Abstract
Carcinogenesis is an evolutionary process that establishes the ‘hallmarks of cancer’ by natural selection of cell clones that have acquired advantageous heritable characteristics. Evolutionary adaptation has also been proposed as a mechanism that promotes drug resistance during systemic cancer therapy. This review summarises the evidence for the evolution of resistance to cytotoxic and targeted anti-cancer drugs according to Darwinian models and highlights the roles of genomic instability and high intra-tumour genetic heterogeneity as major accelerators of this evolutionary process. Clinical implications and strategies that may prevent the evolution of resistance or target the origins of genetic heterogeneity are discussed. New technologies to measure intra-tumour heterogeneity and translational research on serial biopsies of cancer lesions during and after therapeutic intervention are identified as key areas to further the understanding of determinants and mechanisms of the evolution of drug resistance.
Similar content being viewed by others
Main
‘It may be said that natural selection is daily and hourly scrutinising, throughout the world, every variation, even the slightest; rejecting that which is bad, preserving and adding up all that is good; silently and insensibly working, wherever and whenever opportunity offers, at the improvement of each organic being in relation to its organic and inorganic conditions of life.’
Charles Darwin—On the Origin of Species by Means of Natural Selection, 1859
Charles Darwin proposed that natural selection is the mechanism that leads to evolutionary change of species over time. Cancer was first described as an evolutionary process by Nowell (Nowell, 1976), who hypothesised that natural selection occurs in tumours in the form of clonal selection leading to constant evolutionary change and possibly to drug resistance. Evolutionary processes in cancer development and progression have since been studied, which has allowed remarkable insights into the biology of cancer (for summary, see Graves, 2000). In contrast, our understanding of the contribution of evolutionary processes to the development of drug resistance is still in its infancy and, with the major exception of Goldie and Coldman's principles to prevent the evolution of drug resistance through the of use combination chemotherapy at the earliest possible time (Goldie and Coldman, 1984), what is known has hardly translated into clinical strategies to improve the outcome of cancer therapy. This review summarises current evidence for the role of Darwinian evolution of tumours during drug treatment and derives clinical strategies that may help to elucidate this process in detail and ultimately prevent tumour acquisition of drug resistance.
Evolutionary ground rules
Three prerequisites are necessary and sufficient for evolution by natural selection to occur:
-
i)
Individual variations exist in the population
-
ii)
These variations are heritable
-
iii)
Variations in individuals lead to differential survival and reproduction
Evolution by natural selection is the causal consequence because individuals with favourable characteristics are positively selected based on their reproductive advantage, whereas less well-adapted individuals are eliminated from the population through negative selection. On the basis of these principles, the main determinant of the ability of a population to evolve is the extent of heritable variation within the population.
Tumour somatic mutations and genetic intra-tumour heterogeneity
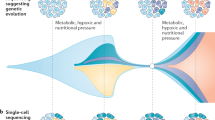
Cancer is caused by DNA and epigenetic alterations and usually arises as a clonal growth from a single founder cell (Fialkow, 1979). Insights into the frequency and pattern of somatic mutations across whole cancer genomes became available recently through the advent of next-generation sequencing technology. For example, sequencing of all protein coding genes in several solid tumours, including glioblastomas, colorectal, pancreatic and breast cancers, revealed approximately 40–80 mutations per tumour, which alter protein sequence (reviewed in Fox et al, 2009). Importantly, the sequencing techniques applied in these studies were not optimised to detect mutations that were only present in a small fraction of tumour cells within the sequenced samples. By the time a cancer reaches detection limits, it is composed of billions of malignant cells, all carrying somatic mutations that were present in the founder cell but also additional mutations acquired by generations of daughter cells during tumour progression, which were passed on to their individual clonal progeny. Thus, although most cancers are of monoclonal origin, the expansion of the population size, which occurs after malignant transformation, coupled with the constant acquisition of mutations promotes the diversion into subclones and a dramatic increase in genetic tumour heterogeneity (Figure 1). Somatic mutation analyses of the immunoglobulin heavy gene locus by ultra-deep sequencing in chronic lymphocytic leukaemia detected multiple subclones in most samples, which supports this model (Campbell et al, 2008). Phylogenetic trees constructed from these data furthermore demonstrated ancestral relationships of subclones and provided evidence for Darwinian evolution by positive selection. Genetic heterogeneity has also been detected in many solid tumours (Marusyk and Polyak, 2010) but is probably significantly under-reported in cancer genome sequencing studies because they overlooked rare mutations. Thus, robust data regarding the total number of mutations and subclones in clinically detectable tumours are unavailable but current estimates are as high as several billions (Klein, 2006). As Darwinian evolution is fuelled by this population heterogeneity, the study of the origin and measurements of the extent of genetic heterogeneity are key steps to understand how cancer drug resistance develops. A further obstacle for the interpretation of large-scale somatic mutation analyses is that fitness effects of the vast majority of mutations are unknown. The RNA interference-based functional genomic screening approaches can experimentally test the phenotypic effect of silencing large numbers of genes individually and may support the interpretation of mutation data sets by identifying genes that influence cellular fitness or drug sensitivity.
