Abstract
Rab2A, a small GTPase localizing to the endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC), regulates COPI-dependent vesicular transport from the ERGIC. Rab2A knockdown inhibited glucose-stimulated insulin secretion and concomitantly enlarged the ERGIC in insulin-secreting cells. Large aggregates of polyubiquitinated proinsulin accumulated in the cytoplasmic vicinity of a unique large spheroidal ERGIC, designated the LUb-ERGIC. Well-known components of ER-associated degradation (ERAD) also accumulated at the LUb-ERGIC, creating a suitable site for ERAD-mediated protein quality control. Moreover, chronically high glucose levels, which induced the enlargement of the LUb-ERGIC and ubiquitinated protein aggregates, impaired Rab2A activity by promoting dissociation from its effector, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), in response to poly (ADP-ribosyl)ation of GAPDH. The inactivation of Rab2A relieved glucose-induced ER stress and inhibited ER stress-induced apoptosis. Collectively, these results suggest that Rab2A is a pivotal switch that controls whether insulin should be secreted or degraded at the LUb-ERGIC and Rab2A inactivation ensures alleviation of ER stress and cell survival under chronic glucotoxicity.
Similar content being viewed by others
Introduction
The endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC), which is characterized by a 53-kDa membrane protein (ERGIC53) and the COPI subunit β-COP, is a tubular vesicular membrane system that links the rough ER and the Golgi1. The ERGIC functions as the first post-ER sorting station for anterograde and retrograde protein traffic1. One mechanism that regulates COPI-mediated anterograde and retrograde transport from the ERGIC is sorting by the small GTPase Rab through various effector proteins. Two Rab proteins with opposing functions are associated with the ERGIC membranes. Whereas Rab1A is involved in membrane tethering at the ERGIC and cis-Golgi during anterograde transport2, Rab2A acts in conjunction with an atypical kinase C iota/lambda and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) to promote the formation of COPI vesicles that are enriched in recycling proteins3,4. GAPDH is a key glycolytic enzyme as well as an effector of Rab2A and its glycolytic activity is known to be regulated by poly (ADP-ribose) polymerase (PARP)-dependent poly (ADP-ribosyl)ation5,6. However, the function of GAPDH in trafficking does not require its glycolytic activity3.
In addition to the protein sorting function of the ERGIC, recent studies have revealed the following additional and novel functions of this organelle: (i) the ERGIC can capture a wide range of incompletely folded proteins that have eluded the quality control stage in the ER and then enable correction of the protein conformation7,8,9,10,11,12,13,14,15, (ii) the ERGIC is a key membrane source for the LC3 lipidation step of autophagosome biogenesis16. Thus, there is a growing body of evidence to show that the ERGIC itself is involved in the quality control of damaged or unnecessary proteins in cells.
One of the responses by which cells deal with misfolded proteins in the ER is ER-associated degradation (ERAD). ERAD is a sequential reaction in which misfolded proteins are transported from the ER lumen into the cytoplasm across the ER membrane by a multistep process called retrotranslocation and are then degraded in a ubiquitin-proteasome dependent manner17,18. When the production of misfolded proteins exceeds the degradation capacity, the proteins often aggregate and accumulate within the cytoplasm19. Such intracellular aggregates are observed in a number of neurodegenerative diseases including Alzheimer's and Huntington's diseases and are frequently ubiquitinated19,20.
Recently, large aggregates of ubiquitinated proteins have also been reported in pancreatic β cells from Zucker diabetic fatty rats and INS 832/13 β cells that have been treated with prolonged high levels of glucose, as well as in humans with type 2 diabetes21,22. Type 2 diabetes is a disease that results from insulin deficiency and resistance to peripheral insulin action, which leads to a chronic hyperglycemic state23. It is characterized by the eventual dysfunction of pancreatic β cells. In a pre-diabetic state, pancreatic β cells can overcome insulin deficiency by increasing insulin secretion. However, pancreatic β cells gradually become unable to compensate for insulin deficiency because chronic hyperglycemia causes their function to deteriorate23. Under hyperglycemic conditions, especially in pancreatic β cells, both oxidative stress and the high burden of biosynthesis of proinsulin, a precursor of insulin, induces the misfolding of proteins in the endoplasmic reticulum (ER), which results in ER stress23,24.
In the present study, we found that silencing of Rab2A inhibited glucose-stimulated insulin secretion (GSIS) and enlarged the aggregates of polyubiquitinated proinsulin, which were formed in the cytoplasmic vicinity of a unique, large spheroidal ERGIC (LUb-ERGIC), where the well-known ERAD machinery was also concentrated. Moreover, we demonstrated that Rab2A activity was diminished by poly (ADP-ribosyl)ation of GAPDH under high glucose condition. This inhibition resulted in the accumulation of ERAD proteins at the LUb-ERGIC and the relief of ER stress caused by chronically high levels of glucose in MIN6 cells. Our findings raise the possibility that Rab2A is a pivotal regulator that switches the function of the ERGIC from “insulin secretion mode” to “protein quality control mode”, depending on the time to which the cells are exposed to glucose and that inactivation of Rab2A ensures cell survival under conditions of chronic glucotoxicity.
Results
Rab2A knockdown inhibits insulin secretion and induces accumulation of proinsulin and the ERAD machinery in a unique large peripheral ERGIC in MIN6 cells
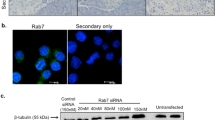
Rab2A is highly involved in COPI-dependent vesicular transport from the ERGIC to the ER and the Golgi25,26. However, the physiological significance of Rab2A function in insulin-secreting cells remains unknown. Consequently, we examined the effect of Rab2A knockdown by RNA interference on GSIS in MIN6 cells, which are derived from mouse pancreatic β cells. Insulin secretion induced by high levels of glucose (25 mM) was inhibited partially in the Rab2A-knockdown cells, in which Rab2A expression was suppressed by more than 90% (Fig. 1a). The decrease of insulin secretion by Rab2A knockdown is expected from previous reports showing that Rab2A controls membrane traffic from the ERGIC to the Golgi25,26. Though the reduction of insulin secretion in Rab2A-knockdown cells appeared to be moderate, this could be explained by the fact that GSIS is dependent not only on the amount of proinsulin newly transported from the ER to the Golgi but also on that of reserved insulin granules in the post-Golgi27, which would not be affected by Rab2A knockdown. Furthermore, fluorescence microscopy using an antibody against ERGIC53, a marker of the ERGIC, revealed that silencing of Rab2A enlarged the ERGIC (data not shown), probably due to the impairment of COPI-dependent vesicle budding from the ERGIC. Interestingly, whereas the diameter of well-known peripheral ERGIC throughout the cell is ~0.8 μm, we concomitantly identified a large, unique ERGIC53-positive spheroidal structure with a diameter of 2–4 μm in the peripheral region of MIN6 cells (Fig. 1b). Rab2A was localized to the large peripheral ERGIC as well as the large irregularly shaped ERGIC53 positive structure near the nucleus, designated as the perinuclear ERGIC (Fig. 1c) and the large peripheral ERGIC existed in both control and Rab2A-knockdown MIN6 cells. Immunofluorescence using an anti-ERGIC53 antibody revealed that the diameter of the large peripheral ERGIC was increased in Rab2A-knockdown cells, ~3.2 μm as compared with ~2.3 μm in control cells (Fig. 1d), which suggests that Rab2A also regulates the morphology of the large peripheral ERGIC through COPI-dependent membrane trafficking. As we expected, proinsulin accumulated in the large peripheral ERGIC after Rab2A knockdown (Fig. 1e).
