on the first 5-min PET images. In this study, a Gaussian filter with 4-mm FWHM was applied to all FBB PET images of ADNI datasets.
Statistical analysis
We assessed the intra-observer agreement of the BAPL score between the sPET20m and PET20m images using Cohen’s weighted kappa. We calculated the accuracy, sensitivity, and specificity for the interpretations of sPET20m images. We assessed the difference of group characteristics using an independent t-test, one-way ANOVA, and chi-squared test. We evaluated the difference in SUVR between sPET20m and PET20m images using an independent t-test or Mann–Whitney U test, and evaluated the relationship of SUVRs between them using the Pearson’s correlation coefficient. We assessed agreement of SUVRs of both PET images using the Bland–Altman 95% limits of agreement. We did the statistical analyses using the MedCalc software version 16.4 (MedCalc Software, Mariakerke, Belgium) and NCSS 12 Statistical Software (NCSS, LLC. Kaysville, Utah, USA). Statistical significance was defined as p < 0.05.
Results
Assessment of image quality
PSNR, SSIM and NRMSE
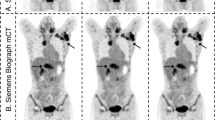
The original PET2m, PET20m and sPET20m images and a synthetic image generated by U-net are shown in Fig. 2.
The input and output of PET images (upper row, BAPL 1; middle row, BAPL 2; lower row, BAPL 3). PET2m image (input image) is very noisy and the image quality is poor (a,e,i). The ground truth with 20-min scanning (b,f,j) and synthetic PET images generated from the proposed deep learning (c,g,k) and the U-net (d,h,l) are shown. The synthetic PET image generated from our model is better in reflecting the underlying anatomical details than is the PET image generated from the U-net. In the BAPL 2 case, a small positive lesion (red arrows, e–h) is equivocal in the PET2m image (e), but clearly shown in sPET20m image (g) as in PET20m image (f).
Both the proposed and the U-net methods significantly reduce noise, but the U-net produces a slightly blurrier image than does the proposed method. For quantitative comparison, we calculated averaged PSNR, SSIM, and NRMSE for all datasets. The results are summarized in Table 2, which shows that the proposed method had the highest PSNR and SSIM, and lowest NRMSE, whereas PET2m images showed the worst performance in internal validation. The proposed model shows similar performance for the temporal validation dataset, in terms of PSNR, SSIM, and NRMSE. As shown in Table 2, our method also improved the image qualities of the 5-min images in external validation.
SUVR
Internal validation dataset
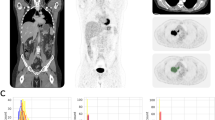
In internal validation, there was no statistically significant difference of SUVR between the PET20m and sPET20m images in the striatum, frontal, parietal, temporal, occipital lobes, and global brain (Fig. 3, Supplementary Table 1 and Fig. 1). In the Bland–Altman analysis, the regional mean difference of SUVR between sPET20m and PET20m images was 0.005 (95% confidence interval (CI) − 0.008, 0.017) in the negative group (Fig. 4a) and 0.024 (95% CI 0.010, 0.037) in the positive group (Fig. 4b). Upper and lower limits of agreement were 0.131 (95% CI 0.110, 0.152) and − 0.121 (95% CI − 0.142, − 0.100) in the negative group, and 0.180 (95% CI 0.157, 0.203) and − 0.133 (95% CI − 0.156, − 0.110) in the positive group, respectively.
Temporal and external validation datasets
In temporal and external validations, we also compared SUVRs of the entire representative areas between sPET20m and PET20m images and found result similar to those of internal validation (Supplementary Tables 2, 3 and Figs. 2, 3). There was a very strong positive correlation between SUVRs of sPET20m and PET20m images in temporal validation (r = 0.988, p < 0.001, Fig. 5a) and external validation (r = 0.987, p < 0.001, Fig. 5c). In the Bland–Altman analysis, the mean difference of SUVR between sPET20m and PET20m images was 0.015 (95% CI 0.009, 0.021) in temporal validation (Fig. 5b). Upper and lower limits of agreement were 0.092 (95% CI 0.081, 0.102) and − 0.062 (95% CI − 0.072, − 0.051). In external validation, the mean difference of SUVR was − 0.035 (95% CI − 0.039, − 0.030) (Fig. 5d). Upper and lower limits of agreement were 0.045 (95% CI 0.038, 0.053) and − 0.115 (95% CI − 0.123, − 0.107).