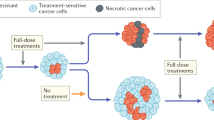
Schematic view of tumour heterogeneity during tumour progression and treatment. Acquired mutations in daughter cells of a single founder cell (left) promote diversion into subclones (different colours reflect different clones). Some new mutations lead to accelerated growth (for example yellow and orange clones). Fitness reducing mutations lead to negative selection (cells with brown cytoplasm). Drug treatment leads to selective survival of a drug resistant clone (pink) and generates an evolutionary bottleneck that reduces genetic heterogeneity transiently. Heterogeneity is re-established rapidly through acquisition of mutations by daughter cells of the resistant clone.
Genomic instability: foundations for genetic heterogeneity in cancer?
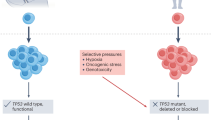
On the basis of the multistep model of carcinogenesis, several mutations need to accumulate for a cancer to arise. Loeb (1991) showed by mathematical modelling that a colorectal cancer with such moderately large numbers of mutations cannot arise within the lifetime of a human under the assumption that mutation rates are similar to those observed in normal tissues. He proposed that cells need to adopt a mutator phenotype, which leads to an elevated rate at which genomes acquire changes, at an early stage of carcinogenesis to acquire the number of mutations necessary for malignant transformation. His results were challenged by Tomlinson et al (1996), who showed that the natural mutation rate is sufficient for carcinogenesis with the proviso that some of these mutations provide a fitness advantage leading to the clonal expansion of a large enough target population to produce the necessary subsequent mutations. The discussion as to whether the natural mutation rate is sufficient to generate the number of mutations observed in advanced cancers or whether genomic instability is mandatory, is still ongoing. Regardless of whether genomic instability is a necessity for carcinogenesis, it has been identified in many advanced tumours and leads to increased genetic intra-tumour heterogeneity (Lengauer et al, 1998). Furthermore, accelerated evolutionary adaptation through genomic instability has been shown by studies linking chromosomal instability (CIN), genomic instability characterised by frequent losses or gains of chromosomes during cell divisions, with the accelerated acquisition of drug resistance phenotypes of cancer cells in vitro (Duesberg et al, 2000) and with early tumour recurrence after drug treatment in animal models (Sotillo et al, 2010). The clinically important concept that increased heterogeneity could accelerate evolutionary adaptation and possibly enhance biological fitness to counter environmental selection pressures could in turn favour the clonal selection of genomically unstable cancer cells by cancer therapies. These potential advantages conferred by genomic instability to the tumour cells must be balanced against the selective disadvantage, which can result from the generation of large numbers of cells with deleterious mutations or tumour lethality conferred by excessive chromosomal instability in animal models (Weaver et al, 2007). Importantly, evolutionary models have revealed that genomic instability can be positively selected on the basis of its proliferation advantage in environments with high DNA damage rates (e.g. during chemotherapy) in which unstable cells avoid cell cycle arrest after DNA damage in contrast to genomically stable cells that are negatively selected because they frequently undergo cell cycle arrest and have a lower proliferation rate (Komarova and Wodarz, 2003).
Thus, it is conceivable that there is an optimal degree of genome instability required to accelerate the development and progression of cancers and that excessive instability is deleterious to the tumour. Results from animal tumour models demonstrating that excessive chromosomal instability may have a tumour-suppressive role leads to the tantalising proposal that exacerbation of tumour genome instability provides an opportunity for therapeutic intervention (Weaver et al, 2007).
