Abstract
Background:
Preliminary data indicate that the molecular epidemiology of localised gastrointestinal stromal tumour (GIST) may be different from that of advanced GIST. We sought to investigate the molecular epidemiology of sarcomas, including GIST, in the Rhone-Alpes region in France.
Patients and methods:
A prospective and exhaustive study in the Rhone-Alpes Region in France to assess the precise incidence of primary sarcomas with systematic centralised pathological review and molecular analysis was conducted for 2 consecutive years.
Results:
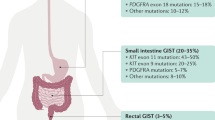
Among 760 patients with a confirmed diagnosis of sarcoma, 131 (17%) had a GIST. The majority of patients had gastric primaries (61%). Mutational analysis could be performed in 106 tumour samples (74%), and 71 (67%) had exon 11 mutations. PDGFRA mutations were found in 16% of cases, which is twice as high as previously reported for advanced GIST.
Conclusion:
Data indicate that PDGFRA mutations in localised GIST may be twice as high as what was previously reported in patients with advanced disease. This finding may have important consequences for patients offered adjuvant imatinib, although most of these tumours are in the low-risk group.
Similar content being viewed by others
Main
Gastrointestinal stromal tumours (GISTs) are rare tumours of mesenchymal origin arising in the gastrointestinal tract. Incidence data obtained from registries indicate an incidence of approximately 12–15 new cases per million inhabitants per year in western countries (Goettsch et al, 2005; Nilsson et al, 2005); however, there remain uncertainties because of variations in diagnostic criteria before the NCI consensus reported in 2002 (Fletcher et al, 2002). Approximately 95% of GISTs stain positive for CD117 (KIT) and 85% of cases harbour activating mutations in the gene of one of two structurally related transmembrane tyrosine-kinase receptors: KIT and PDGFRA (Hirota et al, 1998; Heinrich et al, 2003a, 2003b; Corless et al, 2005). These activating mutations affect primarily the exons 9 and 11 of KIT, but may also be found on exons 8, 13 and 17 of KIT and exons 12, 14 and 18 of PDGFRA (Heinrich et al, 2003a, 2008; Debiec-Rychter et al, 2006). The relative frequency of the different KIT and PDGFRA mutations in patients with advanced GIST has been previously reported (Heinrich et al, 2003a, 2008; Debiec-Rychter et al, 2006). The frequency of KIT and PDGFRA mutations in localised GIST has been reported in a single-institution study from Italy. Other preliminary reports indicate that PDGFRA mutations may be higher in localised than metastatic GIST, which may reflect their more favourable prognosis. Two recent autopsy series have shown that the incidence of GIST may be as high as 50% in stomach specimens (Kawanowa et al, 2006; Agaimy et al, 2007); in one of these series, canonical KIT or PDGFRA mutations were found in 50% of assessable tumours (Agaimy et al, 2007). We sought to assess the precise incidence of GIST and other sarcomas in the Rhone-Alpes region in France. This report focuses exclusively on GIST.
Patients and methods
A prospective and exhaustive study was conducted in the Rhone-Alpes region in France to assess the precise incidence of primary sarcomas and to assess conformity of management with published guidelines. Rhone-Alpes represent 10% of the French population (5 958 320 of 60 825 000 inhabitants) and 10% of the French territory. All cases for which a diagnosis of sarcoma was raised were systematically and centrally reviewed by experts (DR and AVD) to confirm the diagnosis.
Collection of data
All pathologists of the Rhone-Alpes region (n=42) agreed to participate in the study after a first meeting to inform them. They had to prospectively notify their cases, and to address a paraffin-embedded tumour sample for expert review. A financial support was allocated for each notified case and each tumour bloc. Duplications were checked in the database by a comparison of the patient's initials and the date of birth. If one patient was noted to have several diagnostic materials (e.g., biopsy, surgery and metastasis), or if tumour locations were multiple, the patient could be included only once in the study.