Clinical interpretations for internal validation dataset
Turing test
Tests 1 and 2 showed similar results (Table 3). Test 1, a test to decide whether the presented single PET image was real or synthetic, showed that, regardless of the duration of clinical reading experience in nuclear medicine, the overall accuracy was not high (44.8% and 63.8%). In Test 2, a test to select a real PET image out of two PET images of the same patient, the more experienced the physicians were in clinical reading, the more often the real PET image was selected (48.3–60.3%). Overall, however, the clinicians did not seem to distinguish well between generated PET images and real PET images.
BAPL score
The three physicians assessed the sPET20m images according to the BAPL scoring system, and there was no poor or inadequate image that was difficult to interpret. In five, six, and eight patients out of 58 patients, each physician assessed the BAPL score differently from the ground-truth score. Table 4 shows the accuracy, sensitivity, and specificity for the three physicians. Overall, the mean values for accuracy, sensitivity, and specificity were 89.1%, 91.3%, and 83.3%, respectively. The confusion matrices are provided in Table 5.
We evaluated the intra-observer agreement using Cohen’s weighted kappa by comparing the BAPL scores between the sPET20m and PET20m images. Clinicians’ Cohen’s weighted kappa was 0.902 (DY), 0.887 (YJ), and 0.844 (JE), with a mean value of 0.878.
Discussion
In this study, we investigated the feasibility of a deep-learning-based reconstruction approach using short-time acquisition PET scans. We used PET images acquired for 2 and 20 min as input and target images, respectively. Quantitative and qualitative analyses showed that the proposed method produces efficient synthetic PET images from short-scanning PET images. We calculated image-quality metrics (such as PSNR, SSIM, and NRMSE) for model evaluation between the synthetic images and ground-truth images (standard scanning images). Overall, the proposed method improved the image quality by suppressing the noise in short-scanning images. Note that the SSIM index depends on the parameters (\({K}_{1}\) and \({K}_{2}\)). In our study, the average SSIM index for the synthetic images increased from 0.8818 to 0.9939 when \({K}_{1}, {K}_{2}\) increased from \(0.0002\,\mathrm{ to }\,0.0007\) and \(0.01\, \mathrm{to }\,0.03\), respectively. However, in this case, the differences in the SSIM index were very small. Our deep-learning method also improved the image qualities of the 5-min images of the ADNI dataset, even though the test domain significantly differs from our training domain.
We adopted the GAN framework with an additional mean-squared loss between the synthetic sPET20m image and the PET2m image. The performance of the proposed network was compared with that of the conventional U-net. The U-net minimizes only the pixelwise loss between the synthetic PET image and ground-truth (i.e., PET20m) image, resulting in an over-smoothed image, whereas the proposed approach clearly reconstructs the detailed structures of the brain (Fig. 2)25. In terms of quality measurements, such as PSNR, NRMSE, and SSIM, the proposed method outperformed the U-net. The time taken to generate a synthetic single sPET20m image from a PET2m image was within a few milliseconds on the GPU system, which would make the proposed method adequate for clinical use.