Evidence for drug resistance by Darwinian evolution
The harsh clinical reality is that acquired multi-drug resistance almost invariably occurs in advanced and metastatic solid tumours leading to disease progression and eventually death. The following examples highlight how Darwinian evolution driven by intra-tumoural genetic heterogeneity fosters resistance to the selection pressure of systemic cancer therapies. This analysis of drug resistance from a Darwinian perspective should allow better insights into how resistance develops and highlight important strategies to limit tumour acquisition of drug resistance.
Selection conferred by cytotoxic chemotherapy
Inactivating mutations of the MSH6 mismatch repair gene were identified in a sequencing study of recurrent glioblastoma multiforme after treatment with the alkylating agent temozolomide but not in untreated tumours (Hunter et al, 2006). Such mutations confer temozolomide resistance, probably by preventing temozolomide-induced cell death (Yip et al, 2009) and cause a hypermutator phenotype as evidenced by the detection of approximately 200 000 somatic point mutations in MSH6-mutant tumours after temozolomide treatment. The ability to detect this mutational pattern by DNA-sequencing approaches mandates their presence in the majority of cells in the analysed sample. This implies that temozolomide chemotherapy led to positive selection of MSH6-mutant cells and elimination of the majority of daughter clones present in the pre-treatment tumour. This clonal selection of MSH6-mutant cells would be predicted to lead to a transient but dramatic reduction in tumour heterogeneity before the expansion of the resistant clone re-establishes genetic heterogeneity through the acquisition of new mutations (Figure 1). Such an evolutionary bottleneck (Merlo et al, 2006) may provide a therapeutic window of opportunity to target the emerging resistant clone, whereas genetic heterogeneity and hence the potential for drug resistance to evolve is low. The hypermutator state induced by the MSH6 mutation is of great clinical concern as it may facilitate rapid evolution of clones with higher fitness, and thereby contribute to tumour progression and resistance to subsequent lines of treatment.
Inactivating BRCA2 mutations impair the error-free cellular double-strand DNA repair system and carriers of germ line BRCA2 mutations are predisposed to the development of breast, ovarian and other cancers (Rahman and Stratton, 1998). Such cancers are highly sensitive to drugs that induce double-strand breaks such as the DNA-damaging platinum agents and PARP inhibitors that block a complementary DNA repair pathway (Fong et al, 2009). Thus, inactivating BRCA2 mutations cause genomic instability that promotes carcinogenesis but also increase the sensitivity to specific drugs. Secondary mutations of the mutant BRCA2 gene, which restore BRCA2 function and lead to PARP inhibitor resistance have been identified in BRCA2-defective cell lines after they acquired resistance to these drugs (Edwards et al, 2008). The reversal of BRCA2-inactivating point mutations occurred by short deletions that are potentially fostered by BRCA2 dysfunction itself. This may highlight that the increased genetic plasticity induced by genomic instability accelerates the evolution of resistance in these cases.