All diagnosed patients with a first diagnosis of primary sarcoma, between 1 March 2005 and 28 February 2006, and living in the Rhône-Alpes region (as testified by the ZIP code of patients’ address), were included in the study. All subtypes of sarcomas were included: soft tissue, bone and visceral (GISTs and gynaecological sarcomas) sarcomas. Exclusion criteria concerned relapsed tumour, date of diagnosis not matched with the inclusion period, patient living outside of the Rhone-Alpes region and no definitive histology in favour of sarcoma after the review. A validation series of all new cases diagnosed between 1 March 2006 and 28 February 2007 was also collected and analysed using the same methodology.
Pathological review
All cases of suspicious sarcomas were reviewed by regional sarcoma expert pathologists (JYS, DRV and AVD) and national experts (JM Coindre and a panel of pathologists from French Sarcoma group). New tissue sections were prepared from the paraffin-embedded samples provided by the primary pathologist, and immunohistochemical study was systematically performed again by the expert reviewer (Lurkin et al, 2010). Diagnoses were performed in accordance with the 2002 WHO classification.
Mutation analysis
After manual microdissection of tumour or normal tissue, DNA was extracted from paraffin-embedded material, using the MasterPure kit (Epicentre Biotechnologies, Le Parray en Yvelines, France). Exons 9, 11, 13 and 17 of KIT and exons 12, 14 and 18 of PDGFRA were amplified using the primers detailed in Table 1.
Except for exon 9 of KIT, mutation screening was performed by dHPLC analysis with a WAVE system (Transgenomic, Montluçon, France). Mutations were confirmed by sequencing with the same primer as for PCR; PCR products were purified using Qiagen MiniElute PCR (Qiagen, Courtaboeuf, France) purification columns and then sequenced on both strands with the DYEnamic ET Dye terminator kit (Amersham Bioscience, Orsay, France) and analysed on a MegaBACE 1000 automatic sequencer (Amersham Bioscience).
For KIT exon 9, the six-nucleotide duplication was assessed using high-resolution agarose gel electrophoresis (Resophor; Laboratoire Eurobio, Les Ulis, France) of the 47 bp (or 53 bp) PCR product.
Statistical analysis
Data were described using percentages for qualitative variables and median and range for numerical variables. All statistical analyses were performed using the SPSS 12.0 package (SPSS Inc., Chicago, IL, USA).
Results
Epidemiology of GIST
A total of 703 patients for whom a diagnosis of sarcoma was raised were screened; 42 patients (6.1%) were diagnosed outside the study period; 128 patients (18.5%) were found to have relapse (and not primary sarcoma); 44 patients (6.4%) managed in the Rhone-Alpes were found to live outside the predefined region; 33 patients (4.8%) lived and were managed outside the Rhone-Alpes; and 68 patients (9.8%) did not have sarcoma after expert pathologic review. In all, 376 patients (54.4%) conformed to the inclusion criteria: confirmed diagnosis of sarcoma after expert review, initial diagnosis between 1 March 2005 and 28 February 2006 (date of biopsy if there was one/of surgery if no biopsy) and resident in Rhone-Alpes region; 67 patients (17.8%) were diagnosed with GIST. For the second year, the diagnosis of sarcoma was raised in 581 patients, of whom 369 patients had a confirmed diagnosis of incident sarcoma, with 64 of them (16.7% of all sarcomas) identified as incident cases with GIST. Overall, 745 of 1284 patients had a confirmed diagnosis of sarcoma, of which 131 (18%) were GISTs. The crude incidence of GIST was therefore 11.2 per million inhabitant per year.
Patients’ characteristics
The main characteristics of the 131 patients with GIST are described in Table 2. In brief, a majority of patients were female (n=75, 57%), median age was 66 (range 34–91) years and most (61%) tumours originated from the stomach. In all, 117 patients (89%) were diagnosed as having localised GIST, whereas 14 patients (11%) had metastatic disease at initial diagnosis, confined to the abdominal cavity in all cases (liver metastases n=9; mesentery or peritoneum n=8; and 2 patients had lymph node metastases). Four tumours were diagnosed incidentally on surgical specimens obtained for other reasons: two tumours were found in oesophagectomy specimens (one performed after sulphuric acid ingestion and one performed for squamous cell carcinoma), one tumour was found on a gastrectomy specimen performed for gastric adenocarcinoma and one patient had an incidental diagnosis of GIST during cholecystectomy (the surgeon resected a small gastric lesion). Risk stratification was performed according to both the National Institute of Health (NIH) (Fletcher et al, 2002) and the Armed Forces Institute of Pathology (AFIP) (Miettinen and Lasota, 2006) criteria (Table 2).