Some previous studies have also tried to reduce noise and improve image quality using a deep-learning technique in PET imaging5,6,7,8,9. Most of these studies aimed at maintaining the quality of the PET image while reducing the injection dose of radiopharmaceuticals in order to minimize radiation exposure. They showed that the image quality of low-dose PET could be restored like the original PET images obtained with standard protocols while reducing the conventional radiopharmaceutical dose by up to 99%. However, they all used synthesized low-dose data (i.e., a small amount of data selected from the entire acquisition period), which may differ from the measured data obtained from the true low dose. A feasibility study on real data is needed for clinical use. One study restored a low-quality PET image taken in 3 min to match a standard image taken in 12 min7. This study differs from ours in that it used MRI information taken together to restore image quality. Considering the absence of a PET/MRI scanner in most hospitals, the proposed method using PET images only could be used in general clinical practice. Another study reported that using a 5-min PET image, one frame of 20-min data without deep-learning methods, did not relevantly affect the accuracy of disease discrimination26. The advantage of our method is that it can generate PET images like those of full-time scanning images with only 2-min data in any part, regardless of the frame. In our study, no comparison of diagnostic accuracy between PET images obtained by our method and 5-min PET images was done. However, if PET image reconstruction with short-time data is required, we think that our method, along with the PET imaging method using one frame 5-min data, can broaden the range of options that can be selected according to the situation.
Since amyloid PET images are used in hospitals to care for patients with memory impairment, deep-learning-generated images must have an image quality similar enough to the original image that it can be used for interpretation in the clinics. In this study, we used several methods to decide whether generated images could be available clinically. We did tests to find an answer to the following questions: Can physicians distinguish between PET20m and generated sPET20m images? What is the difference in visual interpretation results? What is the difference between quantitative analysis using SUVR in both images?
When PET20m and generated sPET20m images were presented at the same time to three nuclear medicine physicians who were in charge of clinical reading, the accuracy of the selection of the PET20m images was within 40–60%. This suggests that synthetic PET images generated by our method are almost indistinguishable from the real PET image. Next, we did the BAPL scoring test to assess the intra-observer agreement and diagnostic accuracy. In our study, Cohen's weighted kappa was above 0.84, which indicated an almost perfect intra-observer agreement. We also did BAPL scoring on generated PET images, which we compared with the ground-truth scores. In the strong positive cases (BAPL 3), all three physicians showed a 100% accuracy, but in the negative (BAPL 1) and weak positive (BAPL 2) cases, between 5 and 8 of the 58 patients were false-positive or false-negative. It is already known that the amyloid PET study itself, even if obtained according to a conventional protocol, can cause misclassification when visually read. Some studies have reported that about 10% of the results may be inconsistent27, 28. In addition, some errors from the deep-learning algorithm could be added, so we think that the misclassification has increased a little in our study. We also think that the physician’s opinion may have some influence on the interpretation of how much the amyloid uptake is positive in the visual reading that distinguishes BAPL 1 and 2. Few studies have evaluated the accuracy of physicians’ interpretations among studies related to deep learning on a subject similar to ours. One study showed 89% accuracy when read using deep-learning-generated PET images, which is very similar to our result6.
In order to make up for the weak points of the visual reading, SUVR is used as a quantitative indicator in routine practice to infer the severity or prognosis of the disease23. In the generated brain PET images of this study, regional SUVRs were not significantly different from the values of ground-truth images in negative and positive cases (p > 0.05). In the Bland–Altman analysis, the mean of the difference was 0.005 in the negative case and 0.024 in the positive case, and the limits of agreement of each region were small. That is, our deep-learning model can generate images with SUVR values that are comparable to those of the original PET images. We obtained similar results by means of temporal and external validations, which allowed us to reconfirm this fact. Taken together, these results suggest that the synthetic amyloid PET images generated by our deep-learning method could be used for clinical reading purposes.
Our study has some limitations that need to be considered for clinical use. First, our deep-learning model trained on FBB PET with 2-min data should be tested under various acquisition conditions. Using multicenter datasets for training or incorporating domain adaptation techniques could improve image quality, which is a part of our future work29, 30. In this study, in order to avoid overfitting, we evaluated our model using the ADNI data, a completely different dataset, and our hospital data obtained at a different time from the training dataset. Second, in our study, we empirically chose 2-min images as a training dataset for short scanning. However, 2-min PET images may not be optimal. More rigorous analysis may be needed to choose the proper short-scanning image. Third, we generated only trans-axial PET images in this study. Although interpretation guidelines for FBB PET recommend using trans-axial PET images for clinical reading, coronal and sagittal PET images have also been used recently for reading. In the next study, we need to apply our deep-learning model to generate three orthogonal PET images. In addition, the application of a 3-dimensional model and finding the optimal hyperparameters is a problem to be solved in the future.
