Abstract
Background:
Pazopanib has shown clinical activity against multiple tumour types and is generally well tolerated. However, isolated elevations in transaminases and bilirubin have been observed. This study examined polymorphisms in molecules involved in pharmacokinetic and pharmacodynamic pathways of pazopanib and their association with hepatic dysfunction.
Methods:
Twenty-eight polymorphisms in 11 genes were evaluated in pazopanib-treated renal cell carcinoma patients. An exploratory analysis was conducted in 116 patients from a phase II study; a replication study was conducted in 130 patients from a phase III study.
Results:
No polymorphisms were associated with alanine aminotransferase elevation. The Gilbert's uridine-diphosphoglucuronate glucuronosyltransferase 1A1 (UGT1A1) TA-repeat polymorphism was significantly associated with pazopanib-induced hyperbilirubinemia in the phase II study. This association was replicated in the phase III study (P<0.01). Patients with TA6/TA6, TA6/TA7, and TA7/TA7 genotypes experienced median bilirubin increases of 0.31, 0.37, and 0.71 × upper limit of the normal range (ULN), respectively. Of the 38 patients with hyperbilirubinemia (⩾1.5 × ULN), 32 (84%) were either TA7 homozygotes (n=18) or TA7 heterozygotes (n=14). For TA7 homozygotes, the odds ratio (95% CI) for developing hyperbilirubinemia was 13.1 (5.3–32.2) compared with other genotypes.
Conclusions:
The UGT1A1 polymorphism is frequently associated with pazopanib-induced hyperbilirubinemia. These data suggest that some instances of isolated hyperbilirubinemia in pazopanib-treated patients are benign manifestations of Gilbert's syndrome, thus supporting continuation of pazopanib monotherapy in this setting.
Similar content being viewed by others
Main
Pazopanib (Votrient, GlaxoSmithKline), recently approved by the United States Food and Drug Administration (FDA) for the treatment of patients with advanced renal cell carcinoma (RCC) (GlaxoSmithKline, 2009), is an oral angiogenesis inhibitor targeting vascular endothelial growth factor receptors-1, -2, and -3; platelet-derived growth factor receptors-α and -β; and the stem cell factor receptor, c-kit (Sonpavde and Hutson, 2007). It is currently under clinical development for the treatment of multiple tumour types (Altorki et al, 2008; Friedlander et al, 2008; Sleijfer et al, 2009; Taylor et al, 2009; Sternberg et al, 2010). Clinical meaningful efficacy was observed in pazopanib-treated patients with RCC (Hutson et al, 2010; Sternberg et al, 2010). The safety profile of pazopanib was generally acceptable and tolerable. The most common adverse events were diarrhoea, hypertension, hair colour changes, nausea, anorexia, and vomiting (Sternberg et al, 2010). Elevations of liver transaminases were the most common treatment-emergent laboratory abnormalities (Sternberg et al, 2010). Elevations in alanine aminotransferase (ALT) >3 × upper limit of the normal range (ULN) occurred in 18% of the pazopanib-treated RCC patients. Isolated elevations of total bilirubin (TBL) >1.5 × ULN were seen in 17% of pazopanib-treated RCC patients. Elevations in liver enzymes were generally asymptomatic and reversible and concurrent elevations in both ALT and bilirubin were rare. As the aetiology and mechanism of liver enzyme elevation in pazopanib-treated patients remain unknown, recommended management guideliness are based on analysis of data from clinical trials.
Treatment-associated elevations in transaminases and bilirubin have been reported with other tyrosine kinase inhibitors including sunitinib, lapatinib, and erlotinib (Motzer et al, 2007; Loriot et al, 2008; Ryan et al, 2008), and the incidence varies with agent. It is possible that these treatment-associated elevations in liver enzymes observed with tyrosine kinase inhibitors reflect overlapping on-target and off-target class effects; however, specific mechanisms remain to be elucidated. Abnormalities in clinical liver chemistry measurements are important safety signals for liver injury and may lead to treatment discontinuation, thereby compromising the potential treatment benefit to the patient. Understanding the underlying mechanisms of liver chemistry abnormalities may enable better interpretation and clinical management of these safety signals, and in appropriate circumstances, allow patients to benefit from continued anticancer treatment.