Molecular data
Mutation analysis could be performed in 106 of 131 patients (81%), 55 of 67 patients (82%) in the first cohort and 51 of 64 patients (80%) in the second cohort (Table 2). In 16 cases, mutational analysis could not be performed because of the fixative used, and in the 9 remaining cases the sample size was not sufficient to retrieve enough DNA for sequencing (microbiopsies or microGIST).
The majority of samples harboured KIT mutations (n=71 of 106, 67%), and no KIT or PDGFRA mutations was found in 18 patients (17%). Seventeen tumours (16%) harboured PDGFRA mutations, 15 tumours originated from the stomach (14 with PDGFRA exon 18 and one PDGFRA exon 12), one from the peritoneum (omentum, PDGFRA exon 18 mutation) and one from the small bowel (PDGFRA exon 12 mutation). In addition, 10 patients (9%) had tumours with exon 9 mutations, 8 of which originated from the small bowel (including the duodenum), one from the rectum and one from the pelvis. The relative frequencies of KIT exon 11, KIT exon 9 and PDGFRA mutations are described in Table 3. When considering only patients with localised disease for whom molecular data were available (n=94), KIT exon 9, 11, 13 and 17 mutations were found in 9 (10%), 49 (52%), 3 (3%) and 1 (1%) patient, respectively, whereas PDGFRA mutations were found in 14 (15%) patients (exon 18, n=12 and exon 12, n=2), and 18 (19%) patients had KIT and PDGFRA wild-type tumours. Of the 14 PDGFRA exon 18 mutations, 13 (93%) involved codon 842, and 10 were D842V substitutions (11%). Molecular analysis could be performed for only one of the four incidentally diagnosed GISTs and showed a KIT exon 11 mutation. For the three other cases, the amount of DNA was not sufficient to perform molecular analysis.
Correlation of mutation and risk stratification
The aim of this correlative analysis was to determine which patient would be eligible for adjuvant imatinib, and it was therefore conducted only for patients with localised disease. Furthermore, both the NIH and the AFIP risk stratification were simplified into two categories grouping very low and low-risk group on one side and intermediate- and high-risk groups (i.e., those eligible for adjuvant imatinib per EMEA approval) on the other. Results of this correlative analysis are depicted in Table 4. As previously described (Blackstein et al, 2010), approximately 15–20% of patients from the high (intermediate and high) risk group in the NIH classification were reallocated to the low (very low and low) risk group of the AFIP risk stratification. This was true for all mutations types except for PDGFRA mutants in which 50% of the patients were reclassified as low risk, leaving only 7% of patients in the high-risk group of the AFIP classification (vs 57% when using the NIH classification; Table 4). The distribution of PDGFRA mutant tumours between the low- and high-risk categories was significantly different from that of KIT mutant or wild-type tumours when using the AFIP classification (P=0.005, Kruskal–Wallis test), but not when using the NIH classification (P=0.452, Kruskal–Wallis test). This difference of distributions is likely the consequence of the association of PDGFRA mutations with tumours of gastric origin (Lasota et al, 2004).