Rab2A knockdown inhibits glucose-stimulated insulin secretion and induces accumulation of proinsulin in the large peripheral ERGIC.
(a) Upper: MIN6 cells were transfected with scramble or Rab2A siRNA. The cells were lysed and the lysates were immunoblotted with anti-Rab2A antibody, using the GAPDH signal as a loading control. Lower: MIN6 cells were transfected with scramble or Rab2A siRNA. The transfected cells were washed with pre-warmed control KREBS buffer and exposed to 3 mM glucose in control KREBS buffer for 60 min at 37°C. The cells were treated subsequently with 3 mM glucose (Low glu) or 25 mM glucose (High glu) in pre-warmed control KREBS buffer for 60 min at 37°C to induce insulin secretion. The insulin secreted into the buffer was quantified using an AlphaLISA insulin kit (mean ± s.d., n = 6 in each condition). Full-length blots are presented in Supplementary Fig. S10. (b) ERGIC53 and GM130 were subjected to double staining in MIN6 cells. Dotted lines indicate the outline of a cell and arrows indicate the large peripheral ERGIC. Scale bars: 5 μm. (c) ERGIC53 was immunostained with anti-ERGIC53 antibody in MIN6 cells expressing GFP-Rab2A. Images of insets indicate GFP-Rab2A localized to the large peripheral ERGIC. A GFP-Rab2A/ERGIC53-positive compartment in the middle of the cell indicates the perinuclear ERGIC (Arrows). (d) Left: MIN6 cells transfected with scramble or Rab2A siRNA were immunostained with anti-ERGIC53 antibody. Images were derived by three-dimensional reconstruction of the cells (3D reconstruction). Arrows indicate the large peripheral ERGIC in MIN6 cells. Right: the diameter of the large peripheral ERGIC in MIN6 cells transfected with scramble or Rab2A siRNA was determined (mean ± s.d., n = 30 in each condition). Scale bars: 5 μm. (e) Left: MIN6 cells were transfected with scramble or Rab2A siRNA. ERGIC53 and proinsulin were subjected to double staining in the cells. Right: the mean fluorescent intensities of proinsulin at the large peripheral ERGIC in the cells transfected with scramble or Rab2A siRNA were quantified (mean ± s.e., n = 19 in each condition). (c and e) Insets indicate high magnification images of the corresponding white rectangles. Scale bars: 5 μm (low magnification) and 2 μm (high magnification).
Interestingly, immunofluorescence and differential interference contrast (DIC) microscopy revealed that proteins thought to be involved in ERAD, namely subunits of the Sec61 complex (Sec61α1-GFP, HA-Sec61β and γ), Derlin-1, BiP and p9717, were localized to the large peripheral ERGIC (Figs. 2a–2h). In particular, Sec61α1-GFP appeared to be localized solely to the large peripheral ERGIC whereas HA-Sec61β and -γ appeared to be localized also to the other conventional ERGICs. These results indicate that the large peripheral ERGIC might be a suitable site for the effective ERAD of misfolded proteins, while a role of Sec61 complex in ERAD has been controversial28. Consequently, we addressed the role of the large peripheral ERGIC in the quality control of proteins in MIN6 cells.
Proteins associated with the ERAD system are localized to the large peripheral ERGIC.
(a) ERGIC53 was immunostained with anti-ERGIC53 antibody in MIN6 cells expressing Sec61α1-GFP. (b) ERGIC53 was immunostained with anti-ERGIC53 antibody in MIN6 cells expressing HA-Sec61β. (c) ERGIC53 was immunostained with anti-ERGIC53 antibody in MIN6 cells expressing HA-Sec61γ. (d) Derlin-1 was immunostained with anti-Derlin-1 antibody in MIN6 cells expressing GFP-p58. (e) BiP was immunostained with anti-BiP antibody in MIN6 cells expressing GFP-p58. (f) ERGIC53 and p97 were subjected to double staining in MIN6 cells. (g) MIN6 cells expressing GFP-p58, HA-Sec61α1 and Myc-Rab2A were analyzed by immunofluorescence and DIC microscopy. (h) Derlin-1 was immunostained with anti-Derlin-1 antibody in MIN6 cells expressing GFP-p58 and Myc-Rab2A and the cells were analyzed by immunofluorescence and DIC microscopy. (a–h) Insets indicate high magnification images of the corresponding white rectangles. Scale bars: 5 μm (low magnification) and 2 μm (high magnification).
Polyubiquitinated proinsulin aggregates in the immediate vicinity of the LUb-ERGIC
To our surprise, dual-immunofluorescence staining revealed that large aggregates of ubiquitinated proteins were almost always associated with the large peripheral ERGIC, which we refer to as the Large Ubiquitinated protein-associated ERGIC (LUb-ERGIC), in MIN6 cells (Fig. 3a). The large aggregates of ubiquitinated proteins associated with the LUb-ERGIC were also observed in INS-1E cells, which are another insulin-secreting cell line (Supplementary Figs. S1a and S1b). In these cell lines, the LUb-ERGIC was observed under both low and high levels of glucose, but the LUb-ERGIC was not observed in HeLa cells even under high levels of glucose (data not shown). Such aggregates were not observed in any region of the cytoplasm except the LUb-ERGIC (data not shown). γ-tubulin was not localized to the LUb-ERGIC (Supplementary Fig. S1c), which indicates that the aggregates of ubiquitinated proteins at the LUb-ERGIC differ from the well-known aggresome.
Polyubiquitinated proinsulin aggregates at the LUb-ERGIC.
