Abstract
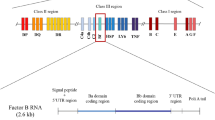
The proteases of the lectin pathway of complement activation, MASP-1 and MASP-2, are encoded by two separate genes. The MASP1 gene is located on chromosome 3q27, the MASP2 gene on chromosome 1p36.23–31. The genes for the classical complement activation pathway proteases, C1r and C1s, are linked on chromosome 12p13. We have shown that the MASP2 gene encodes two gene products, the 76 kDa MASP-2 serine protease and a plasma protein of 19 kDa, termed MAp19 or sMAP. Both gene products are components of the lectin pathway activation complex. We present the complete primary structure of the human MASP2 gene and the tight cluster that this locus forms with non-complement genes. A comparison of the MASP2 gene with the previously characterised C1s gene revealed identical positions of introns separating orthologous coding sequences, underlining the hypothesis that the C1s and MASP2 genes arose by exon shuffling from one ancestral gene.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Whaley K, Schwaeble W Complement and complement deficiencies Semin Liver Dis 1997 17 297–310
Lu JH, Thiel S, Wiedemann H, Timpl R, Reid KB Binding of the pentamer/hexamer forms of mannan-binding protein to zymosan activates the proenzyme C1r2C1s2 complex, of the classical pathway of complement, without involvement of C1q J Immunol 1990 144 2287–2294
Matsushita M, Endo Y, Fujita T Complement-activating complex of ficolin and mannose-binding lectin-associated serine protease J Immunol 2000 164 2281–2284
Hansen S, Holmskov U Structural aspects of collectins and receptors for collectins Immunobiology 1998 199 165–189
Hoppe H, Reid K Collectins--soluble proteins containing collagenous regions and lectin domains--and their roles in innate immunity Protein Sci 1994 3 1143–1158
Thiel S, Petersen S, Vorup-Jensen T et al Interaction of C1q and Mannan-Binding Lectin (MBL) with C1r, C1s, MBL-associated serine proteases 1 and 2, and the MBL-associated protein MAp19 J Immunol 2000 165 878–887
Vorup-Jensen T, Petersen S, Hansen A et al Distinct pathways of mannan-binding lectin (MBL)- and C1 complex autoactivation revealed by reconstitution of MBL with recombinant MBL-associated serine protease-2 J Immunol 2000 165 2093–2100
Matsushita M, Thiel S, Jensenius J, Terai I, Fujita T Proteolytic activities of two types of mannose-binding lectin-associatedserine protease J Immunol 2000 165 2637–2642
Takahashi M, Miura S, Ishii N et al An essential role of MASP-1 in activation of the lectin pathway Immunopharmacology 2000 49 3 (Abstract 003)
Stover C, Thiel S, Thelen M, Lynch N, Vorup-Jensen T, Jensenius J, Schwaeble W Two constituents of the initiation complex of the Mannan-Binding Lectin activation pathway of complement are encoded by a single structural gene J Immunol 1999 162 3481–3490
Takahashi M, Endo Y, Fujita T, Matsushita M A truncated form of mannose-binding lectin-associated serine protease (MASP)-2 expressed by alternative polyadenylation is a component of the lectin complement pathway Int Immunol 1999 11 859–863
Stover C,Thiel S, Lynch N, Schwaeble WThe rat and mouse homologues of MASP-2 and MAp19, components of the mannan-binding lectin activation pathway of complement J Immunol 1999 63 6848–6859
Stover C,Schwaeble W, Lynch N, Thiel S, Speicher MAssignment of the gene encoding Mannan-Binding Lectin-associated serine protease-2 (MASP-2) to human chromosome 1p36.2–3 by in situ hybridization and somatic cell hybrid analysis Cytogenet Cell Genet 1999 84 148–149
Cooke C, Alwine J The cap and the 3′ splice site similarly affect polyadenylation efficiency Mol Cell Biol 1996 16 2579–2584
Thiel S, Vorup-Jensen T, Stover C et al A second serine protease associated with mannan-binding lectin that activates complement Nature 1997 386 506–510
Endo Y, Takahashi M, Nakao M et al Two lineages of mannose-binding lectin-associated serine protease (MASP) in vertebrates J Immunol 1998 161 4924–4930
Endo Y, Sato T, Matsushita M, Fujita T Exon structure of the gene encoding the human mannose-binding protein-associated serine protease light chain: comparison with complement C1r and C1s genes Int Immunol 1996 8 1355–1358