Discussion
This study is the first to date to report incidence with molecular analysis of GIST based on a prospectively collected exhaustive population sample (5 958 320 inhabitants in the Rhône-Alpes region). Overall, our data are in agreement with previously reported series in terms of primary tumour location, size and gender (Braconi et al, 2008). One of the key findings is the differences in the frequency of PDGFRA mutations that seem almost twice as high as that reported in patients with advanced disease (Debiec-Rychter et al, 2006; Braconi et al, 2008; Braggio et al, 2008; Du et al, 2008; Heinrich et al, 2008) (Table 5), including in patients with localised disease. These differences may be because of the more indolent behaviour of tumours bearing mutant PDGFRA (Lasota et al, 2004; Dematteo et al, 2008), whereas some KIT exon 11 and KIT exon 9 mutations have a higher risk of relapse after surgical excision. Other recently reported series have found somewhat lower frequencies of PDGFRA mutations: Braggio et al (2008) reported frequencies of 7.3% in patients with completely resected GIST in a specific population from Brazil. In both their series and ours, the small numbers preclude any definitive conclusion and these observations must be confirmed by larger studies. Emile et al (2009) recently reported a large series of GIST with molecular epidemiology and found that PDGFRA mutation were twice as high in localised GIST than that previously reported for patients with advanced disease. This series, which used different inclusion criteria (expert pathology referral), confirms what was found in our series. However, unlike Emile et al (2009), we could not find any significant difference in age between patients with PDGFRA-mutated tumours and those with other mutation type (Kruskal–Wallis nonparametric test P=0.295 for PDGFRA mutants vs other mutation types). Furthermore, in contrast with what Emile et al (2009) found, the frequency of KIT exon 9 mutations in patients with localised disease in our series was comparable to that previously reported in patients with advanced disease (approximately 10%; Heinrich et al, 2003a; Debiec-Rychter et al, 2006)
Although the predictive role of KIT mutations have been studied and reported in patients with advanced disease (Heinrich et al, 2003a; Debiec-Rychter et al, 2006; Van Glabbeke et al, 2007), the precise role of mutations in the biology of localised GISTs and their correlation with prognosis remains debated, although PDGFRA mutations seem associated with a rather indolent course (Lasota et al, 2004), whereas deletions of KIT exon 11 encompassing codons 557 and 558 were found to have poor prognosis (Martin et al, 2005). More data regarding the prognostic significance of different kinase mutations will likely be generated from the control arms of the adjuvant trials that have now finished accrual (Dematteo et al, 2009; Gronchi et al, 2009).
These observations are of importance considering the recent introduction of adjuvant imatinib treatment in high-risk GIST. PDGFRA exon 18 mutations (D842V), which affect the activation loop of the PDGFRα kinase, are biochemically less sensitive to imatinib than the more common KIT exon 11 mutations. As they may also have a better prognosis, the molecular characterisation may prove to be useful in selecting those patients in whom adjuvant imatinib is truly required. It is noteworthy that although 57% of the tumours with PDGFRA mutations in our series were classified as intermediate or high risk according to the NIH consensus (Fletcher et al, 2002), most of these tumours were classified as low or very low risk according to the AFIP risk classification (Miettinen and Lasota, 2006). The main reason for this shift is likely the gastric origin of most of the tumours harbouring PDGFRA mutations (15 of 17, 88%). Similarly, the identification of exon 9 may be needed to propose the adequate dose of adjuvant imatinib (Van Glabbeke et al, 2007).
Finally, the absolute incidence of GIST may be largely underestimated as suggested by the reports of Agaimy et al (2007) and Kawanowa et al (2006), which reported the so-called ‘microGIST’ in up to 50% of specimens of gastrectomies performed for other causes. Conversely, most of these tumours never become ‘clinically significant’, as the rate of clinically detected GIST is 12–15 per million inhabitants in Western countries as shown by our data and others (Nilsson et al, 2005; Monges et al, 2007). It is noteworthy that three patients in the 2005 cohort and one in the 2006 cohort had a diagnosis of GIST after surgery for another cause. Furthermore, new diagnostic or imaging procedures, such as endoscopic ultrasound, may lead to an increase in the diagnosis of small GIST. The ‘true incidence’ of GIST may therefore be a moving concept in the future.