We sought to identify genetic markers in selected candidate genes involved in pazopanib metabolism and pharmacodynamics that may predict risk of ALT and/or bilirubin elevation on treatment. Pazopanib is a substrate for p-glycoprotein and breast cancer-resistant protein, an inhibitor of the human uptake transporter OATP1B1, and an inhibitor of uridine-diphosphoglucuronate glucuronosyltransferase 1A1 (UGT1A1). It is predominately metabolised by CYP3A4 with a minor contribution from CYP2C8 and CYP1A2. Functional polymorphisms in these genes as well as in pazopanib target genes were evaluated using data from two clinical studies that evaluated the efficacy and safety of pazopanib in patients with advanced metastatic RCC (Hutson et al, 2010; Sternberg et al, 2010).
Materials and Methods
The protocol and informed consent forms were reviewed and approved by Institutional Review Boards/Independent Ethics Committees according to local guidelines. Clinical studies were conducted in accordance with the Declaration of Helsinki. Written informed consent for participation in the clinical studies was obtained from all patients, and an additional informed consent for pharmacogenetics (PGx) research was obtained for participation in the genetic study.
Patients
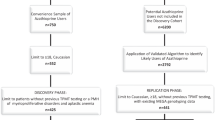
The phase II RCC study (VEG102616, Study 1) had 225 participants (Hutson et al, 2010) and the phase III RCC study (VEG105192, Study 2) had 435 participants (Sternberg et al, 2010). All patients from Study 1 and 290 patients from Study 2 received pazopanib (800 mg daily); the remaining 145 patients in Study 2 were randomised to the placebo arm. To minimise the effect of genetic heterogeneity among ethnic groups on the statistical analyses, the present PGx analyses were performed using the self-reported ‘white’ patients of European heritage, which represented the largest subgroup in each of the clinical studies. There were 156 self-reported white patients in Study 1, of whom 116 provided written informed consent for genetic research and had sufficient DNA for genotyping. There were 204 self-reported white patients in Study 2 who received pazopanib, of whom 130 provided written informed consent for genetic research and had sufficient DNA for genotyping. The present PGx analysis population, therefore, consisted of data from the 116 white patients from Study 1 and 130 white patients from Study 2 who received pazopanib and had genotypic data for at least one of the genetic markers evaluated (Figure 1).
Patient disposition. Of the 225 pazopanib-treated patients in Study 1 (VEG102616) and the 290 pazopanib-treated patients in Study 2 (VEG105192), 112 and 124 patients, respectively, were included in the UGT1A1 pharmacogenetics (PGx) analysis. These patients were self-reported to be white, provided consent for the PGx analysis, had sufficient DNA for genotyping and valid genotypic data for the UGT1A1 TA-repeat polymorphism, and had baseline and on-treatment total bilirubin (TBL) data.
Liver chemistry measurements
Alanine aminotransferase and bilirubin measurements were performed by local institutional laboratories. Both ALT and TBL values were converted to ULN by dividing the laboratory value by the institutional ULN. None of the patients from either study had a baseline ALT level >3 × ULN or baseline bilirubin level >1.5 × ULN.
Genetic polymorphisms and genotyping
Twenty-eight genetic polymorphisms in 11 genes involved in the pharmacokinetics and pharmacodynamics of pazopanib were selected (Table 1). The selection was based on reported associations or assumed functional changes of the polymorphisms to the expression or activity of the proteins.
The DNA was extracted from blood using the Qiagen (Valencia, CA, USA) QiAmp DNA Blood kit. The UGT1A1 TA-repeat polymorphism (rs8175347) was genotyped using the FDA-approved Third Wave Invader Assay, which called two alleles: the TA6 (*1) allele and the TA7 (*28) allele. In the rare instance when a patient had a TA-repeat number that was not 6 or 7 (<1%), the genotype call for that patient was treated as missing data. The remaining polymorphisms were genotyped using Illumina (San Diego, CA, USA) GoldenGate platform (Fan et al, 2003), a single base chain extension assay modified by GlaxoSmithKline (Research Triangle Park, NC, USA) (Taylor et al, 2001), TaqMan SNP Genotyping assays (Applied Biosystems, Foster City, CA, USA; Livak et al, 1995), or sequencing.