(a) ERGIC53 and ubiquitinated proteins (Ub-proteins) were subjected to double staining in MIN6 cells. Dotted lines indicate the outline of a cell. (b) ERGIC53 and proinsulin were subjected to double staining in MIN6 cells (Intact) or MIN6 cells treated with 1% Triton X-100 alone (1% Tx) or sequentially with 1% Tx, DNase I and 2 M NaCl (LUb-ERGIC enriched fraction) (See Methods and Supplementary Fig. S2 in detail). High mag indicates high magnification images of the white rectangles in the upper hand panels. Arrowhead indicates proinsulin localized to the LUb-ERGIC. Arrows indicate high intensity dots of proinsulin in the LUb-ERGIC. (c) ERGIC53 and proinsulin were subjected to double staining in MIN6 cells expressing ubiquitin tagged with RFP (RFP-Ubiquitin). (d) MIN6 cells were treated with 10 μg/ml CHX or 1 μM MG132 for 2 hr. Left: Ub-proteins and proinsulin in the 1% Tx soluble fraction (1% Tx) were immunoblotted to evaluate the effect of the inhibitors, using the GAPDH signal as a loading control. Right: proinsulin in the 1% Tx/2 M NaCl resistant fraction (LUb-ERGIC enriched fraction) was immunoblotted with anti-proinsulin antibody, using the actin signal as a loading control. Ub1 and Ubn represent mono and polyubiquitinated proinsulin, respectively. (e) MIN6 cells were transfected with scramble or insulin siRNA. The cells were lysed and the lysates were immunoblotted with anti-proinsulin antibody, using the actin signal as a loading control. (f) Left: 36 hr after MIN6 cells were transfected with scramble or insulin siRNA, the cells were exposed to low glucose for 24 hr and then low glucose (Low glu) or high glucose (High glu) for 12 hr and immunostained with antibodies against ERGIC53 and Ub-proteins. Images were derived by three-dimensional reconstruction of the cells (3D reconstruction). Arrows indicate the LUb-ERGIC. Right: the mean fluorescent intensities of Ub-proteins at the LUb-ERGIC were quantified (mean ± s.d., n = 18 in each condition). Indicated P values were calculated with Student's t-test, followed by the Bonferroni correction. Scale bars: 5 μm. (a and c) Insets show high magnification images of the corresponding white rectangles. Scale bars: 5 μm (low magnification) and 2 μm (high magnification). Full-length blots are presented in Supplementary Fig. S10.
Next, we investigated the composition of the aggregates. Under hyperglycemic conditions, the high burden of biosynthesis of proinsulin induces misfolding of proteins in the ER, which results in ER stress23,24. This excessive biosynthesis of proinsulin can easily result in the accumulation of misfolded proteins in the ER, which are retrotranslocated to the cytoplasm by the ERAD machinery and subsequently ubiquitinated29. Thus we assumed that the most promising candidate for the ubiquitinated protein detected in MIN6 cells was proinsulin. To test this, we dual-immunostained ERGIC53 and proinsulin in MIN6 cells and found that a large punctate proinsulin signal was associated with the LUb-ERGIC (Fig. 3b, Intact and High mag). Next, to examine the characteristic features of the ubiquitinated protein aggregates, we adapted a nuclear matrix protein isolation procedure30,31. By immunofluorescence, we confirmed that ubiquitinated protein aggregates and ERGIC53 at the LUb-ERGIC were resistant to treatment with 1% Triton X-100/2 M NaCl (Supplementary Fig. S2 in detail). So, we referred 1% Triton X-100/2 M NaCl resistant fraction as LUb-ERGIC enriched fraction hereafter. Using this method, we determined that the punctate proinsulin signal was resistant to either 1% Triton X-100 or 1% Triton X-100/2 M NaCl (Fig. 3b, 1% Tx and LUb-ERGIC enriched fraction). We also observed that, in cells that expressed ubiquitin tagged with red fluorescence protein (RFP-Ubiquitin), an insoluble aggregate of proinsulin at the LUb-ERGIC colocalized with RFP-Ubiquitin and ERGIC53 (Fig. 3c). Moreover, on western blot analysis of LUb-ERGIC enriched fractions, proinsulin was observed mainly in bands of approximately 10, 17 and 27 kDa (Fig. 3d, right), which probably correspond to proinsulin, proinsulin conjugated with one molecule of ubiquitin and proinsulin conjugated with two molecules of ubiquitin, respectively. The intensity of these bands decreased upon pretreatment with cycloheximide (CHX), an inhibitor of protein synthesis and increased upon treatment with MG132, a proteasome inhibitor (Fig. 3d, right), which indicates that the bands corresponded to the aggregation of polyubiquitinated proinsulin in the LUb-ERGIC because the proteasome specifically degrades polyubiquitinated proteins. We also confirmed that MG132 treatment increased the amount of ubiquitinated protein in extracts from cells treated with 1% Triton X-100, whereas the total amount of proinsulin was unaltered (Fig. 3d, left).
Furthermore, we investigated the effect of high levels of glucose on the amount of ubiquitinated protein in the LUb-ERGIC, because high glucose levels induce the expression of insulin in MIN6 cells23. As shown in Supplementary Fig. S3, exposure of MIN6 cells to a high glucose concentration (25 mM) for 12–18 hr increased the amount of polyubiquitinated protein in the LUb-ERGIC, but exposure to low glucose (5.6 mM) or CHX treatment did not (data not shown). This result is consistent with the previous report that aggregates of ubiquitinated proteins form in INS 832/13 β cells after exposure to high glucose for 8–72 hr21. Furthermore, the induced increase in ubiquitinated aggregates was not observed in insulin-knockdown cells in which the expression of proinsulin was suppressed by more than 90% (Figs. 3e and 3f).
Collectively, these results suggested that one component of the insoluble aggregates at the LUb-ERGIC was polyubiquitinated proinsulin and that the increase in the size of ubiquitinated protein aggregates by high glucose is dependent on synthesis of proinsulin. Moreover, the LUb-ERGIC probably provides a suitable site for the accumulation of the ERAD machinery and its substrates.
The LUb-ERGIC is a long-lived stationary structure that has similar properties to the conventional ERGIC
Next, we examined closely the characteristic features of the LUb-ERGIC in MIN6 cells. Time-lapse microscopy of cells labeled with GFP-p58 (a rat homologue of ERGIC53), which also localized to the ERGIC53-positive LUb-ERGIC (data not shown), revealed that the LUb-ERGIC originated from homotypic fusions among punctate peripheral ERGICs distributed within the cytoplasm and remained stationary (Supplementary Fig. S4a; Supplementary Movie S1). By getting the time lapse imaging for more than 30 min, we found that 98% of the LUb-ERGICs exist for longer than 30 min, indicating that LUb-ERGIC is a stable compartment (Supplementary Fig. S4b). These results strongly suggested that the LUb-ERGIC is neither a fixation artifact nor an incidentally generated compartment. Collectively, the LUb-ERGIC is one of the long-lived stationary ERGICs that are involved in vesicular transport between the ER and the Golgi apparatus, but with a unique morphology. We also observed the vesicular transport of VSVGts045-GFP, a well-known cargo protein of anterograde transport, from the ER to the Golgi apparatus via the LUb-ERGIC as well as via the conventional ERGIC (Supplementary Fig. S4c). Moreover, Yip1A, another marker protein of the ERGIC32, was also localized to the LUb-ERGIC (data not shown). MIN6 cells contained approximately one LUb-ERGIC per cell (Supplementary Fig. S4d) and the LUb-ERGIC was distinguishable from the ER owing to its sensitivity to a high concentration of H89, an inhibitor of protein kinase A (PKA)33 and from the Golgi apparatus due to its insensitivity to brefeldin A (BFA), an inhibitor of ADP-ribosylation factor (ARF)34 (Supplementary Figs. S4d and S4e). Although BFA had no effect on the number of the LUb-ERGIC, the diameter of the LUb-ERGIC increased by BFA treatment (Supplementary Fig. S4e). This result suggests that the enlargement might result from the dissociation of β-COP from the ERGIC membrane by BFA, possibly leading to inhibition of vesicle budding from the ERGIC. Therefore, we concluded that the LUb-ERGIC has similar properties to the conventional ERGIC regarding vesicular transport and is distinguishable from the ER and the Golgi.