In conclusion, we presented an image-restoration method using a deep-learning technique to yield a clinically acceptable amyloid brain PET image with short-time data. Qualitative and quantitative analysis by means of internal, temporal, and external validations showed that the image quality and quantitative value of the generated PET images were very similar to those of the original images. Although more evaluation and validation are needed, we found that applying deep-learning techniques to amyloid brain PET images can reduce acquisition time and provide clinically equivalent interpretable images as standard images.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Mallik, A., Drzezga, A. & Minoshima, S. Clinical amyloid imaging. Semin. Nucl. Med. 47, 31–43 (2017).
Minoshima, S. et al. SNMMI procedure standard/EANM practice guideline for amyloid PET imaging of the brain 1.0. J. Nucl. Med. 57, 1316–1322 (2016).
Duffy, I. R., Boyle, A. J. & Vasdev, N. Improving PET imaging acquisition and analysis with machine learning: a narrative review with focus on Alzheimer’s disease and oncology. Mol. Imaging 18, 1536012119869070 (2019).
Zhu, G. et al. Applications of deep learning to neuro-imaging techniques. Front. Neurol. 10, 869 (2019).
Gatidis, S. et al. Towards tracer dose reduction in PET studies: simulation of dose reduction by retrospective randomized undersampling of list-mode data. Hell. J. Nucl. Med. 19, 15–18 (2016).
Chen, K. T. et al. Ultra-low-dose 18F-florbetaben amyloid PET imaging using deep learning with multi-contrast MRI inputs. Radiology 290, 649–656 (2019).
Xiang, L. et al. Deep auto-context convolutional neural networks for standard-dose PET image estimation from low-dose PET/MRI. Neurocomputing 267, 406–416 (2017).
Ouyang, J. et al. Ultra-low-dose PET reconstruction using generative adversarial network with feature matching and task-specific perceptual loss. Med. Phys. 46, 3555–3564 (2019).
Gong, K., Guan, J., Liu, C. & Qi, J. PET image denoising using a deep neural network through fine tuning. IEEE Trans. Radiat. Plasma Med. Sci. 3, 153–161 (2019).
Kang, E., Min, J. & Ye, J. C. A deep convolutional neural network using directional wavelets for low-dose X-ray CT reconstruction. Med. Phys. 44, 360–375 (2017).
Chen, H. et al. Low-dose CT denoising via convolutional neural network. Biomed. Opt. Express 8, 679–694 (2017).
Jeong, Y. J., Yoon, H. J. & Kang, D. Y. Assessment of change in glucose metabolism in white matter of amyloid-positive patients with Alzheimer disease using F-18 FDG PET. Medicine 96, e9042 (2017).
Goodfellow, I. et al. Generative adversarial nets. in NIPS 2014 (2014).
Wolterink, J. M., Leiner, T., Viergever, M. A. & Isgum, I. Generative adversarial networks for noise reduction in low-dose CT. IEEE Trans. Med. Imaging. 36, 2536–2545 (2017).
Park, H. S. et al. Unpaired image denoising using a generative adversarial network in X-ray CT. IEEE Access. 7, 110414–110425 (2019).
Ye, J. C., Han, Y. & Cha, E. Deep convolutional framelets: a general deep learning framework for inverse problems. SIAM J. Imaging Sci. 11, 991–1048 (2017).
Ioffe, S. & Szegedy, C. Batch normalization: accelerating deep network training by reducing internal covariate shift. Preprint at arXiv:1502.03167 (2017).
Nair, V. & Hinton, G.E. Rectified linear units improve restricted Boltzmann machines. in ICML. 807–814 (2010).
Chui, C. K. An Introduction to Wavelets (Elsevier, Amsterdam, 2014).