Change history
29 March 2012
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Agaimy A, Wunsch PH, Hofstaedter F, Blaszyk H, Rummele P, Gaumann A, Dietmaier W, Hartmann A (2007) Minute gastric sclerosing stromal tumors (GIST tumorlets) are common in adults and frequently show c-KIT mutations. Am J Surg Pathol 31: 113–120
Blackstein ME, Corless CL, Ballman KV, Antonescu CR, Blanke CD, Demetri GD, von-Mehren M, Maki RG, Pisters PW, Dematteo RP (2010) Risk assessment for tumor recurrence after surgical resection of localized primary gastrointestinal stromal tumor (GIST): North American Intergroup phase III trial ACOSOG Z9001. 2010 Gastrointestinal Cancer Symposium. Abstract 6. 2010. Orlando, FL
Braconi C, Bracci R, Bearzi I, Bianchi F, Costagliola A, Catalani R, Mandolesi A, Ranaldi R, Galizia E, Cascinu S, Rossi G, Giustini L, Latini L, Valeri N, Cellerino R (2008) KIT and PDGFRalpha mutations in 104 patients with gastrointestinal stromal tumors (GISTs): a population-based study. Ann Oncol 19: 706–710
Braggio DA, Braggio E, Small IA, Bacchi CE, Lopes LF, Valadao M, Portella S, Romano S, Guimaraes DP, Ferreira CG (2008) The profile of platelet-derived growth factor receptor alpha (PDGFRA) gene alterations in GIST patients (pts) from Brazil. J Clin Oncol (Meeting Abstracts) 26: 10561
Corless CL, Schroeder A, Griffith D, Town A, McGreevey L, Harrell P, Shiraga S, Bainbridge T, Morich J, Heinrich MC (2005) PDGFRA mutations in gastrointestinal stromal tumors: frequency, spectrum and in vitro sensitivity to imatinib. J Clin Oncol 23: 5357–5364
Debiec-Rychter M, Sciot R, Le CA, Schlemmer M, Hohenberger P, van Oosterom AT, Blay JY, Leyvraz S, Stul M, Casali PG, Zalcberg J, Verweij J, Van GM, Hagemeijer A, Judson I (2006) KIT mutations and dose selection for imatinib in patients with advanced gastrointestinal stromal tumours. Eur J Cancer 42: 1093–1103
Dematteo RP, Ballman KV, Antonescu CR, Maki RG, Pisters PW, Demetri GD, Blackstein ME, Blanke CD, von MM, Brennan MF, Patel S, McCarter MD, Polikoff JA, Tan BR, Owzar K (2009) Adjuvant imatinib mesylate after resection of localised, primary gastrointestinal stromal tumour: a randomised, double-blind, placebo-controlled trial. Lancet 373: 1097–1104
Dematteo RP, Gold JS, Saran L, Gonen M, Liau KH, Maki RG, Singer S, Besmer P, Brennan MF, Antonescu CR (2008) Tumor mitotic rate, size, and location independently predict recurrence after resection of primary gastrointestinal stromal tumor (GIST). Cancer 112: 608–615
Du S, Zhou Y, Shi Y, Fu H, Zhao G (2008) The analysis of status and clinical implication of KIT and PDGFRa mutations in gastrointestinal stromal tumor (GIST). J Clin Oncol (Meeting Abstracts) 26: 10562
Emile JF, Brahimi S, Coindre JM, Monges G, Scoazec JY, Blay JY, Bouche O, Agamah E (2009) Molecular epidemiology of GISTs: incidence of PDGFRA and KIT exon 9 mutations in the large French population-based study molecGIST. J Clin Oncol (Meeting Abstracts) 27: 10535
Fletcher CD, Berman JJ, Corless C, Gorstein F, Lasota J, Longley BJ, Miettinen M, O’Leary TJ, Remotti H, Rubin BP, Shmookler B, Sobin LH, Weiss SW (2002) Diagnosis of gastrointestinal stromal tumors: a consensus approach. Hum Pathol 33: 459–465
Goettsch WG, Bos SD, Breekveldt-Postma N, Casparie M, Herings RM, Hogendoorn PC (2005) Incidence of gastrointestinal stromal tumours is underestimated: results of a nation-wide study. Eur J Cancer 41: 2868–2872