Reese MG, Eeckman FH Novel Neural Network Algorithms for Improved Eukaryotic Promoter Site Recognition. The 7th international Genome sequencing and analysis conference, Hilton Head Island, South Carolina, 16–20 September 1995
Bucher P Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences J Mol Biol 1990 212 563–578
Prestridge DS SIGNAL SCAN: A computer program that scans DNA sequences for eukaryotic transcriptional elements CABIOS 1991 7 203–206
Ghosh D New developments of a transcription factors database Trends Biochem Sci 1991 16 445–447
Kunz, J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva N, Hall M Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression Cell 1993 73 585–596
Cruz M, Cavallo L, Gorlach J et al Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in cryptococcus neoformans Mol Cell Biol 1999 19 4101–4112
Sabatini D, Erdjument-Bromage H, Lui M, Tempst P, Snyder S RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs Cell 1994 78 35–43
Brown E, Albers M, Shin T et al A mammalian protein targeted by G1-arresting rapamycin-receptor complex Nature 1994 369 756–758
Vilella-Bach M, Nuzzi P, Fang Y, Chen J The FKBP12-rapamycin-binding domain is required for FKBP12-rapamycin-associated protein kinase activity and G1 progression J Biol Chem 1999 274 4266–4272
Onyango P, Lubyova B, Gardellin P, Kurzbauer R, Weith A Molecular cloning and expression analysis of five novel genes in chromosome 1p36 Genomics 1998 50 187–198
Lench N, Macadam R, Markham A The human gene encoding FKBP-rapamycin associated protein (FRAP) maps to chromosomal band 1p36.2 Hum Genet 1997 99 547–549
Moore P, Rosen C, Carter K Assignment of the human FKBP12-rapamycin-associated protein (FRAP) gene to chromosome 1p36 by fluorescence in situ hybridization Genomics 1996 33 331–332
Gelpi C, Algueró A, Angeles Martinez M, Vidal S, Juarez C, Rodriguez-Sanchez J Identification of protein components reactive with anti-PM/Scl autoantibodies Clin Exp Immunol 1990 81 59–64
Ge Q, Wu Y, Trieu E, Targoff I Analysis of the specificity of anti-PM-Scl autoantibodies Arthritis Rheum 1994 37 1445–1452
Briggs M, Burkard K, Butler J Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation J Biol Chem 1998 273 13255–13263
Bliskovski V, Liddell R, Ramsay E, Miller M, Mock B Structure and localization of mouse Pmscl1 and Pmscl2 genes Genomics 2000 64 106–110
Ou SH, Wu F, Harrich D, Garcia-Martinez L, Gaynor R Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs J Virol 1995 69 3584–3596
Peek R, van Gelderen BE, Bruinenberg M, Kijlstra A Molecular cloning of a new angiopoietin-like factor from the human cornea Invest Ophthalmol Vis Sci 1998 39 1782–1788
Tosi M, Duponchel C, Meo T, Couture-Tosi E Complement genes C1r and C1s feature an intronless protease domain closely related to haptoglobin J Mol Biol 1989 208 709–714
Kusumoto H, Hirosawa S, Salier J, Hagen F, Kurachi K Human genes for complement C1r and C1s in a close tail-to-tail arrangement Proc Natl Acad Sci 1988 85 7307–7311
Tosi M, Duponchel C, Meo T, Julier C Complete cDNA sequence of human complement C1s and close physical linkage of the homologous genes C1s and C1r Biochemistry 1987 26 8516–8524
Van Cong N, Tosi M, Gross M et al Assignment of the complement serine protease genes C1r and C1s to chromosome 12 region 12p13 Hum Genet 1988 78 363–368
Dang Q, DiCera E Residue 225 determines the Na+-induced allosteric regulation of catalytic activity in serine proteases Proc Natl Acad Sci USA 1996 93 10653–10656
Lawson P, Reid K A novel PCR-based technique using expressed sequence tags and gene homology for murine genetic mapping: localization of the complement genes Int Immunology 1999 12 231–240
Irwin D Evolution of an active-site codon in serine proteases Nature 1988 336 429–430
Long M, Rosenberg C, Gilbert W Intron phase correlations and the evolution of the intron/exon structure of genes Proc Natl Acad Sci USA 1995 92 12495–12499
Gilbert W, de Souza S, Long M Origin of genes Proc Natl Acad Sci USA 1997 94 7698–7703