Ronneberger, O., Fischer, P. & Brox, T. U-Net: convolutional networks for biomedical image segmentation. in MICCAI 2015 (2015).
Gu, J. et al. Recent advances in convolutional neural networks. Pattern Recogn. 77, 354–377 (2018).
Zhang, Y. C. & Kagen, A. C. Machine learning interface for medical image analysis. J. Digit. Imaging 30, 615–621 (2017).
Bullich, S. et al. Optimized classification of 18F-Florbetaben PET scans as positive and negative using an SUVR quantitative approach and comparison to visual assessment. Neuroimage Clin. 15, 325–332 (2017).
Barthel, H. & Sabri, O. Florbetaben to trace amyloid-β in the Alzheimer brain by means of PET. J. Alzheimers Dis. 26, 117–121 (2011).
Johnson, J., Alahi, A. & Fei-Fei, L. Perceptual Losses for Real-Time Style Transfer and Super-Resolution. in ECCV 2016 (2016).
Tiepolt, S. et al. Influence of scan duration on the accuracy of β-amyloid PET with florbetaben in patients with Alzheimer’s disease and healthy volunteers. Eur. J. Nucl. Med. Mol. Imaging. 40, 238–244 (2013).
Oh, M. et al. Clinical significance of visually equivocal amyloid PET findings from the Alzheimer’s disease neuroimaging initiative cohort. NeuroReport 29, 553–558 (2018).
Yamane, T. et al. Inter-rater variability of visual interpretation and comparison with quantitative evaluation of 11C-PiB PET amyloid images of the Japanese Alzheimer’s Disease Neuroimaging Initiative (J-ADNI) multicenter study. Eur. J. Nucl. Med. Mol. Imaging 44, 850–857 (2017).
Gao, Y., Li, Y., Ma, K. & Zheng, Y. A universal intensity standardization method based on a many-to-one weak-paired cycle generative adversarial network for magnetic resonance images. IEEE Trans. Med. Imaging. 38, 2059–2069 (2019).
Chen, J. et al. Generative adversarial networks for video-to-video domain adaptation. in AAAI (2020).
Acknowledgements
This research was supported by the National Research Foundation (NRF) of Korea funded by the Ministry of Science, ICT & Future Planning (NRF-2018. R1A2B2008178). Hyoung Suk Park and Kiwan Jeon authors were supported by the National Institute for Mathematical Sciences (NIMS) grant funded by the Korean government (No. NIMS-B20900000). Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense Award No. W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. Data used in the current study were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the study design and implementation of the database and/or provided data, but did not participate in analysis or writing of this manuscript. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Author information
Authors and Affiliations
Contributions
Y.J.J., D.Y.K., and H.S.P. conceived and designed the study. Y.J.J. and J.E.J. did the data collection. Y.J.J., D.Y.K., and J.E.J. interpreted PET images. H.J.Y. analyzed the SUVR of PET images. H.S.P. and K.J. conducted synthetic PET image generation using deep learning methods. K.C. did the statistical analysis. Y.J.J. and H.S.P. wrote the manuscript, and all authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jeong, Y.J., Park, H.S., Jeong, J.E. et al. Restoration of amyloid PET images obtained with short-time data using a generative adversarial networks framework. Sci Rep 11, 4825 (2021). https://doi.org/10.1038/s41598-021-84358-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-84358-8
This article is cited by
-
Deep learning-based PET image denoising and reconstruction: a review
Radiological Physics and Technology (2024)
-
Deep learning-based image reconstruction and post-processing methods in positron emission tomography for low-dose imaging and resolution enhancement
European Journal of Nuclear Medicine and Molecular Imaging (2022)
-
Applications of Generative Adversarial Networks (GANs) in Positron Emission Tomography (PET) imaging: A review
European Journal of Nuclear Medicine and Molecular Imaging (2022)
-
A Brief History of Nuclear Medicine Physics, Instrumentation, and Data Sciences in Korea
Nuclear Medicine and Molecular Imaging (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.