Rab2A knockdown accelerates accumulation of the ERAD machinery and aggregation of polyubiquitinated proinsulin in the LUb-ERGIC
Misfolded proteins often escape from the ER retention system and are transported into the ERGIC, where they are potentially subject to quality control7,8,9,10,11,12,13,14,15. Given that Rab2A knockdown inhibited the insulin secretion and induced enlargement of the LUb-ERGIC (Figs. 1a and 1d), we assumed that Rab2A was a promising regulator of the accumulation in the LUb-ERGIC of the ERAD machinery and protein substrates that escape from the ER.
First, we determined by western blotting that the membrane components of ERAD, namely Derlin-1 and Sec61α1 and a cytoplasmic protein, p97, accumulated in the LUb-ERGIC fraction after knockdown of Rab2A (Figs. 4a and 4b). The increased accumulation of these proteins (Derlin-1 and Sec61α1) might be induced by the impairment of their retrieval from the LUb-ERGIC to the ER. This is suggested by the fact that Rab2A knockdown resulted in the dissociation of β-COP, a subunit of COPI, from the LUb-ERGIC membrane (Supplementary Fig. S5a) and also impaired the retrograde transport of ERGIC53 (Supplementary Fig. S5b), which is retrieved from the ERGIC to the ER in a COPI-dependent manner1. Rab2A knockdown also caused the accumulation of proinsulin (one of ERAD substrates at the LUb-ERGIC) at both the ERGIC (Supplementary Fig. S5c) and the LUb-ERGIC (Fig. 1e), decreased the amount of insulin-GFP and insulin secretion (Supplementary Fig. S5d; Fig. 1a) and induced the accumulation of VSVGts045-GFP at the LUb-ERGIC (Supplementary Fig. S5e). Collectively, these results suggest that Rab2A knockdown induces the inhibition of vesicular transport from the LUb-ERGIC to the ER and the Golgi and thereby enables the ERAD machinery to accumulate in the LUb-ERGIC as a result of normal protein influx from the ER.
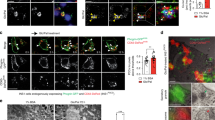
Rab2A knockdown induces accumulation of ERAD machinery and aggregation of polyubiquitinated proinsulin in the LUb-ERGIC.
(a–c and f) MIN6 cells were transfected with scramble or Rab2A siRNA. (a) Derlin-1 and Sec61α1 in the 1% Tx soluble (1% Tx) and LUb-ERGIC enriched fractions were immunoblotted with anti-Derlin-1 and Sec61α1 antibodies. Indicated values show the level of Derlin-1 or Sec61α1 normalized to that of GAPDH or actin. (b) p97 in the 1% Tx soluble (1% Tx) and LUb-ERGIC enriched fractions was immunoblotted with anti-p97 antibody. Indicated values show the level of p97 normalized to that of GAPDH or actin. (c) Left: ERGIC53 and Ub-proteins were subjected to double staining in the cells. Right: the mean fluorescent intensities of Ub-proteins at the LUb-ERGIC in the cells were quantified (mean ± s.e., n = 19 in each condition). (d) Left: MIN6 cells were transfected with wild-type GFP-Rab2A (WT) or dominant-positive GFP-Rab2A Q65L (Q65L). ERGIC53 and Ub-proteins were subjected to double staining in the transfected cells. Right: the mean fluorescent intensities of Ub-proteins at the LUb-ERGIC in the cells expressing GFP alone, GFP-Rab2A or GFP-Rab2A Q65L were quantified (mean ± s.e.; GFP: n = 20; WT: n = 16; Q65L: n = 19). (e) Left: ERGIC53 and Ub-proteins were subjected to double staining in MIN6 cells. Images were derived by three-dimensional reconstruction of the cells (3D reconstruction). Arrows indicate the LUb-ERGIC. Scale bars: 5 μm. Right: the size of the LUb-ERGIC and the mean fluorescent intensities of Ub-proteins at the LUb-ERGIC were measured and plotted (n = 30, x-axis: diameter of the LUb-ERGIC, y-axis: mean fluorescent intensity of Ub-proteins at the LUb-ERGIC). The line indicates the linear regression line, y = 113.2x-155.26 and the R2 value is 0.7234. (f) Proinsulin in the 1% Tx soluble (1% Tx) and LUb-ERGIC enriched fractions was immunoblotted with anti-proinsulin antibody, using the GAPDH and actin signals, respectively, as loading controls. Ub1 or and Ubn represent mono and polyubiquitinated proinsulin, respectively. (c and d) Insets indicate high magnification images of the corresponding white rectangles. Scale bars: 5 μm (low magnification) and 2 μm (high magnification). Full-length blots are presented in Supplementary Fig. S10.
Next, we investigated the effect of Rab2A function on the aggregation of ubiquitinated proteins at the LUb-ERGIC using immunofluorescence microscopy. As expected, the fluorescence intensity of ubiquitinated proteins at the LUb-ERGIC increased significantly by 2.26 fold in Rab2A-knockdown cells (Fig. 4c). By using a nuclear matrix protein isolation procedure as described above, we confirmed biochemically that polyubiquitinated protein increased in the LUb-ERGIC enriched fraction upon Rab2A knockdown (data not shown). In contrast, polyubiquitinated protein decreased significantly by 0.50 fold in cells expressing GFP-Rab2A Q65L, which is a dominant active form of Rab2A (Fig. 4d). Also, the size of the LUb-ERGIC in MIN6 cells that expressed GFP-Rab2A Q65L decreased as compared to that in MIN6 cells that expressed GFP-Rab2A WT (data not shown), which was consistent with the result showing a positive correlation between the size of the LUb-ERGIC and fluorescent intensity of ubiquitinated proteins at the LUb-ERGIC (Fig. 4e). More interestingly, after Rab2A knockdown, the amount of polyubiquitinated proinsulin was increased in the LUb-ERGIC fraction, but not in the soluble fraction (Fig. 4f).
The fact that the amount of Derlin-1, Sec61α1, p97 and ubiquitinated protein in the LUb-ERGIC fraction was increased in Rab2A-knockdown cells suggested that the retrotranslocation of misfolded proteins containing proinsulin from the LUb-ERGIC into the cytoplasm would be accelerated by Rab2A knockdown. We could eliminate the possibility that the accelerated aggregation results from inhibition of the ubiquitin-proteasome or the autophagy system, because both the increase in ubiquitinated protein upon MG132 treatment and the modification of LC3 upon the induction of autophagy occurred normally in Rab2A-knockdown cells (Supplementary Figs. S6a and S6b). Thus, we hypothesized that Rab2A knockdown induces the accumulation of ERAD machinery in the LUb-ERGIC and accelerates the aggregation of polyubiquitinated proinsulin at the LUb-ERGIC.