The rapid acquisition of tumours to non-cross resistant cytotoxics, which the tumour has not yet been exposed to, is a major cause of treatment failure in patients who relapse after chemotherapy (Gottesman et al, 2002). The selection of a single sub-clone harbouring a mutation that confers resistance specifically to the challenging chemotherapy drug does not sufficiently explain how cross-resistance to structurally and mechanistically unrelated drugs arises. Several mechanisms including the expression of multidrug efflux pumps, degree of apoptosis proficiency, and increased activity of drug detoxification mechanisms have been identified as potential mechanism of multidrug resistance (reviewed in Gottesman et al, 2002). Genomic instability is also likely to have a vital role in this process by increasing genetic intra-tumour heterogeneity and fostering the simultaneous presence of multiple drug-resistant clones or the provision of means through which these clones can be developed. The frequent losses and gains of whole chromosomes during cell divisions in CIN cancer cells trigger rapid alterations in gene dosage (Duesberg et al, 2000). In vitro studies have confirmed that CIN cells acquire multi-drug resistance at an accelerated rate compared with diploid cells resulting from the selection pressure influenced by drug exposure that is likely to be ‘catalysed’ by whole chromosome mis-segregation events (Duesberg et al, 2000). Clinical evidence substantiating a role for chromosomal instability in driving multi-drug resistance is still limited, principally because methods to detect differences in tumour karyotype from cell to cell are poorly developed and not suitable to high throughput approaches. However, an RNA expression signature reflecting chromosomal instability has been shown to be associated with poor prognosis in six different cancer types (Carter et al, 2006) and with taxane resistance in ovarian cancers (Swanton, 2009), supporting the hypothesis that CIN is associated with poor prognosis and resistance to therapy. In contrast, CIN ovarian cancers seem to be more likely to respond to platinum agents, indicating that this tumour-specific phenotype might be targetable. The reason for this increased sensitivity is unclear but the inability to tolerate excessive DNA damage analogous to the effect of PARP inhibitors on BRCA mutation carriers (Fong et al, 2009) is a possibility. Therefore, genomic instability phenotypes, exemplified by CIN, increase the likelihood of drug-specific and multidrug resistance mechanisms occurring simultaneously, or as has been suggested ‘a selected phenotype’, namely drug-specific resistance catalysed by a putative gene or gene on a mis-segregated chromosome, is associated with ‘multiple unselected phenotypes’ or multi-drug resistance, conferred by dramatic alterations in gene dosage brought about by the selected whole chromosome mis-segregation event (Duesberg et al, 2000).
Selection conferred by targeted drugs
The BCR–ABL translocation is the characteristic driver mutation in chronic myeloid leukaemia (CML). Targeted treatment with the BCR–ABL inhibitor imatinib leads to complete cytogenetic remissions in a high proportion of patients. The emergence of a subclone of leukaemic cells carrying an imatinib resistance mutation in the BCR–ABL kinase domain is the main cause of relapse in patients treated with imatinib (Gorre et al, 2001). The BCR–ABL mutations T315I and E255K, which both confer imatinib resistance, have been shown to pre-exist in minority sub-populations of CML cells from many treatment naive patients and can be detected by sensitive PCR-based methods (Roche-Lestienne et al, 2002). Both mutations have a higher leukaemogenic potential than the non-mutated BCR–ABL clones. Alternative resistance mutations (Y253H, M351T, F317L and T315A) with a lower leukaemogenic potential than the unmutated BCR–ABL gene have also been identified in imatinib-resistant CML (Skaggs et al, 2006). Thus, the selection of a resistant clone by cancer treatment can influence disease biology and potentially clinical behaviour by selecting for more or less aggressive clones. Nilotinib and dasatinib are second generation BCR–ABL inhibitors specifically designed to retain activity against most imatinib-resistant BCR–ABL mutants (Druker, 2006). Whether such drugs can delay the occurrence of resistance when used in the first line setting by preemptively targeting the resistance mechanism that will most likely evolve is currently being investigated in clinical trials. If this strategy is successful, similarly designed second-generation targeted drugs that inhibit the primary driver mutation but also potential resistance mutations may improve the outcomes of many other cancer types with known targetable pathways. However, several co-existing CML clones carrying different imatinib resistance mutations were identified in a number of patients before treatment who subsequently developed resistance to imatinib (Shah et al, 2002). The occurrence of polyclonal resistance in genetically heterogeneous tumour cell populations is of clinical importance because a single drug may be insufficient to target all resistant BCR–ABL variants. This demonstrates the need to develop specific drugs for various resistance mutations and to tailor them to individual patients based on the profile of resistance clones present in the pre-treatment tumour or emerging during treatment.
The EGF receptor tyrosine kinase inhibitor (TKI) gefitinib has high activity in patients with non-small cell lung cancers that harbour activating somatic mutations of the EGF receptor (Lynch et al, 2004). In contrast, the secondary EGFR mutation T790M causes insensitivity to gefitinib and is detectable in 50% of patients after the acquisition of drug resistance (Kosaka et al, 2006). Analogous to the pre-existence of imatinib resistance mutations in some treatment-naive CML patients, the presence of the T790M mutation was also detected in a minority population of cancer cells in some TKI-naive patients who subsequently develop resistance (Inukai et al, 2006). The presence of resistance mutations before treatment initiation is associated with a significantly shorter response duration and the prevalence of the T790M allele in CTCs has been shown to increase over time during gefitinib treatment, which provides direct evidence for clonal expansion of resistant clones on treatment (Maheswaran et al, 2008). The detection of changes in clonal composition by CTC analyses during drug therapy further demonstrate that real-time tumour genotyping is possible with this technique. Surprisingly, this study also detected the emergence of distinct EGFR-activating mutations from CTCs, which were different from the activating EGFR mutations present in the primary tumours. This is a further example of how drug treatment can shape tumour biology by negatively selecting the initially dominant clone and promoting selection of a resistant clone with a distinct driver mutation.