In vitro UGT1A1 inhibition
The activity of human UGT1A1 was measured in the absence and presence of pazopanib. Human UGT1A1 Supersomes (BD Gentest, BD Biosciences, San Jose, CA, USA) were preincubated in duplicate for 5 min at 37°C in the presence of the pore-former alamethicin, the UGT1A1 substrate 7-hydroxy-4-(trifluoromethyl) coumarin (HFC), and pazopanib concentrations ranging from 0 to 250 μ M. At the end of preincubation, the enzymatic reaction was initiated by adding the cofactor uridine 5′-diphosphoglucuronic acid (UDPGA). The reaction was terminated after an additional 10 min by adding acetonitrile/acetic acid (94/6 [v/v]), followed by centrifugation to sediment-precipitated protein. The production of the HFC metabolite 4-trifluoromethylumbelliferyl glucuronide (HFC-gluc) in each supernatant was quantified by HPLC-UV, and the half-maximal inhibitory concentration (IC50) value for inhibition of UGT1A1 activity was determined. Incubations without pazopanib were performed in the presence and absence of UDPGA to confirm production of the UDPGA-dependent glucuronide metabolite HFC-gluc. Incubations without probe substrate (HFC) were performed to determine any potential assay interference by pazopanib. The IC50 of curcumin, a positive-control inhibitor, was determined in parallel incubations in which pazopanib was replaced with appropriate concentrations of curcumin.
In vitro OATP1B1 inhibition
Inhibition of uptake of the OATP1B1 probe substrate [3H]-estradiol 17β-D-glucuronide ([3H]-EG) by pazopanib was measured using a stably transfected CHO cell line. The CHO-OATP1B1 cells were cultured to confluency, trypsinised, and seeded into 24-well assay plates (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) at a density of 70 000 cells cm–2 in Dulbecco's Modified Eagle Medium (DMEM) with GlutaMAX, 10% (v/v) foetal bovine serum, 0.5% (v/v) penicillin/streptomycin 10 000 units ml–1, 0.1% (v/v) L-proline 50 mg ml–1, and 0.7% (v/v) geneticin 50 mg ml–1. The cell monolayers were used 2 days after seeding and induced for at least 24 h before use with the addition of DMEM containing sodium butyrate (final concentration, 5 mM). In the inhibition studies, CHO-OATP1B1 monolayers were preincubated (37°C) for 15–30 min in 1 ml of Dulbecco's Phosphate-Buffered Saline (DPBS) with the appropriate concentration of pazopanib. Triplicate wells were used for each concentration of test compound. After removal of preincubation solution, 400 μl of DPBS containing the radiolabelled probe substrate and the appropriate concentrations of pazopanib was added to the wells and the cells were incubated at 37°C for 5 min. The solution was then removed and the wells rinsed rapidly three times using 800 μl cold (4°C) DPBS before solubilisation with 500 μl of 1% (v/v) Triton X-100. Total radioactivity was determined by scintillation counting.
Statistical analysis
The data were analysed both as continuous variables in a quantitative trait analysis (QTA) and as discrete values according to predefined thresholds in a case–control analysis. All analyses were performed in SAS version 9.1.3 (SAS Institute, Cary, NC, USA). The QTA was performed using an analysis of covariance model to assess the effect of genetic factors on baseline, maximum on-treatment, and change from baseline (delta) values for ALT and TBL. In the analyses of maximum on-treatment ALT and maximum on-treatment TBL, baseline was included in the model as a covariate. A log10-transformation was performed on the ALT and TBL values and a rank transformation was performed on the change-from-baseline values to correct for skewness of the data.
For case–control association analysis, a patient was defined as an ALT ‘case’ if one or more ALT measurements were ⩾3 × ULN during dosing with pazopanib. A patient was classified as an ALT ‘control’ if all ALT measurements were within the normal range (⩽1 × ULN). A patient was defined as a TBL ‘case’ if one or more TBL measurements were ⩾1.5 × ULN during exposure to pazopanib, and a TBL ‘control’ if all measurements were within the normal range (⩽1 × ULN). The effect of a genetic polymorphism on ALT or TBL was assessed using Fisher's exact test.
The initial exploratory analysis was performed using data from the phase II study (Study 1; VEG102616) and the replication analysis was conducted using data from the phase III study (Study 2; VEG105192). Correction for multiple tests was not performed. Instead, a threshold of P<0.01 was used to determine statistical significance in each study. A marker was ‘replicated’ when it was identified to be statistically significant in Study 1 followed by replication in Study 2. For replicated markers, a combined analysis using data from both studies was carried out to evaluate the overall effect.
Results
Demographic and baseline characteristics for patients included in the present PGx investigation from Study 1 and Study 2 are shown in Table 2. None of the patients included in the PGx analysis had a baseline ALT level >3 × ULN or baseline bilirubin level >1.5 × ULN. Exploratory analyses of the effects of the 28 genetic polymorphisms on maximum ALT and bilirubin were first examined using data from Study 1. None of the markers were significantly associated with maximum ALT (P<0.01), and three markers in the UGT1A1 and CYP1A2 loci were significantly associated with maximum bilirubin (P<0.01). The markers from the UGT1A1 locus were the TA-repeat polymorphism (UGT1A1*28) and the –3279T/G polymorphism (UGT1A1*60), and the marker from the CYP1A2 locus was the –163C/A polymorphism (Table 1).