Prolonged exposure to high glucose levels decreases Rab2A activity in a manner dependent on PARP activity and GAPDH
We assumed that prolonged treatment with high levels of glucose affects the activity of Rab2A at the LUb-ERGIC and results in aberrant accumulation of the ERAD machinery and its substrates. Consequently, we investigated the kinetics of Rab2A activity by a GTP loading assay (See Methods). After incubation in low glucose medium for 24 hr, the cells were transferred to high glucose medium. Rab2A activity increased at first from 100% to 118% at 1 hr after exposure, when GSIS was induced. It then decreased, reaching 60% after 15 hr, after which it plateaued at approximately 52% (Fig. 5a). The transient increase in Rab2A activity at 1 hr after stimulation, when a large amount of insulin is secreted (Supplementary Fig. S7), appears to contribute to normal insulin secretion because Rab2A is required for vesicular transport from the ER to the Golgi25. The diameter of the LUb-ERGIC was measured under the same conditions and was found to increase steadily from 2.04 μm to 2.64 μm during the first 12 hr after exposure and then reached a plateau (Fig. 5b).
Prolonged exposure to high glucose levels decreases Rab2A activity in a manner dependent on PARP activity and GAPDH.
(a, b, e and f) MIN6 cells were exposed to low glucose (5.6 mM) for 24 hr and then high glucose (25 mM) for the indicated times. (a) Upper: the cells were analyzed using the GTP loading assay for Rab2A (See Methods). Lower: Rab2A activity was quantified by calculating the amount of GTP-bound Rab2A normalized to that of input Rab2A (mean ± s.e., n = 4). (b) Upper: the cells were immunostained with anti-ERGIC53 antibody. Representative images of the LUb-ERGIC at each indicated time were shown. Scale bars: 2 μm. Lower: the diameter of the LUb-ERGIC was determined (mean ± s.e., n = 30 or 31). Statistical analyses were performed to compare the data obtained at 0 hr and 12 hr and at 0 hr and 24 hr. Indicated P values were calculated with Student's t-test, followed by the Bonferroni correction. (c and d) MIN6 cells were transfected with scramble or GAPDH siRNA. The cells were analyzed by immunoblotting with anti-GAPDH and actin antibodies in (c) and by the GTP loading assay for Rab2A in (d). The right bar graph in (d) indicates mean ± s.e. (n = 3). (e) Upper: the whole cell lysates (input) were immunoprecipitated with anti-poly (ADP-ribose) (PAR) antibody. The immunoprecipitates were immunoblotted with anti-GAPDH and PAR antibodies. Lower: the degree of poly (ADP-ribosyl)ated GAPDH was quantified by calculating the amount of immunoprecipitated GAPDH normalized to that of input GAPDH (mean ± s.e., n = 3). (f) Upper: the whole cell lysates (input) were immunoprecipitated with anti-GAPDH antibody. The immunoprecipitates were immunoblotted with anti-Rab2A and GAPDH antibodies. Lower: the amount of Rab2A interacting with GAPDH was quantified by calculating the amount of immunoprecipitated Rab2A normalized to that of GAPDH (mean ± s.e., n = 3). (g and h) MIN6 cells were exposed to low glucose for 24 hr and then high glucose with PJ34 at the indicated concentrations for 15 hr. In (g), the cells were analyzed by immunoprecipitation as described in (f). In (h), the cells were analyzed using the GTP loading assay for Rab2A. Full-length blots are presented in Supplementary Fig. S10.
To identify regulators of Rab2A activity upon exposure to high levels of glucose, we focused on GAPDH, which is an effector of Rab2A3 and is known to be poly (ADP-ribosyl)ated by PARP under hyperglycemic conditions in aortic endothelial cells6. In GAPDH-knockdown cells in which expression was suppressed by more than 80% (Fig. 5c), Rab2A activity decreased by more than 60% as compared with control cells (Fig. 5d), which indicated that GAPDH could be a promising regulator of Rab2A.
Next, we investigated the extent of poly (ADP-ribosyl)ation of GAPDH upon exposure to high levels of glucose. By immunoprecipitation, we found that the amount of poly (ADP-ribosyl)ated GAPDH slightly decreased at 2 hr after exposure to high levels of glucose, but had increased 1.7 fold at 15 hr after exposure (Fig. 5e). Poly (ADP-ribosyl)ated GAPDH was detected as a single band as described in previous reports5,35. The amount of Rab2A interacting with GAPDH had slightly increased 1.2 fold at 2 hr after exposure, but had decreased 0.42 fold at 15 hr (Fig. 5f). These findings indicate that poly (ADP-ribosyl)ation of GAPDH upon exposure to high levels of glucose causes the dissociation of GAPDH from Rab2A and results in its inactivation. Accordingly, PJ34, an inhibitor of PARP, increased Rab2A activity and the amount of Rab2A interacting with GAPDH in a dose-dependent manner (Figs. 5g and 5h). However, the enlargement of the LUb-ERGIC that is normally induced by prolonged treatment with high levels of glucose was not observed in PJ34-treated cells (Supplementary Fig. S8). Moreover, we confirmed that (i) the diameter of the LUb-ERGIC was enlarged, (ii) the amount of ubiquitinated protein at the LUb-ERGIC increased and (iii) insulin secretion decreased in GAPDH-knockdown cells (Supplementary Figs. S9a–S9c), as had been observed in Rab2A-knockdown cells; all of which also suggest that GAPDH could be a promising regulator of Rab2A.
Knockdown of Rab2A or GAPDH decreases ER stress whereas PJ34 increases ER stress
Given that ERAD is one of the cellular responses to ER stress, it is possible that ubiquitinated proteins aggregate at the LUb-ERGIC to alleviate ER stress. Consequently, we investigated the unfolded protein response (UPR) to ER stress, which is induced by chronically high levels of glucose, in pancreatic β cells36.
First, we analyzed the kinetics of the UPR in MIN6 cells exposed to high levels of glucose by quantifying the phosphorylation states of PERK (protein kinase RNA-like endoplasmic reticulum kinase), eIF2α and IRE1α, which are all involved in the UPR. Interestingly, as shown in Fig. 6a, the phosphorylation levels of PERK, eIF2α and IRE1α were higher before stimulation with high glucose (time point at 0 hr after incubation with low glucose medium for 24 hr) than at 3 hr after exposure (Fig. 6a, right, 0 hr versus 3 hr). It has been reported that activation of PERK and eIF2α in cells in which insulin secretion is not needed is required to reduce insulin biosynthesis through translational inhibition37; however, the role of the activation of IRE1α remains unclear at this time.
Knockdown of Rab2A or GAPDH decreases ER stress whereas PJ34 increases ER stress.
