Abstract
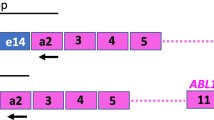
The chromosomal translocation t(8;21)(q22;q22) is one of the most frequent karyotypic aberrations in acute myeloid leukemia (AML) and results in a chimeric fusion transcript AML1/MTG8. Since AML1/MTG8 fusion transcripts remain detectable by RT-PCR in t(8;21) AML patients in long-term hematological remission, quantitative assessment of AML1/MTG8 transcripts is necessary for the monitoring of minimal residual disease (MRD) in these patients. Competitive RT-PCR and recently real-time RT-PCR are increasingly used for detection and quantification of leukemia specific fusion transcripts. For the direct comparison of both methods we cloned a 42 bp DNA fragment into the original AML1/MTG8 sequence. The resulting molecule was used as an internal competitor for our novel competitive nested RT-PCR for AML1/MTG8 and as an external standard for the generation of AML1/MTG8 standard curves in a real-time PCR assay. Using this standard molecule for both PCR techniques, we compared their sensitivity, linearity and reproducibility. Both methods were comparable with regard to all parameters tested irrespective of analyzing serial dilutions of plasmids, cell lines or samples from t(8;21) positive AML patients at different stages of the disease. Therefore, both techniques can be recommended for the monitoring of MRD in these particular AML patients. However, the automatization of the real-time PCR technique offers some technical advantages
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Berger R, Flandrin G, Bernheim A, Le Coniat M, Vecchione D, Pacot A, Derre J, Daniel MT, Valensi F, Sigaux F, Ochoa Noguera ME . Cytogenetic studies on 519 consecutive de novo acute nonlymphocytic leukemias Cancer Genet Cytogenet 1987 29: 9–21
Fenaux P, Preudhomme C, Lai JL, Morel P, Beuscart R, Bauters F . Cytogenetics and their prognostic value in de novo acute myeloid leukemia: a report on 283 cases Br J Haematol 1989 73: 61–67
Erickson P, Gao J, Chang KS, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H . Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt Blood 1992 80: 1825–1831
Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, Kamada N, Ohki M . The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript EMBO J 1993 12: 2715–2721
Tighe JE, Daga A, Calabi F . Translocation breakpoints are clustered on both chromosome 8 and chromosome 21 in the t(8;21) of acute myeloid leukemia Blood 1993 81: 592–596
Bitter MA, Le Beau MM, Rowley JD, Larson RA, Golomb HM, Vardiman JW . Associations between morphology, karyotype and clinical features in myeloid leukemias Hum Pathol 1987 18: 211–225
Bloomfield CD, Lawrence D, Byrd JC, Carrol A, Pettenati MJ, Tantravahi R, Patil SR, Davey FR, Berg DT, Schiffer CA, Arthur DC, Mayer RJ . Frequency of prolonged remission duration after high-dose cytarabine intensification in acute myeloid leukemia varies by cytogenetic subtype Cancer Res 1998 58: 4173–4179
Heil G, Krauter J, Raghavachar A, Hoelzer D, Hossfeld D, Mertelsmann R, Noens L, Schlimok G, Port M, Schwab G, Ganser A . Risk-adapted induction and consolidation therapy including autologous peripheral blood stem cell transplantation (PBSCT) in adult de novo AML patients Blood 1997 90: 508a (Abstr.)