Replication analyses of the three significant TBL markers identified in Study 1 were performed using data from Study 2. Only the TA-repeat polymorphism from the UGT1A1 gene was replicated (P<0.01). This polymorphism was not associated with maximum ALT measurements in either study.
Data from both studies were subsequently combined to determine the overall effect of the UGT1A1 TA-repeat polymorphism on bilirubin levels. Of the 246 patients included in this PGx analysis, data for bilirubin and the UGT1A1 TA-repeat marker were obtained for 236 patients (Figure 1). Of the remaining 10 patients, 5 had missing UGT1A1 genotype data and 5 had missing log10-transformed maximum or baseline TBL data. As expected, a significant association between the UGT1A1 TA-repeat polymorphism and maximum bilirubin was observed (P=1.6 × 10−6 and P=1.8 × 10−8 for the QTA and case–control analysis, respectively). The median values of maximum bilirubin were 0.75 × ULN, 0.87 × ULN, and 1.40 × ULN for patients with the TA6/TA6, TA6/TA7, and TA7/TA7 genotypes, respectively. Moreover, this polymorphism was significantly associated with bilirubin increases from baseline (delta) during pazopanib treatment (P=4.5 × 10−5). The median bilirubin increases were 0.31 × ULN, 0.37 × ULN, and 0.71 × ULN for pazopanib-treated patients with the TA6/TA6, TA6/TA7, and TA7/TA7 genotypes, respectively. Thus, the median on-treatment bilirubin increase for TA7 homozygotes was 2.3-fold greater than the increase observed for the TA6 homozygotes (Figure 2).
The distribution of baseline total bilirubin (TBL) and maximum TBL by UGT1A1 TA-repeat genotype for pazopanib-treated white patients from both Study 1 and Study 2. The maximum bilirubin value for one patient who had the TA7/TA7 genotype was truncated from 19 × upper limit of normal (ULN) to 6 × ULN for viewing purposes. The number of patients with each genotype, and the median values for baseline TBL, maximum TBL, and maximum change-from-baseline (delta) TBL are provided. The variation in baseline TBL, maximum TBL, and change-from-baseline TBL by genotype is statistically significant for each end point: P=4.0 × 10−3, 1.6 × 10−6, and 4.5 × 10−5, respectively.
Compared with the TA6/TA6 and TA6/TA7 genotypes, the odds ratio (95% confidence interval), positive predictive value, and negative predictive value for TA7/TA7 genotype were 13.1 (5.3–32.2), 0.49, and 0.90, respectively. The incidence of hyperbilirubinemia was 49% (18 of 37) for patients with the TA7/TA7 genotypes and 12% (14 of 113) for patients with the TA6/TA7 genotypes (Figure 3). In contrast, pazopanib-related incidence of hyperbilirubinemia was only 7% (6 of 86) for patients with the TA6/TA6 genotype. Of the 38 cases of TBL elevation, 32 patients (84%) were either TA7 homozygotes (n=18, 47%) or TA7 heterozygotes (n=14, 37%).
Cumulative incidence of hyperbilirubinemia (⩾1.5 × upper limit of normal [ULN]) by UGT1A1 TA-repeat genotype for pazopanib-treated white patients from both Study 1 and Study 2. The cumulative incidence of total bilirubin (TBL) ⩾1.5 × ULN for each genotype group at a time point (weeks) is the proportion of patients from the group who had at least one on-treatment TBL value of 1.5 × ULN or greater on or before the time point.
The ability of pazopanib to inhibit the two major determinants of serum bilirubin levels, UGT1A1 and OATP1B1, was measured. Pazopanib was shown to be a potent inhibitor of UGT1A1 as well as OATP1B1, with IC50 of 1.2 and 0.79 μ M, respectively (Figure 4).