(a) Left: MIN6 cells were exposed to low glucose (5.6 mM) and then high glucose (25 mM) for the indicated times. The cells were lysed and the lysates were immunoblotted with antibodies against phospho-PERK (pPERK), PERK, phospho-eIF2α (peIF2α), eIF2α, phospho-IRE1α (pIRE1α), IRE1α and actin. Right: the activity of PERK, eIF2α and IRE1α at 0, 0.5, 1, 3, 6, 12 and 18 hr after stimulation with high glucose was quantified by calculating the level of each phosphorylated protein normalized to that of the respective total protein (mean ± s.e., n = 3). Data from Fig. 5a were also added to the same graph and indicated that the activity of the ER stress markers remained stationary whereas Rab2A activity continued to decrease over the course of exposure to high glucose. (b) Left: MIN6 cells were transfected with scramble or Rab2A siRNA. One day after siRNA transfection, the cells were exposed to low glucose for 24 hr and then high glucose for 15 hr. The cells were lysed and the lysates were immunoblotted with antibodies against pPERK, PERK, peIF2α, eIF2α, pIRE1α, IRE1α and actin. Right: the activities of PERK, eIF2α and IRE1α were quantified by determining the level of each protein in the phosphorylated state normalized to the total level of each (mean ± s.d., n = 4). (c) A similar experiment to that described in (b) was performed for MIN6 cells transfected with scramble or GAPDH siRNA. The bar graphs on the right show the mean ± s.d. (n = 4). (d) Left: MIN6 cells were exposed to low glucose for 24 hr and then high glucose without (Control) or with 15 μM PJ34 for 15 hr. The level of each of the phosphorylated ER stress markers in the cells was analyzed by immunoblotting as described in (b). Right: bar graphs show the mean ± s.d. (n = 4). Full-length blots are presented in Supplementary Fig. S10.
Upon stimulation with high glucose, GSIS occurred and then ceased within ~1 hr (Supplementary Fig. S7). Under the same conditions, the levels of PERK and eIF2α phosphorylation decreased rapidly to the lower steady state level within 1 hr (Fig. 6a, right). The level of IRE1α phosphorylation had increased markedly at 1 hr and then decreased subsequently to a lower level (Fig. 6a, right). The initial increase of the UPR by phosphorylation of IRE1α might be due to a requirement for the transient biosynthesis of insulin in response to high glucose levels38. To examine the involvement of Rab2A in the UPR, we analyzed the levels of PERK, eIF2α and IRE1α phosphorylation in Rab2A-knockdown cells during the 15 hr after stimulation with high glucose. In Rab2A-knockdown cells, the levels of PERK, eIF2α and IRE1α phosphorylation were decreased by 0.63, 0.76 and 0.71 fold, respectively, as compared with the control cells (Fig. 6b). Similarly, in GAPDH-knockdown cells, the levels of PERK, eIF2α and IRE1α phosphorylation were decreased by 0.48, 0.84 and 0.50 fold, respectively, as compared with the control cells (Fig. 6c), which again indicated that GAPDH could be a regulator of Rab2A. In contrast, in cells treated with PJ34, the levels of PERK, eIF2α and IRE1α phosphorylation were increased by 1.22, 1.32 and 1.28 fold, respectively, as compared with the control cells (Fig. 6d). These results suggest that a decrease in Rab2A activity that is induced by chronically high glucose might relieve ER stress in a PARP- and GAPDH-dependent manner. To confirm this idea, we examined the ability of Rab2A-knockdown cells to resist apoptosis induced by thapsigargin- or tunicamycin-mediated ER stress. As shown in Fig. 7, the ability to resist apoptosis was increased remarkably in Rab2A-knockdown cells as compared with control cells.
Rab2A knockdown inhibits ER stress-induced apoptosis.
MIN6 cells transfected with scramble or Rab2A siRNA were treated with 1 μM thapsigargin or 10 μg/ml tunicamycin for 6, 12 or 24 hr. Left: control cells or Rab2A-knockdown cells treated with thapsigargin or tunicamycin for 24 hr were subjected to staining with Hoechst 33342 and TUNEL. Scale bars: 5 μm. Right: TUNEL-positive cells were determined (n = 3, we observed 300 cells in each experiment). Statistical analyses were performed to compare the data from the control cells and Rab2A-knockdown cells treated with thapsigargin or tunicamycin for 24 hr.
Discussion
The LUb-ERGIC in MIN6 cells provides a suitable site for ERAD-mediated protein quality control
In the present study, we found that the ERAD machinery and its ubiquitinated substrates accumulated in the LUb-ERGIC in MIN6 cells (Figs. 2 and 3a). This accumulation was augmented under high glucose conditions, when the production of misfolded proinsulin is enhanced in the ER. This result indicates that the LUb-ERGIC is a potential site for ERAD under conditions that induce the UPR. Although the ERAD system was considered originally to be specific to the ER, it is probable that the ERGIC, including the LUb-ERGIC, has a similar ERAD function to the ER. Recently, it has been reported that the ERGIC, as well as the ER, is involved in the conformation-based quality control of proteins1. For example, important components of the protein quality control machinery including a number of chaperone proteins and UDP-glucose:glycoprotein glucosyltransferase, which is a protein folding sensor and glycosyltransferase, are reported to localize to the ERGIC39,40. Moreover, even when VSVGts045 is overexpressed in Chinese Hamster Ovary cells at the non-permissive temperature, at which VSVGts045 cannot be transported from the ER, some translocates to the ERGIC and cis-Golgi, rather than being retained in the ER7. This indicates that misfolded proteins that escape from the ER quality control pathway due to system overload or failed recognition can be caught by a subsequent checkpoint in the ERGIC. Thus, ER-resident proteins such as chaperones and misfolded proteins could recycle between the ER and the ERGIC. However, it remains less clear whether protein components of the ERAD machinery, including the membrane proteins Derlin-1 and the Sec61 complex, recycle between the two organelles, because none of the membrane proteins have ER retrieval signals such as a cytosolic di-lysine or di-arginine motif. These motifs are recognized directly by COPI subunits and are thought to function in the retrieval of proteins to the ER41. Nonetheless, it has been reported that the Sec61 complex is located in both the ER and the ERGIC in COS1 cells as well as MIN6 cells, even though the retrieval mechanism is still unknown42. This mechanism needs to be elucidated in the near future.
Previous studies have shown that ERAD substrates accumulate in a specialized perinuclear compartment, referred to as the ER-derived quality control compartment (ERQC), before they are delivered to the proteasome43,44,45,46. In addition to misfolded proteins, the lectin chaperone calnexin and calreticulin also concentrate in the ERQC, while BiP remains distributed throughout the ER43,44. Not only ERAD machinery components such as HRD1, p97 and Derlin-1 but also UPR sensors such as PERK and IRE1α accumulate in ERQC45, which suggests a possible close correlation between UPR and ERAD in the ERQC. Although the LUb-ERGIC is also the domain where ERAD proteins accumulated and may have the similar function of the ERQC, several aspects of the LUb-ERGIC are different from those of ERQC with regard to intracellular localization, morphology and localization of ERGIC53. We speculate that MIN6 cells might develop such a specialized compartment to trap excess proinsulin and enable its degradation during transport to the plasma membrane. Additional experiments would be required to elucidate the exact difference between LUb-ERGIC and ERQC.