Fenaux P, Lai JL, Preudhomme C, Jouet JP, Deminatti M, Bauters F . Is translocation (8;21) a ‘favorable’ cytogenetic rearrangement in acute myeloid leukemia? Nouv Rev Fr Hematol 1990 32: 179–182
Chang KS, Fan YH, Stass SA, Estey EH, Wang G, Trujillo JM, Erickson P, Drabkin H . Expression of AML1-ETO fusion transcripts and detection of minimal residual disease in t(8;21)-positive acute myeloid leukemia Oncogene 1993 8: 983–988
Downing JR, Head DR, Curcio Brint AM, Hulshof MG, Motroni TA, Raimondi SC, Carroll AJ, Drabkin HA, Willman C, Theil KS, Civin CI, Erickson P . An AML1/ETO fusion transcript is consistently detected by RNA-based polymerase chain reaction in acute myelogenous leukemia containing the (8;21)(q22;q22) translocation Blood 1993 81: 2860–2865
Nucifora G, Larson RA, Rowley JD . Persistence of the 8;21 translocation in patients with acute myeloid leukemia type M2 in long-term remission Blood 1993 82: 712–715
Jurlander J, Caligiuri MA, Ruutu T, Baer MR, Strout MP, Oberkircher AR, Hoffmann L, Ball ED, Frei Lahr DA, Christiansen NP, Block AM, Knuutila S, Herzig GP, Bloomfield CD . Persistence of the AML1/ETO fusion transcript in patients treated with allogeneic bone marrow transplantation for t(8;21) leukemia Blood 1996 88: 2183–2191
Miyamoto T, Nagafuji K, Akashi K, Harada M, Kyo T, Akashi T, Takenaka K, Mizuno S, Gondo H, Okamura T, Dohy H, Niho Y . Persistence of multipotent progenitors expressing AML1/ETO transcripts in long-term remission patients with t(8;21) acute myelogenous leukemia Blood 1996 87: 4789–4796
Elmaagacli AH, Beelen DW, Stockova J, Trzensky S, Kroll M, Schaefer UW, Stein C, Opalka B . Detection of AML1/ETO fusion transcripts in patients with t(8;21) acute myeloid leukemia after allogeneic bone marrow transplantation or peripheral blood progenitor cell transplantation Blood 1997 90: 3230–3231
Kusec R, Laczika K, Knobl P, Friedl J, Greinix H, Kahls P, Linkesch W, Schwarzinger I, Mitterbauer G, Purtscher B, Haas O, Lechner K, Jaeger K . AML 1/ETO fusion mRNA can be detected in remission blood samples of all patients with t(8;21) acute myeloid leukemia after chemotherapy or autologous bone marrow transplantation Leukemia 1994 8: 735–739
Muto A, Mori S, Matsushita H, Awaya N, Ueno H, Takayama N, Okamoto S, Kizaki M, Ikeda Y . Serial quantification of minimal residual disease of t(8;21) acute myelogenous leukaemia with RT-competitive PCR assay Br J Haematol 1996 95: 85–94
Tobal K, Yin JA . Monitoring of minimal residual disease by quantitative reverse transcriptase-polymerase chain reaction for AML 1-MTG8 transcripts in AML-M2 with t(8;21) Blood 1996 88: 3704–3709
Tobal K, Liu Yin JA . Molecular monitoring of minimal residual disease in acute myeloblastic leukemia with t(8;21) by RT-PCR Leuk Lymphoma 1998 31: 115–120
Siebert PD, Larrick JW . Competitive PCR Nature 1992 359: 557–558
Cross NCP, Feng L, Chase A, Bungey J, Hughes TP, Goldman JM . Competitive polymerase chain reaction to estimate the number of of bcr-abl transcripts in chronic myeloid leukemia patients after bone marrow transplantation Blood 1993 82: 1929–1936
Meijerink JP, Smetsers TF, Raemaekers JM, Bogman MJ, De Witte T, Mensink EJ . Quantitation of follicular non-Hodgkin's lymphoma cells carrying t(14;18) by competitive polymerase chain reaction Br J Haematol 1993 84: 250–256
Brisco MJ, Condon J, Hughes E, Neoh SH, Sykes PJ, Seshadri R, Toogood I, Waters K, Tauro G, Ekert H, Morley AA . Outcome prediction in childhood acute lymphoblastic leukaemia by molecular quantification of residual disease at the end of induction Lancet 1994 343: 196–200
Cross NCP . Quantitative PCR techniques and applications Br J Haematol 1995 89: 693–697
Holland PM, Abramson RD, Watson R, Gelfand DH . Detection of specific polymerase chain products by utilizing the 5′ to 3′ exonuclease activity of termus aquaticus DNA polymerase Proc Natl Acad Sci USA 1991 88: 7276–7280
Förster VT . Zwischenmolekulare Energiewanderung und Fluoreszenz Ann Physics (Leipzig) 1948 2: 55–75
Marcucci G, Livak KJ, Bi W, Strout MP, Bloomfield CD, Caligiuri MA . Detection of minimal residual disease in patients with AML1/ETO-associated acute myeloid leukemia using a novel quantitative reverse transcription polymerase chain reaction assay Leukemia 1998 12: 1482–1489