Discussion
This study shows that both maximum on-treatment bilirubin concentration and bilirubin increase from baseline were strongly associated with the UGT1A1 TA-repeat polymorphism. None of the genetic markers evaluated were predictive of ALT elevation. Bilirubin is metabolised by UGT1A1 for elimination. The UGT1A1 genetic variant TA7 is known to cause reduced expression of UGT1A1 (Bosma et al, 1995), and the TA7/TA7 (*28/*28) genotype predisposes individuals to Gilbert's syndrome, a benign form of episodic jaundice (Bosma et al, 1995; Raijmakers et al, 2000). This UGT1A1 TA-repeat polymorphism has also been reported to be associated with hyperbilirubinemia induced by several drugs, such as tranilast, nilotinib, and indinavir (Zucker et al, 2001; Danoff et al, 2004; Singer et al, 2007). Pazopanib is a potent inhibitor of UGT1A1 activity in vitro, with an IC50 of 1.2 μ M. Thus, this study suggests that pazopanib-induced hyperbilirubinemia may be the result of inhibition of UGT1A1 activity combined with genetic defects of the UGT1A1 gene. This would presumably result in higher levels of unconjugated hyperbilirubinemia, usually associated with a benign clinical course. It is possible that bilirubin elevation associated with other tyrosine kinase inhibitors such as sunitinib, lapatinib, and erlotinib may also be related to UGT1A1 genotype. However, to our knowledge, data from genetic investigations for drug-induced hyperbilirubinemia for these compounds are not yet available.
We observed that 6 (16%) of the 38 patients who had isolated hyperbilirubinemia had the TA6/TA6 genotype, suggesting that additional factors may contribute to bilirubin elevation in pazopanib-treated patients. Concurrent elevations of transaminases were not seen in these six patients. A recent genome-wide association study identified OATP1B1 (alternative symbol SLCO1B1) as one of the top loci associated with bilirubin levels (Johnson et al, 2009). Genetic markers in the OATP1B1 gene were also evaluated in this study and were not found to be associated with pazopanib-related bilirubin elevation. As pazopanib is also a potent inhibitor of OATP1B1 (in vitro), we cannot exclude the possibility that pazopanib-induced hyperbilirubinemia may be the result of inhibition of both OATP1B1 and UGT1A1 activity combined with genetic defects of the UGT1A1 gene. The lack of association with genetic markers in the OATP1B1 gene may stem from the fact that pazopanib inhibited hepatic uptake of bilirubin to the same level in both wild type and variant forms.
Among the 246 patients evaluated in this study, concurrent elevation of ALT and bilirubin was observed in one patient (0.4%). This patient was heterozygous (TA6/TA7) for the UGT1A1 TA-repeat polymorphism and had direct bilirubin of 0.5 mg l–1, representing 22% of TBL (2.3 mg dl–1). Reduced UGT1A1 activity, both as a result of pazopanib inhibition of UGT1A1 and the genetic defect of the UGT1A1 gene, may have contributed to the bilirubin elevation observed in this patient. Concurrent elevations in ALT and bilirubin levels after pazopanib treatment could indicate liver toxicity (Kaplowitz, 2006); therefore, it is important to rule out this possibility to allow continued treatment. This is particularly important because continued anticancer treatment may allow for more favourable clinical outcomes and should be feasible in patients with benign hyperbilirubinemia, as might be the case for most pazopanib-treated patients. It follows that, for each patient, the clinical management of hepatotoxicity needs to be tailored according to the severity and impact of this adverse effect on the patient. In addition, the mechanisms involved in the observed elevation in transaminases and bilirubin should also be considered.
This study focused on data from trials evaluating pazopanib monotherapy in patients with RCC. The observed elevation in ALT or bilirubin levels was not associated with liver metastasis for patients included in the present PGx analysis. The full implications of these findings for regimens combining pazopanib with other agents are currently unknown. Clearly, it is important for clinicians to be aware of potential drug–drug interactions, particularly with compounds known to be metabolised and eliminated predominantly through the UGT1A1 pathway. Therefore, concomitant administration of pazopanib and UGT1A1 substrates that have narrow therapeutic index should be undertaken with caution. Pazopanib itself is not known to be subject to significant glucuronidation by UGT1A1. It is, therefore, unlikely that defective UGT1A1 would have a direct impact on the exposure of pazopanib and, therefore, exposure-related toxicities.
The practical implication of our finding is for clinicians to recognise that isolated hyperbilirubinemia in pazopanib-treated patients, after exclusion of other causes such as haemolysis, seems to have a benign aetiology. Given the life-threatening nature of cancer and the benign clinical course of unconjugated hyperbilirubinemia, the data do not call for population-based prospective UGT1A1 screening to exclude patients from pazopanib monotherapy. Bilirubin fractionation should be conducted in patients who develop hyperbilirubinemia during pazopanib treatment, which could distinguish unconjugated hyperbilirubinemia from conjugated hyperbilirubinemia. The latter would require further investigation. This may help to elucidate the nature of the bilirubin elevation and might enable differentiation of the risk of progression of drug-induced liver injury. A limitation of this study is the lack of bilirubin fractionation data to allow full assessment between the UGT1A1 polymorphism and unconjugated bilirubin levels.