Rab2A at the LUb-ERGIC is a pivotal switch for selecting either insulin secretion or ERAD
We propose that Rab2A, which is a resident of the ERGIC, plays a crucial role in regulating ERAD at the LUb-ERGIC under prolonged high glucose conditions (Fig. 8). Our results indicated that the amount of β-COP, a subunit of COPI, in the LUb-ERGIC fraction decreased in Rab2A-knockdown cells (Supplementary Fig. S5a), whereas polyubiquitinated proinsulin, the Sec61 complex, Derlin-1 and p97 accumulated at the LUb-ERGIC (Fig. 4). Although it was difficult to observe β-COP at the LUb-ERGIC by indirect immunofluorescence (data not shown), we also showed that Rab2A, which recruits β-COP to ERGIC membrane, was localized to the LUb-ERGIC (Fig. 1c) and BFA treatment, which detaches β-COP from the membrane, enlarged the size of the LUb-ERGIC (Supplementary Fig. S4e). These results suggest that vesicular transport from the LUb-ERGIC to the ER and the Golgi would be performed in a COPI-dependent manner and would be abrogated through the decrease of Rab2A activity upon exposure to high glucose. The inhibition of vesicular transport from the LUb-ERGIC would result in the accumulation of the above-mentioned ERAD machinery in the LUb-ERGIC owing to normal protein influx from the ER into the LUb-ERGIC. Consequently, the LUb-ERGIC would be a suitable site for ERAD because the accumulated Sec61 complex and Derlin-1 would facilitate the retrotranslocation of misfolded proinsulin, while a role of Sec61 complex in ERAD has been controversial28. Rab2A activity at the LUb-ERGIC probably forms part of the regulatory system against glucotoxicity in MIN6 cells by facilitating ERAD, relieving glucotoxic ER stress and avoiding apoptosis (Figs. 6 and 7).
A schematic model of the formation of ubiquitinated proinsulin aggregates in the vicinity of the LUb-ERGIC by prolonged high glucose treatment.
Rab2A, which regulates vesicular transport of proinsulin from the LUb-ERGIC to the Golgi, is required for GSIS under transient high glucose exposure. When the condition of high glucose continues, GAPDH is poly (ADP-ribosyl)ated by PARP (1) and dissociates from Rab2A (2), resulting in the disruption of COPI vesicle budding from the LUb-ERGIC. So, prolonged high glucose treatment enlarges the LUb-ERGIC and induces the accumulation of proinsulin and ERAD machinery in the LUb-ERGIC owing to the inhibition of COPI-dependent vesicle transport from the LUb-ERGIC whereas the protein influx from the ER normally occurs. Thereby, misfolded proinsulin can be retrotranslocated from its lumen into the cytoplasm and aggregates of ubiquitinated proinsulin can be formed near the LUb-ERGIC (3). The poly (ADP-ribosyl)ation-mediated inactivation of Rab2A upon high glucose treatment could ensure alleviation of ER stress and ER stress-induced apoptosis, possibly through accelerating the retrotranslocation or ERAD of proinsulin in the LUb-ERGIC.
GAPDH regulates Rab2A activity via poly (ADP-ribosyl)ation in response to prolonged exposure to high glucose
According to the previous biochemical experiments, GAPDH is recruited to membrane-bound Rab247. This suggests that GAPDH is actually a downstream effector of Rab2A in terms of the recruitment of Rab2A to the ERGIC membranes. In contrast, as shown in Fig. 5d, we found that the amounts of Rab2A in the GTP-bound form, which generally corresponds to membrane-bound form of Rab2A, was lower in GAPDH-knockdown cells than control cells, which suggests that GAPDH is an upstream factor of Rab2A, at least, in terms of the recruitment of Rab2A to the ERGIC membranes. One possible explanation is that a variety of mutual interactions between Rab2A and GAPDH might be involved in the recruitment of the two proteins to the membranes. In that case, poly(ADP-ribosyl)lation could be one of regulatory post-translational modifications. As our experimental system is different from theirs, more extensive studies will be needed to clarify it.
Our results also suggest that GAPDH, a key glycolytic enzyme, which modifies Rab2A activity via poly (ADP-ribosyl)ation of GAPDH itself, regulates ERAD in the LUb-ERGIC in response to prolonged exposure to high glucose (Figs. 5 and 8; Supplementary Figs. S9a and S9b). This in turn implies that the early secretory pathway is linked closely to glucose metabolism via Rab2A and GAPDH. In aortic endothelial cells under conditions of hyperglycemia, GAPDH is poly (ADP-ribosyl)ated by PARP, which is activated by DNA strand breaks that are produced by mitochondrial ROS and the glycolytic activity of the enzyme is inhibited6. Analogous to this, one possible feedback mechanism for the inactivation of Rab2A under high glucose levels is the production of ROS in the ER. As mentioned above, glucose induces the biosynthesis of proinsulin in the ER. Given that proinsulin contains three disulphide bonds per molecule, this imposes a significant oxidative burden on pancreatic β cells, because the creation of disulphide bonds imposes oxidative stress on the cell, the degree of which correlates with the number of disulphide bonds formed48. As an adaptive response to the oxidative stress, poly (ADP-ribosyl)ation of GAPDH by PARP also inhibits Rab2A activity. This feedback mechanism ensures ER homeostasis by eliminating misfolded proteins through the ERAD system at the LUb-ERGIC as well as ER stress.
Our current findings suggest a possible mechanism for ensuring protein quality control in differentiating cells, in which the excessive production of secreted protein is a constant possibility, through regulation of the vesicular transport system. In insulinoma (MIN6) cells, the early secretory pathway is probably linked closely to glucose metabolism via Rab2A and GAPDH to ensure cell survival.
Methods
Cell culture
MIN6 cells (a gift from Dr. Susumu Seino, Kobe University Graduate School of Medicine, Kobe, Japan) were grown in Dulbecco's modified Eagle medium (DMEM; Wako) with 10% fetal calf serum (FCS), penicillin/streptomycin (Gibco), 72 μM β-mercaptoethanol and 25 mM glucose in humidified 5% CO2 at 37°C. INS-1E cells (a gift from Dr. Claes B. Wollheim, University Medical Center, Geneva, Switzerland) were grown in RPMI-1640 medium (Nissui) supplemented with 10 mM HEPES, 5% FCS, 1 mM sodium pyruvate, 50 μM β-mercaptoethanol and penicillin/streptomycin in humidified 5% CO2 at 37°C. Unless we mentioned glucose condition, we used MIN6 and INS-1E cells under normal cell culture condition (25 mM glucose) as described above.