Darwinian evolution of drug resistance: complexities and limitations
These examples demonstrate the applicability of Darwinian concepts to the development of cancer cell intrinsic drug resistance phenotypes. However, Darwinian evolution results from the complex interactions of heritable phenotypes with multiple environmental and microenvironmental influences that need to be considered in the drug resistance process. For example, the selection pressure resulting from imatinib treatment in CML is modulated by microenvironmental influences as exemplified by studies in CML mouse models that demonstrate that cytokine secretion from the tumour stroma can precipitate imatinib resistance (Williams et al, 2007). Furthermore, re-modelling of the microenvironment through recruitment of bone marrow-derived cells, including macrophages and suppressor cells can establish resistance to anti-angiogenic agents (Shojaei and Ferrara, 2008). It remains unclear whether the acquisition of mutations in cancer cells or co-evolving mutations or epigenetic modification of stromal cells control such microenvironmental changes conferring drug resistance. The current intense study of cancer cell–stroma interactions will undoubtedly provide further insights into these processes.
Furthermore, non-mutational drug resistance mechanisms, illustrated by the survival of naturally treatment resistant cancer stem cells or quiescent cells that are protected from the effects of chemotherapy drugs that target rapidly proliferating cells, are likely to contribute to therapeutic failure and require further intense study.
Finally, CML and non-small cell lung cancers with activating EGF receptor mutations may be exceptional examples due to their dependence on a single driver mutation that can be targeted directly by tyrosine kinase inhibitors. Nonetheless, the discovery of a minority population of cells in newly diagnosed patients with acute lymphoblastic leukaemia, which corresponds to the relapse clone emerging after chemotherapy indicates that pre-existent drug-resistant clones are also relevant for the development of resistance to classic cytotoxic chemotherapy regimens (Mullighan et al, 2008). Further research is necessary to define if Darwinian concepts of drug resistance can be applied to all tumour entities and cancer drugs.
Strategies based on evolutionary theory
Comprehensive sampling techniques and measuring heterogeneity
Single biopsies are likely to be sub-optimal to provide detailed information about the global genomics of heterogeneous tumours. New tumour-sampling techniques, CTC collection or the analysis of circulating tumour DNA, may allow a more comprehensive analysis of the clonal composition of a cancer provided that cancer cells are shed into the circulation proportional to their clonal frequency. These are areas that require further research. Technologies that can detect pre-existing resistance mutations in heterogeneous tumours before treatment initiation may allow individualised drug therapy approaches avoiding the rapid selection of existing resistant clones. Tools to measure and characterise the genetic heterogeneity of tumours and to detect and quantify genomic instability are also poorly developed, which precludes establishing the exact contribution of these phenomena to accelerated tumour progression and the development of drug resistance. The development of technologies to address these issues should be given high priority to improve the currently predominant monoclonal model of advanced cancers with a more realistic model incorporating genetic heterogeneity.
Monitoring dynamic changes of clonal composition to guide drug therapy
Monitoring changes in clonal composition during cancer drug treatment is crucial to further the understanding of the Darwinian evolution of drug resistance. Techniques to perform serial biopsies of tumours and to analyse CTCs or circulating tumour DNA during drug therapy should be further developed for these purposes. This may provide direct evidence of positive and negative clonal selection and the detection of emerging clones carrying drug-resistance mutations may allow early modification of treatment strategies before clinical progression occurs. The adjustment of the therapeutic strategy based on the resistance profile of residual cancer cells after induction treatment has significantly decimated the population size and with it genomic heterogeneity, may also prevent or delay the development of resistance.