Mensink E, van de Locht A, Schattenberg A, Linders E, Schaap N, Geurts van Kessel A, De Witte T . Quantitation of minimal residual disease in Philadelphia chromosome positive chronic myeloid leukemia using real-time quantitative RT-PCR Br J Haematol 1998 102: 768–774
Luthra R, McBride JA, Cabanillas F, Sarris A . Novel 5′ exonuclease based real time PCR assay for the detection of t(14;18)(q32;q21) in patients with follicular lymphoma Am J Pathol 1998 153: 63–68
Gerard CJ, Olsson K, Ramanathan R, Reading C, Hanania EG . Improved quantitation of minimal residual disease in multiple myeloma using real-time polymerase chain reaction and plasmid-DNA complementarity determinig region III standards Cancer Res 1998 58: 3957–3964
Pongers-Willemse MJ, Verhagen OJ, Tibbe GJ, Wijkhuijs AJ, de Haas V, Roovers E, van der School CE, van Dongen JJ . Real-time quantitative PCR for the detection of minimal residual disease in acute lymphoblastic leukemia using junctional region specific TaqMan probes Leukemia 1998 12: 2006–2014
Gibson UEM, Heid CA, Williams MP . A novel method for real time quantitative RT-PCR Genome Res 1996 6: 995–1001
Heid CA, Stevens J, Livak KJ, Williams MP . Real time quantitative PCR Genome Res 1996 6: 986–994
Asou H, Tashiro S, Hamamoto K, Otsuji A, Kita K, Kamada N . Establishment of a human acute myeloid leukemia cell line (Kasumi-1) with 8;21 chromosome translocation Blood 1991 77: 2031–2036
Lanotte M, Martin-Thouvenin V, Najman S, Balerini P, Valensi F, Berger R . NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3) Blood 1991 77: 1080–1086
Krauter J, Peter W, Pascheberg U, Heinze B, Bergmann L, Hoelzer D, Lübbert M, Schlimok G, Arnold R, Kirchner H, Port M, Ganser A, Heil G . Detection of karyotypic aberrations in acute myeloblastic leukemia: a prospective comparison between PCR/FISH and standard cytogenetics in 140 patients with de novo AML Br J Haematol 1998 103: 72–78
Miller WH, Levine K, DeBlasio A, Frankel SR, Dmitrovsky E, Warrell RP . Detection of minimal residual disease in acute promyelocytic leukemia by a reverse transcription polymerase chain reaction assay for the PML/RAR-alpha fusion mRNA Blood 1993 82: 1689–1694
Miyamoto T, Nagafuji K, Harada M, Eto T, Fujisaki T, Kubota A, Akashi K, Mizuno S, Takenaka K, Kanaji T et al. Quantitative analysis of AML1/ETO transcripts in peripheral blood stem cell harvests from patients with t(8;21) acute myelogenous leukaemia Br J Haematol 1995 91: 132–138
Becker AM, Hahlbrock . K Absolute mRNA quantification using the polymerase chain reaction (PCR): a novel approach by a PCR aided transcript titration assay (PATTY) Nucleic Acids Res 1989 17: 9437–9446
Desjardin LE, Chen Y, Perkins MD, Teixeira L, Cave MD, Eisenach KD . Comparison of the ABI 7700 system (TaqMan) and competitive PCR for quantification of IS6110 DNA in sputum during treatment of tuberculosis J Clin Microbiol 1998 36: 1964–1968
Acknowledgements
This work was supported by the Deutsche Krebshilfe [10-1217-He1] and a grant from the Dr Wilhelm-Kempe-Stiftung. The authors wish to thank Kerstin Görlich, Elvira Lux and Dagmar Reile for their excellent technical assistance.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Wattjes, M., Krauter, J., Nagel, S. et al. Comparison of nested competitive RT-PCR and real-time RT-PCR for the detection and quantification of AML1/MTG8 fusion transcripts in t(8;21) positive acute myelogenous leukemia. Leukemia 14, 329–335 (2000). https://doi.org/10.1038/sj.leu.2401679
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.leu.2401679
Keywords
This article is cited by
-
Molecular detection and targeting of EWSR1 fusion transcripts in soft tissue tumors
Medical Oncology (2013)
-
Prognostic value of real-time quantitative PCR (RQ-PCR) in AML with t(8;21)
Leukemia (2005)
-
Detection of minimal residual disease in hematologic malignancies by real-time quantitative PCR: principles, approaches, and laboratory aspects
Leukemia (2003)
-
Incomplete DJH rearrangements as a novel tumor target for minimal residual disease quantitation in multiple myeloma using real-time PCR
Leukemia (2003)
-
Monitoring of minimal residual disease (MRD) by real-time quantitative reverse transcription PCR (RQ-RT-PCR) in childhood acute myeloid leukemia with AML1/ETO rearrangement
Leukemia (2003)