References
Aklillu E, Carrillo JA, Makonnen E, Hellman K, Pitarque M, Bertilsson L, Ingelman-Sundberg M (2003) Genetic polymorphism of CYP1A2 in Ethiopians affecting induction and expression: characterization of novel haplotypes with single-nucleotide polymorphisms in intron 1. Mol Pharmacol 64 (3): 659–669
Altorki N, Heymach J, Guarino M, Lee P, Felip E, Bauer T, Swann S, Roychowdhury D, Ottesen LH, Yankelevitz D (2008) Phase II study of Pazopanib (GW786034) given preoperatively in stage i-ii non-small cell lung cancer (NSCLC): a proof-of-concept study. Ann Oncol 19 (suppl 8): viii89
Aono S, Yamada Y, Keino H, Hanada N, Nakagawa T, Sasaoka Y, Yazawa T, Sato H, Koiwai O (1993) Identification of defect in the genes for bilirubin UDP-glucuronosyl-transferase in a patient with Crigler-Najjar syndrome type II. Biochem Biophys Res Commun 197 (3): 1239–1244
Bahadur N, Leathart JB, Mutch E, Steimel-Crespi D, Dunn SA, Gilissen R, Houdt JV, Hendrickx J, Mannens G, Bohets H, Williams FM, Armstrong M, Crespi CL, Daly AK (2002) CYP2C8 polymorphisms in Caucasians and their relationship with paclitaxel 6alpha-hydroxylase activity in human liver microsomes. Biochem Pharmacol 64 (11): 1579–1589
Bosma PJ, Chowdhury JR, Bakker C, Gantla S, de Boer A, Oostra BA, Lindhout D, Tytgat GN, Jansen PL, Oude Elferink RP, Chowdhury NR (1995) The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. N Engl J Med 333 (18): 1171–1175
Dai D, Zeldin DC, Blaisdell JA, Chanas B, Coulter SJ, Ghanayem BI, Goldstein JA (2001) Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics 11 (7): 597–607
Danoff TM, Campbell DA, McCarthy LC, Lewis KF, Repasch MH, Saunders AM, Spurr NK, Purvis IJ, Roses AD, Xu CF (2004) A Gilbert's syndrome UGT1A1 variant confers susceptibility to tranilast-induced hyperbilirubinemia. Pharmacogenomics J 4 (1): 49–53
Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, Hansen M, Steemers F, Butler SL, Deloukas P, Galver L, Hunt S, McBride C, Bibikova M, Rubano T, Chen J, Wickham E, Doucet D, Chang W, Campbell D, Zhang B, Kruglyak S, Bentley D, Haas J, Rigault P, Zhou L, Stuelpnagel J, Chee MS (2003) Highly parallel SNP genotyping. Cold Spring Harb Symp Quant Biol 68: 69–78
Friedlander M, Hancock KC, Benigno B, Rischin D, Messing M, Stringer CA, Hodge JP, Ma B, Matthys G, Lager JJ (2008) Pazopanib (GW786034) is active in women with advanced epithelial ovarian, fallopian tube, and peritoneal cancers: results of a phase II study. Ann Oncol 19 (suppl 8): viii211
GlaxoSmithKline (2009) FDA approves GlaxoSmithKline's Votrient for advanced renal cell cancer. www.gsk.com/media/pressreleases/2009/2009_pressrelease_10113.htm
Han XM, Ouyang DS, Chen XP, Shu Y, Jiang CH, Tan ZR, Zhou HH (2002) Inducibility of CYP1A2 by omeprazole in vivo related to the genetic polymorphism of CYP1A2. Br J Clin Pharmacol 54 (5): 540–543
Hutson TE, Davis ID, Machiels JP, De Souza PL, Rottey S, Hong B, Epstein RJ, Baker KL, McCann L, Crofts T, Liu Y, Pandite L, Figlin RA (2010) Efficacy and safety of pazopanib in patients with metastatic renal cell carcinoma. J Clin Oncol 28 (3): 475–480
Johnson AD, Kavousi M, Smith AV, Chen MH, Dehghan A, Aspelund T, Lin JP, van Duijn CM, Harris TB, Cupples LA, Uitterlinden AG, Launer L, Hofman A, Rivadeneira F, Stricker B, Yang Q, O′Donnell CJ, Gudnason V, Witteman JC (2009) Genome-wide association meta-analysis for total serum bilirubin levels. Hum Mol Genet 18 (14): 2700–2710
Jounaidi Y, Hyrailles V, Gervot L, Maurel P (1996) Detection of CYP3A5 allelic variant: a candidate for the polymorphic expression of the protein? Biochem Biophys Res Commun 221 (2): 466–470