Therapeutic strategies exploiting clonal competition
Evolutionary modelling approaches led to the proposal of a new therapeutic strategy that aims to maintain a stable tumour population instead of trying to achieve maximal cell kill (Gatenby et al, 2009). This prevents the elimination of sensitive tumour clones which should, in theory, suppress the growth of therapy resistant clones in a competitive manner. A basic assumption of this strategy is that resistant clones have a lower fitness than sensitive clones because they commit more resources to maintain the resistant phenotype. This strategy has been tested in animal models of ovarian cancer treated with carboplatin but proof of principle in humans is not yet available.
Exploiting genomic instability
Intra-tumour genetic heterogeneity fosters the development of cancer drug resistance by Darwinian evolution and genomic instability mechanisms contribute to such heterogeneity in many tumour types. Thus, genomic heterogeneity may be an attractive therapeutic target to prevent evolution of more aggressive or resistant clones. Synthetic lethality approaches, as exemplified by the use of PARP inhibitors in BRCA-mutant cancers, can specifically target genomically unstable cancer cells by increasing instability to intolerable levels. Similar approaches should be explored to target CIN and other cancer-associated forms of genomic instability. The molecular basis responsible for genome instability mechanisms such as CIN in vivo are still largely unknown and further research to identify genes that promote or sustain CIN should be given high priority to support such developments.
Conclusion
Darwinian evolution contributes to clinical resistance to traditional chemotherapy and targeted drugs. A main driver of evolutionary adaptation during drug treatment is the genetic heterogeneity present in clinically detectable tumours, which is itself determined by population size and fostered by genomic instability. New tools are necessary to study heterogeneity and to analyse changes in heterogeneity and clonal composition during drug treatment. This would allow new insights into these processes and provide the basis to improve therapeutic outcomes based on evolutionary theory and the specific targeting of distinct genomic instability mechanisms.
References
Campbell PJ, Pleasance ED, Stephens PJ, Dicks E, Rance R, Goodhead I, Follows GA, Green AR, Futreal PA, Stratton MR (2008) Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci USA 105: 13081–13086
Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z (2006) A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet 38: 1043–1048
Druker BJ (2006) Circumventing resistance to kinase-inhibitor therapy. N Engl J Med 354: 2594–2596
Duesberg P, Stindl R, Hehlmann R (2000) Explaining the high mutation rates of cancer cells to drug and multidrug resistance by chromosome reassortments that are catalyzed by aneuploidy. Proc Natl Acad Sci USA 97: 14295–14300
Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A (2008) Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451: 1111–1115
Fialkow PJ (1979) Clonal origin of human tumors. Annu Rev Med 30: 135–143
Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS (2009) Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 361: 123–134
Fox EJ, Salk JJ, Loeb LA (2009) Cancer genome sequencing – an interim analysis. Cancer Res 69: 4948–4950
Gatenby RA, Silva AS, Gillies RJ, Frieden BR (2009) Adaptive therapy. Cancer Res 69: 4894–4903
Goldie JH, Coldman AJ (1984) The genetic origin of drug resistance in neoplasms: implications for systemic therapy. Cancer Res 44: 3643–3653
Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL (2001) Clinical resistance to STI-571 cancer therapy caused by BCR–ABL gene mutation or amplification. Science 293: 876–880
Gottesman MM, Fojo T, Bates SE (2002) Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer 2: 48–58
Graves M (2000) Cancer: The Evolutionary Legacy. Oxford University Press: Oxford
Hunter C, Smith R, Cahill DP, Stephens P, Stevens C, Teague J, Greenman C, Edkins S, Bignell G, Davies H, O’Meara S, Parker A, Avis T, Barthorpe S, Brackenbury L, Buck G, Butler A, Clements J, Cole J, Dicks E, Forbes S, Gorton M, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Kosmidou V, Laman R, Lugg R, Menzies A, Perry J, Petty R, Raine K, Richardson D, Shepherd R, Small A, Solomon H, Tofts C, Varian J, West S, Widaa S, Yates A, Easton DF, Riggins G, Roy JE, Levine KK, Mueller W, Batchelor TT, Louis DN, Stratton MR, Futreal PA, Wooster R (2006) A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res 66: 3987–3991