Gronchi A, Judson I, Nishida T, Poveda A, Martin J, Reichardt P, Casali PG, Cesne AL, Hohenberger P, Blay JY (2009) Adjuvant treatment of GIST with imatinib: solid ground or still quicksand? A comment on behalf of the EORTC Soft Tissue and Bone Sarcoma Group, the Italian Sarcoma Group, the NCRI Sarcoma Clinical Studies Group (UK), the Japanese Study Group on GIST, the French Sarcoma Group and the Spanish Sarcoma Group (GEIS). Eur J Cancer 45: 1103–1106
Heinrich MC, Corless CL, Demetri GD, Blanke CD, von MM, Joensuu H, McGreevey LS, Chen CJ, Van den Abbeele AD, Druker BJ, Kiese B, Eisenberg B, Roberts PJ, Singer S, Fletcher CD, Silberman S, Dimitrijevic S, Fletcher JA (2003a) Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol 21: 4342–4349
Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A, Demetri GD, Fletcher CD, Fletcher JA (2003b) PDGFRA activating mutations in gastrointestinal stromal tumors. Science 299: 708–710
Heinrich MC, Owzar K, Corless CL, Hollis D, Borden EC, Fletcher CD, Ryan CW, von MM, Blanke CD, Rankin C, Benjamin RS, Bramwell VH, Demetri GD, Bertagnolli MM, Fletcher JA (2008) Correlation of kinase genotype and clinical outcome in the North American Intergroup Phase III Trial of imatinib mesylate for treatment of advanced gastrointestinal stromal tumor: CALGB 150105 Study by Cancer and Leukemia Group B and Southwest Oncology Group. J Clin Oncol 26: 5360–5367
Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, Muhammad TG, Matsuzawa Y, Kanakura Y, Shinomura Y, Kitamura Y (1998) Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science 279: 577–580
Kawanowa K, Sakuma Y, Sakurai S, Hishima T, Iwasaki Y, Saito K, Hosoya Y, Nakajima T, Funata N (2006) High incidence of microscopic gastrointestinal stromal tumors in the stomach. Hum Pathol 37: 1527–1535
Lasota J, Dansonka-Mieszkowska A, Sobin LH, Miettinen M (2004) A great majority of GISTs with PDGFRA mutations represent gastric tumors of low or no malignant potential. Lab Invest 84: 874–883
Lurkin A, Ducimetiere F, Ranchere VD, Decouvelaere AV, Cellier D, Gilly FN, Salameire D, Biron P, de Laroche G, Blay JY, Ray-Coquard I (2010) Epidemiological evaluation of concordance between initial diagnosis and central pathology review in a comprehensive and prospective series of sarcoma patients in the Rhone-Alpes region. BMC Cancer 10: 150
Martin J, Poveda A, Llombart-Bosch A, Ramos R, Lopez-Guerrero JA, Garcia del MJ, Maurel J, Calabuig S, Gutierrez A, Gonzalez de Sande JL, Martinez J, De JA, Lainez N, Losa F, Alija V, Escudero P, Casado A, Garcia P, Blanco R, Buesa JM (2005) Deletions affecting codons 557–558 of the c-KIT gene indicate a poor prognosis in patients with completely resected gastrointestinal stromal tumors: a study by the Spanish Group for Sarcoma Research (GEIS). J Clin Oncol 23: 6190–6198
Miettinen M, Lasota J (2006) Gastrointestinal stromal tumors: pathology and prognosis at different sites. Semin Diagn Pathol 23: 70–83
Monges G, Coindre J, Scoazec J, Bouvier A, Blay J, Loria-Kanza Y, Mathieu-Boue A, Bisot-Locard S (2007) Incidence of gastrointestinal stromal tumors (GISTs) in France: results of the PROGIST survey conducted among pathologists. J Clin Oncol (Meeting Abstracts) 25: 10047
Nilsson B, Bumming P, Meis-Kindblom JM, Oden A, Dortok A, Gustavsson B, Sablinska K, Kindblom LG (2005) Gastrointestinal stromal tumors: the incidence, prevalence, clinical course, and prognostication in the preimatinib mesylate era – a population-based study in western Sweden. Cancer 103: 821–829