Kaplowitz N (2006) Rules and laws of drug hepatotoxicity. Pharmacoepidemiol Drug Saf 15 (4): 231–233
Kerb R (2006) Implications of genetic polymorphisms in drug transporters for pharmacotherapy. Cancer Lett 234 (1): 4–33
Livak KJ, Marmaro J, Todd JA (1995) Towards fully automated genome-wide polymorphism screening. Nat Genet 9 (4): 341–342
Loriot Y, Boudou-Rouquette P, Billemont B, Ropert S, Goldwasser F (2008) Acute exacerbation of hemorrhagic rectocolitis during antiangiogenic therapy with sunitinib and sorafenib. Ann Oncol 19 (11): 1975
Mizuarai S, Aozasa N, Kotani H (2004) Single nucleotide polymorphisms result in impaired membrane localization and reduced atpase activity in multidrug transporter ABCG2. Int J Cancer 109 (2): 238–246
Morisaki K, Robey RW, Ozvegy-Laczka C, Honjo Y, Polgar O, Steadman K, Sarkadi B, Bates SE (2005) Single nucleotide polymorphisms modify the transporter activity of ABCG2. Cancer Chemother Pharmacol 56 (2): 161–172
Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, Oudard S, Negrier S, Szczylik C, Kim ST, Chen I, Bycott PW, Baum CM, Figlin RA (2007) Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med 356 (2): 115–124
Nozawa T, Nakajima M, Tamai I, Noda K, Nezu J, Sai Y, Tsuji A, Yokoi T (2002) Genetic polymorphisms of human organic anion transporters OATP-C (SLC21A6) and OATP-B (SLC21A9): allele frequencies in the Japanese population and functional analysis. J Pharmacol Exp Ther 302 (2): 804–813
Raijmakers MT, Jansen PL, Steegers EA, Peters WH (2000) Association of human liver bilirubin UDP-glucuronyltransferase activity with a polymorphism in the promoter region of the UGT1A1 gene. J Hepatol 33 (3): 348–351
Rodriguez-Antona C, Sayi JG, Gustafsson LL, Bertilsson L, Ingelman-Sundberg M (2005) Phenotype-genotype variability in the human CYP3A locus as assessed by the probe drug quinine and analyses of variant CYP3A4 alleles. Biochem Biophys Res Commun 338 (1): 299–305
Ryan Q, Ibrahim A, Cohen MH, Johnson J, Ko CW, Sridhara R, Justice R, Pazdur R (2008) FDA drug approval summary: lapatinib in combination with capecitabine for previously treated metastatic breast cancer that overexpresses HER-2. Oncologist 13 (10): 1114–1119
Schneider BP, Wang M, Radovich M, Sledge GW, Badve S, Thor A, Flockhart DA, Hancock B, Davidson N, Gralow J, Dickler M, Perez EA, Cobleigh M, Shenkier T, Edgerton S, Miller KD (2008) Association of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in a trial of paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG 2100. J Clin Oncol 26 (28): 4672–4678
Singer JB, Shou Y, Giles F, Kantarjian HM, Hsu Y, Robeva AS, Rae P, Weitzman A, Meyer JM, Dugan M, Ottmann OG (2007) UGT1A1 promoter polymorphism increases risk of nilotinib-induced hyperbilirubinemia. Leukemia 21 (11): 2311–2315
Sleijfer S, Ray-Coquard I, Papai Z, Le Cesne A, Scurr M, Schoffski P, Collin F, Pandite L, Marreaud S, De Brauwer A, van Glabbeke M, Verweij J, Blay JY (2009) Pazopanib, a multikinase angiogenesis inhibitor, in patients with relapsed or refractory advanced soft tissue sarcoma: a phase II study from the European organisation for research and treatment of cancer-soft tissue and bone sarcoma group (EORTC study 62043). J Clin Oncol 27 (19): 3126–3132
Sonpavde G, Hutson TE (2007) Pazopanib: a novel multitargeted tyrosine kinase inhibitor. Curr Oncol Rep 9 (2): 115–119
Sternberg CN, Davis ID, Mardiak J, Szczylik C, Lee ES, Wagstaff J, Barrios CH, Salman P, Gladkov OA, Kavina A, Zarba JJ, Chen M, McCann L, Pandite L, Roychowdhury D, Hawkins RE (2010) Pazopanib in locally advanced and/or metastatic renal cell carcinoma: results of a randomized phase III trial. J Clin Oncol 28 (6): 1061–1068