Inukai M, Toyooka S, Ito S, Asano H, Ichihara S, Soh J, Suehisa H, Ouchida M, Aoe K, Aoe M, Kiura K, Shimizu N, Date H (2006) Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Res 66: 7854–7858
Klein CA (2006) Random mutations, selected mutations: a PIN opens the door to new genetic landscapes. Proc Natl Acad Sci USA 103: 18033–18034
Komarova NL, Wodarz D (2003) Evolutionary dynamics of mutator phenotypes in cancer: implications for chemotherapy. Cancer Res 63: 6635–6642
Kosaka T, Yatabe Y, Endoh H, Yoshida K, Hida T, Tsuboi M, Tada H, Kuwano H, Mitsudomi T (2006) Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin Cancer Res 12: 5764–5769
Lengauer C, Kinzler KW, Vogelstein B (1998) Genetic instabilities in human cancers. Nature 396: 643–649
Loeb LA (1991) Mutator phenotype may be required for multistage carcinogenesis. Cancer Res 51: 3075–3079
Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139
Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, Digumarthy S, Muzikansky A, Irimia D, Settleman J, Tompkins RG, Lynch TJ, Toner M, Haber DA (2008) Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 359: 366–377
Marusyk A, Polyak K (2010) Tumor heterogeneity: causes and consequences. Biochim Biophys Acta 1805: 105–117
Merlo LM, Pepper JW, Reid BJ, Maley CC (2006) Cancer as an evolutionary and ecological process. Nat Rev Cancer 6: 924–935
Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, Downing JR (2008) Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science 322: 1377–1380
Nowell PC (1976) The clonal evolution of tumor cell populations. Science 194: 23–28
Rahman N, Stratton MR (1998) The genetics of breast cancer susceptibility. Annu Rev Genet 32: 95–121
Roche-Lestienne C, Soenen-Cornu V, Grardel-Duflos N, Lai JL, Philippe N, Facon T, Fenaux P, Preudhomme C (2002) Several types of mutations of the Abl gene can be found in chronic myeloid leukemia patients resistant to STI571, and they can pre-exist to the onset of treatment. Blood 100: 1014–1018
Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL (2002) Multiple BCR–ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell 2: 117–125
Shojaei F, Ferrara N (2008) Refractoriness to antivascular endothelial growth factor treatment: role of myeloid cells. Cancer Res 68: 5501–5504
Skaggs BJ, Gorre ME, Ryvkin A, Burgess MR, Xie Y, Han Y, Komisopoulou E, Brown LM, Loo JA, Landaw EM, Sawyers CL, Graeber TG (2006) Phosphorylation of the ATP-binding loop directs oncogenicity of drug-resistant BCR–ABL mutants. Proc Natl Acad Sci USA 103: 19466–19471
Sotillo R, Schvartzman JM, Socci ND, Benezra R (2010) Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature 464: 436–440
Swanton C (2009) Chromosomal instability determines taxane response. Proc Natl Acad Sci USA 106: 8671–8676
Tomlinson IP, Novelli MR, Bodmer WF (1996) The mutation rate and cancer. Proc Natl Acad Sci USA 93: 14800–14803
Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW (2007) Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11: 25–36
Williams RT, den Besten W, Sherr CJ (2007) Cytokine-dependent imatinib resistance in mouse BCR–ABL+, Arf-null lymphoblastic leukemia. Genes Dev 21: 2283–2287
Yip S, Miao J, Cahill DP, Iafrate AJ, Aldape K, Nutt CL, Louis DN (2009) MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res 15: 4622–4629
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Gerlinger, M., Swanton, C. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br J Cancer 103, 1139–1143 (2010). https://doi.org/10.1038/sj.bjc.6605912
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6605912
Keywords
This article is cited by
-
Prognostic value of textural features obtained from F-fluorodeoxyglucose (F-18 FDG) positron emission tomography/computed tomography (PET/CT) in patients with locally advanced cervical cancer undergoing concurrent chemoradiotherapy
Annals of Nuclear Medicine (2023)
-
Acquired ABC-transporter overexpression in cancer cells: transcriptional induction or Darwinian selection?
Naunyn-Schmiedeberg's Archives of Pharmacology (2021)
-
A preliminary PET radiomics study of brain metastases using a fully automatic segmentation method
BMC Bioinformatics (2020)
-
Usefulness of a novel device to divide core needle biopsy specimens in a spatially matched fashion
Scientific Reports (2020)
-
LPAR1 regulates the development of intratumoral heterogeneity in ovarian serous cystadenocarcinoma by activating the PI3K/AKT signaling pathway
Cancer Cell International (2019)