De Souza S, Long M, Klein R, Roy S, Lin S, Gilbert W Toward a resolution of the introns early/late debate: only phase zero introns are correlated with the structure of ancient proteins Proc Natl Acad Sci 1998 95 5094–5099
Saccone S, De Sario A, Della Valle G, Bernardi G The highest gene concentrations in the human genome are in telomeric bands of metaphase chromosomes Proc Natl Acad Sci 1992 89 4913–4917
van der Drift P, Chan A, Zehetner G, Westerveld A, Versteeg R Multiple MSP pseudogenes in a local repeat cluster on 1p36 2: an expanding genomic graveyard? Genomics 1999 62 74–81
Romani M, Baldini A, Volpi E, Casciano I, Nobile C, Muresu R, Siniscalco M Concurrent mapping of an adeovirus 5/SV40 integration site and the U1 snRNA cluster (RNU1) within 400 kb of the chromosome 1p36.1 Cytogenet Cell Genet 1994 67 37–40
White P, Maris J, Beltinger C et al A region of consistent deletion in neuroblastoma maps within human chromosome 1p36.2–36.3 Genetics 1995 92 5520–5524
Guinto ER, Caccia S, Rose T, Futterer K, Waksman G, Di Cera E Unexpected crucial role of residue 225 in serine proteases Proc Natl Acad Sci USA 1999 96 1852–1857
Emi M, Nakamura Y, Ogawa M et al Cloning, characterization and nucleotide sequences of two cDNAs encoding human pancreatic trypsinogens Gene 1986 41 305–310
Saier M Families of transmembrane sugar transport proteins Mol Microbiol 2000 35 699–710
Jentsch TJ, Friedrich T, Schriever A, Yamada H The CLC chloride channel family Pflugers Arch 1999 437 783–795
Fuchs P, Strehl S, Dworzak M, Himmler A, Ambros P Structure of the human TNF receptor 1 (p60) gene (TNFR1) and localization to chromosome 12p13 Genomics 1992 13 219–224
Kemper O, Derre J, Cherif D, Engelmann H, Wallach D, Berger R The gene for the type II (p75) tumor necrosis factor receptor (TNF-RII) is localized on band 1p36.2-p36.3 Hum Genet 1991 87 623–624
Chirgwin J, Przybyla A, MacDonald R, Rutter W Isolation of biologiocally active ribonucleic acid from sources enriched in ribonucleases Biochemistry 1979 18 5294–5299
Acknowledgements
We wish to thank The International Institute for the Advancement of Medicine, University of Leicester for kindly providing the non-tumorous human liver specimen used in this study. We acknowledge the expertise of Cathy Summer (Research Genetics, Huntsville, Alabama, USA) in isolating the MASP-2 specific human BAC clone RP11–99P18 (AJ300188). The authors also wish to thank the Sanger Mapping and Sequence groups for the sequence production of RP4–635E18 (AL109811). Guelnihal Yueksekdag is acknowledged for excellent technical support. We especially thank Dr Jim Kaufman (Institute of Animal Health, Compton, England) for his inspiring comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work was supported by the Wellcome Trust, the German Research Foundation (Deutsche Forschungsgemeinschaft), the Japanese Society for the Promotion of Science, and the EMDO Foundation, Switzerland.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession numbers AL109811, AB033742, AJ297949, AJ299718, and AJ300188.
Rights and permissions
About this article
Cite this article
Stover, C., Endo, Y., Takahashi, M. et al. The human gene for mannan-binding lectin-associated serine protease-2 (MASP-2), the effector component of the lectin route of complement activation, is part of a tightly linked gene cluster on chromosome 1p36.2–3 . Genes Immun 2, 119–127 (2001). https://doi.org/10.1038/sj.gene.6363745
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.gene.6363745
Keywords
This article is cited by
-
Effects of hMASP-2 on the formation of BCG infection-induced granuloma in the lungs of BALB/c mice
Scientific Reports (2017)
-
Mannose-Binding Lectin (MBL) and MBL-associated serine protease-2 (MASP-2) in women with malignant and benign ovarian tumours
Cancer Immunology, Immunotherapy (2014)
-
Deficiency of mannan-binding lectin associated serine protease-2 due to missense polymorphisms
Genes & Immunity (2007)
-
Mannan-binding-lectin-associated serine proteases, characteristics and disease associations
Springer Seminars in Immunopathology (2005)
-
Organization of the MASP2 locus and its expression profile in mouse and rat
Mammalian Genome (2004)