Sugatani J, Yamakawa K, Yoshinari K, Machida T, Takagi H, Mori M, Kakizaki S, Sueyoshi T, Negishi M, Miwa M (2002) Identification of a defect in the UGT1A1 gene promoter and its association with hyperbilirubinemia. Biochem Biophys Res Commun 292 (2): 492–497
Tanimoto K, Yoshiga K, Eguchi H, Kaneyasu M, Ukon K, Kumazaki T, Oue N, Yasui W, Imai K, Nakachi K, Poellinger L, Nishiyama M (2003) Hypoxia-inducible factor-1alpha polymorphisms associated with enhanced transactivation capacity, implying clinical significance. Carcinogenesis 24 (11): 1779–1783
Taylor JD, Briley D, Nguyen Q, Long K, Iannone MA, Li MS, Ye F, Afshari A, Lai E, Wagner M, Chen J, Weiner MP (2001) Flow cytometric platform for high-throughput single nucleotide polymorphism analysis. Biotechniques 30 (3): 661–666, 668–669
Taylor SK, Chia S, Dent S, Clemons M, Grenci P, Wang L, Oza AM, Ivy P, Pritchard K, Leighl N (2009) A phase II study of GW786034 (pazopanib) in patients with recurrent or metastatic invasive breast carcinoma: results after completion of stage I: a trial of the Princess Margaret Hospital Phase II Consortium. J Clin Oncol 27 (15s): abstract 1133
Wong M, Balleine RL, Collins M, Liddle C, Clarke CL, Gurney H (2004) CYP3A5 genotype and midazolam clearance in Australian patients receiving chemotherapy. Clin Pharmacol Ther 75 (6): 529–538
Zucker SD, Qin X, Rouster SD, Yu F, Green RM, Keshavan P, Feinberg J, Sherman KE (2001) Mechanism of indinavir-induced hyperbilirubinemia. Proc Natl Acad Sci USA 98 (22): 12671–12676
Acknowledgements
We thank the investigators and their patients who made this study possible. We acknowledge Leigh Ragone, Morlisa Dixon, Bo Zheng, Nan Bing, and Keith Nangle for their excellent technical support. We thank Melissa Stutts, Vicki Goodman, Theresa Bryant, and Chris Abissi for their contribution in evaluating patients with potential hepatotoxicity; Lauren McCann and Shawn Liu for discussion of the laboratory liver chemistry data. We are grateful to Rafael Amado, Pamela StJean, Matthew Nelson, Anne Marie Martin, and Howard Ball, and the GSK hepatotoxicity board, in particular Christine Hunt and Beena Koshy, for their review and discussion of the data. Financial support for this study and medical editorial assistance was provided by GlaxoSmithKline Pharmaceuticals, Philadelphia, Pennsylvania. We thank Jerome Sah, PhD, ProEd Communications, Inc., for his medical editorial assistance with this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Xu, CF., Reck, B., Xue, Z. et al. Pazopanib-induced hyperbilirubinemia is associated with Gilbert's syndrome UGT1A1 polymorphism. Br J Cancer 102, 1371–1377 (2010). https://doi.org/10.1038/sj.bjc.6605653
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6605653
Keywords
This article is cited by
-
Adverse effects of tyrosine kinase inhibitors in cancer therapy: pathophysiology, mechanisms and clinical management
Signal Transduction and Targeted Therapy (2023)
-
Severe tyrosine-kinase inhibitor induced liver injury in metastatic renal cell carcinoma patients: two case reports assessed for causality using the updated RUCAM and review of the literature
BMC Gastroenterology (2022)
-
Pazopanib interacts with irinotecan by inhibiting UGT1A1-mediated glucuronidation, but not OATP1B1-mediated hepatic uptake, of an active metabolite SN-38
Cancer Chemotherapy and Pharmacology (2019)
-
Influence of UGT1A1 polymorphism on etoposide plus platinum-induced neutropenia in Japanese patients with small-cell lung cancer
International Journal of Clinical Oncology (2019)
-
Harnessing soft tissue sarcoma with low-dose pazopanib – a matter of blood levels
BMC Cancer (2018)