Van Glabbeke MM, Owzar K, Rankin C, Simes J, Crowley J, GIST Meta-analysis Group (2007) Comparison of two doses of imatinib for the treatment of unresectable or metastatic gastrointestinal stromal tumors (GIST): a meta-analysis based on 1640 patients (pts). J Clin Oncol (Meeting Abstracts) 25: 10004
Acknowledgements
We thank Mrs Sylvie Baldassini for her precious collaboration in the assessment of KIT and PDGFRA mutations. We also thank all the pathologists from the Rhône-Alpes region who participated in this study: Prof. F Berger, Prof. E Brambilla, Prof. M Devouassoux, Prof. D Seigneurin, Prof. F Thivolet-Bejui and Drs C Agard, F Alliasmontmayeur, R Angonin, M Augros Monavon, B Balme, B Bancel, R Barnoud, N Ben-Lagha, L Bensaadi, N Berger Dutrieux, I Beschet, F Billard, V Blanc, N Bottero, J Bourloux, J Boutonnat, R Bouvier, C Bozon, B Bringeon, A Buenemd, M Buyck-Mabrut, B Cantero, C Cavailles, L Chalabreysse, P Chalabreysse, M-L Chambonniere, J Chanoz, G Chanoz-Poulard, M Chevalier, B Chouvet, A Ciapa, C Clarte-Tourner, A Clemenson, S Collardeaufrachon, A Corrand-Faure, L Corsois, F Crozes, I Cruel, R Dardelin, C David, M Decaussin, J-F Denier, J Depardon-Dolce, P Derolland, B Descombes-Thivolet, A Dieny, F Dijoud, C Donne, A-V Donsbeck, J-P Donzel, C Douchet, J Dumollard, A Economides, N Elbaz, B Fabre, M Faisant, C Faure, M Faysse, P Felman, C Feutry, M Ffrench, M Fior-Golzan, L Frappart, F Gasnier, A Gentil-Perret, A Glehen, W Godard, J Godeneche, C Gouarderes, F Gouzygrosjean, A Griot, C Guillaubey, C Guillaud, C Herve, V Hervieu, S Isaac-Pinet, L Istier, M Jouffre-Cottier, A Jouvet, J Kanitakis, P Kermanac’h, A Khaddage, J-F Knopf, M-H Koeb, M Labadie, B Lamouliatte, S Lantuejoul, I Laurent, C Lauro-Colleaux, M-H Laverriere, F Le Breton, F Le Marc’hadour, P Lucht-Versini, B Mac Gregor, J-P Machayekhi, M Maisonneuve, H Martin, F Megelechevallier, P Mesguich, D Meyronet, I Morand-Dusserre, J-L Morcillo, F Morel, A Morens, B Muller, C Muller, M Ney, M Neyra, B Pasquier, D Pasquier, C Paulin, M Peoc’h, G Perrot, J Pialat, E Piaton, N Picchetti, N Pienel, P Pocachard, G Pugens, I Remy, J Richard, V Rouault-Plantaz, J-J Roux, M-G Roux-Gilly, L Saint-Genis, G Saint-Pierre, D Salameire, M Salle, C Salon, F Serain, L Siche, M-S Soubeyrand, N Streichenberger, N Sturm, Y Suignard, P Terdjman, S Vancina, B Vaunois, A Vercherin, F Vittetat, M Vochk-Bonnet, L Zappatini.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Cassier, P., Ducimetière, F., Lurkin, A. et al. A prospective epidemiological study of new incident GISTs during two consecutive years in Rhône Alpes region: incidence and molecular distribution of GIST in a European region. Br J Cancer 103, 165–170 (2010). https://doi.org/10.1038/sj.bjc.6605743
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6605743
Keywords
This article is cited by
-
A New Online Dynamic Nomogram: Construction and Validation of a Predictive Model for Distant Metastasis Risk and Prognosis in Patients with Gastrointestinal Stromal Tumors
Journal of Gastrointestinal Surgery (2023)
-
Gastrointestinal stromal tumours
Nature Reviews Disease Primers (2021)
-
Nationwide evaluation of mutation-tailored treatment of gastrointestinal stromal tumors in daily clinical practice
Gastric Cancer (2021)
-
Preoperative adjuvant therapy for locally advanced and recurrent/metastatic gastrointestinal stromal tumors: a retrospective study
World Journal of Surgical Oncology (2020)
-
Imatinib in combination with phosphoinositol kinase inhibitor buparlisib in patients with gastrointestinal stromal tumour who failed prior therapy with imatinib and sunitinib: a Phase 1b, multicentre study
British Journal of Cancer (2020)