Antibodies, reagents and plasmids
The following antibodies were used for the immunofluorescence experiments: rabbit anti-ERGIC53/p58 antibody (Sigma Aldrich); mouse anti-GM130 antibody (BD Transduction Laboratories); rabbit anti-Yip1A antibody32; rabbit anti-Derlin-1 antibody (Novus Biologicals); rabbit anti-BiP antibody (Abcam); mouse anti-p97 antibody (Abcam); mouse anti-mono and polyubiquitinated conjugates antibody (FK2; Enzo Life Sciences); mouse anti-polyubiquitinated conjugates antibody (FK1; Enzo Life Sciences); mouse anti-proinsulin/insulin antibody (Cell Signaling); rabbit anti-TGN46 antibody (Abcam); mouse anti-γ-tubulin antibody (Abcam); Alexa 488-conjugated anti-rabbit IgG antibody (Life Technologies); Alexa 568-conjugated anti-rabbit IgG antibody (Invitrogen); Cy3-conjugated anti-mouse IgG antibody (Chemicon); Alexa 488-conjugated anti-mouse IgG antibody (Invitrogen); Cy5-conjugated anti-rabbit IgG antibody (Abcam); Cy5-conjugated anti-mouse IgG antibody (Rockland). The following antibodies were used for the western blotting experiments: mouse anti-polyubiquitinated conjugates (FK1; Enzo Life Sciences); mouse anti-proinsulin/insulin antibody (Cell Signaling); rabbit anti-Derlin-1 antibody (Novus Biologicals); goat anti-Sec61α1 antibody (Abcam); mouse anti-p97 antibody (Abcam); mouse anti-GAPDH antibody (Millipore); rabbit anti-actin antibody (Sigma Aldrich); mouse anti-Rab2A antibody (Abcam); rabbit anti-Rab2A antibody (Abcam); mouse anti-β-tubulin antibody (Sigma Aldrich); rabbit anti-β-COP antibody (ABR); mouse anti-LC3 antibody (Nanotools); rabbit anti-GFP antibody (MBL); mouse anti-poly (ADP-ribose) antibody (Enzo Life Sciences); rabbit anti-phospho-PERK (Thr980) antibody (Cell Signaling); rabbit anti-PERK antibody (Cell Signaling); rabbit anti-phospho-eIF2α (Ser51) antibody (Cell Signaling); rabbit anti-eIF2α antibody (Cell Signaling); rabbit anti-phospho-IRE1α (Ser724) antibody (Thermo Scientific); rabbit anti-IRE1α antibody (Cell Signaling); HRP-conjugated anti-mouse IgG antibody (Promega); HRP-conjugated anti-rabbit IgG antibody (Cell Signaling); HRP-conjugated anti-goat IgG antibody (Santa Cruz). The following reagents were used: Cycloheximide (Sigma Aldrich); MG132 (Calbiochem); PJ34 (Millipore); Bafilomycin A1 (Wako); Hoechst 33342 (Wako). The following plasmids were used: GFP-p5832; Sec61α1-GFP; HA-Sec61β (the cDNA for mouse Sec61 β was amplified from a mouse brain cDNA library using the forward primer, 5′-GCGAATTCTGATGCCGGGTCCAACG-3′ and the reverse primer, 5′-GCGTCGACCTATGATCGCGTGTACTTGCC-3′; the amplified cDNA was ligated into the vector pCMV-HA purchased from Clontech between the EcoR I and Sal I sites); HA-Sec61γ (the cDNA for mouse Sec61γ was amplified from a mouse brain cDNA library using the forward primer, 5′-GCGTCGACCATGCATCAGGTAATGCAGTTTGTGG-3′ and the reverse primer, 5′-GCAGATCTTCAGCCACCCACAATAATGTTATTAATGG-3′; the amplified cDNA was ligated into pCMV-HA between the Sal I and Bgl II sites); RFP-Ubiquitin (a gift from Dr. Nobuyuki Nukina, RIKEN Brain Institute and CREST/JST, Saitama, Japan); GFP-Rab2A (the cDNA for mouse Rab2A was amplified from a mouse brain cDNA library using the forward primer, 5′-GCGAATTCTGATGGCGTACGCCTATCTCTTCAAGTAC-3′ and the reverse primer, 5′-GCGTCGACTAACACCAGACTCAACAGCAGCCTCCCCC-3′; the amplified cDNA was ligated into the vector pEGFP-C3 purchased from Clontech between the EcoR I and Sal I sites); GFP-Rab2A Q65L (glutamine 65 of GFP-Rab2A was replaced by leucine using a QuikChange Site-directed Mutagenesis Kit from Agilent Technologies, together with the forward primer, 5′-CCAGATCTGGGATACAGCAGGGCTGGAGTCCTTTCG-3′ and the reverse primer, 5′-CGAAAGGACTCCAGCCCTGCTGTATCCCAGATCTGG-3′); VSVGts045-GFP32.
DNA transfection
Plasmids were transfected into MIN6 cells using GenCarrier-2 (Epoch Life Science) in accordance with the manufacturer's instructions.
RNA interference
A small interfering RNA (siRNA) against mouse Rab2A (Sigma Aldrich, ID: SASI_Mm01_00059142), mouse GAPDH (Sigma Aldrich, ID: SASI_Mm01_00189762), mouse insulin-2 (Sigma Aldrich, ID: SASI_Mm02_00313770) or a scrambled siRNA (Ambion) was transfected into MIN6 cells using Lipofectamine RNAiMAX (Invitrogen) in accordance with the manufacturer's instructions. We used 50 nM siRNA (final concentration) for the siRNA transfection.
Immunofluorescence
MIN6 cells were fixed with 3% paraformaldehyde that contained 0.1 M PIPES, 3.6 mM CaCl2 and 2 mM MgCl2 for 30 min at room temperature and then permeabilized with 0.2% Triton X-100 in PBS for 20 min at room temperature. After blocking with 3% BSA in PBS for 30 min, the cells were incubated first with appropriate primary antibodies for 2 hr and then with fluorescently labelled secondary antibodies for 1 hr at room temperature. Ubiquitinated proteins were stained as described previously49. Briefly, MIN6 cells were washed with PBS and then fixed with 2.5% paraformaldehyde in PBS for 10 min at 37°C. The cells were washed three times with PBS for a total time of at least 60 min, after which they were permeabilized by incubation with 0.2% saponin (Sigma Aldrich) in PBS that contained 10% FCS (SS-PBS) for 10 min at room temperature. Primary antibodies were diluted in SS-PBS and incubated with the cells for 20 min at room temperature. The cells were washed three times with PBS for a total time of 20 min. Then the cells were labelled with secondary antibodies diluted in SS-PBS for 20 min at room temperature and washed three times with PBS for a total time of 20 min. The cells were observed by using an LSM510 confocal laser scanning microscope with a Plan-Apochromat 63×/1.4 Oil Objective. To quantify the fluorescence intensity of proinsulin and Ub-proteins at the LUb-ERGIC, Z-stack images were taken every 0.5–0.8 μm and reconstructed using LSM software. The fluorescence intensity of proinsulin and Ub-proteins at the LUb-ERGIC was defined as [the mean fluorescence intensity at the LUb-ERGIC, which was observed as spheroidal or aggregated structure]-[the mean fluorescence intensity in the cytoplasmic region (Ub-proteins) or in cytoplasmic region except for the perinuclear region (proinsulin)]. The fluorescence intensity in control cells was assigned as 1.
Western blotting
Proteins in cell lysates were separated by SDS-PAGE and transferred to PVDF membrane using a semi-dry blotting system (Bio-Rad). After the membrane had been blocked with 3% BSA in TBS that contained 0.1% Tween 20 (TBST) for 60 min at room temperature, it was incubated first with appropriate primary antibodies overnight at 4°C and then with HRP-conjugated secondary antibodies for 1 hr at room temperature. The proteins were detected by enhanced chemiluminescence (Western Lighting Plus-ECL, Perkin Elmer).
TUNEL staining
MIN6 cells that had been transfected with scramble or Rab2A siRNA were treated with 1 μM thapsigargin (Sigma) or 10 μg/ml tunicamycin (Sigma) for the indicated times. The cells were subjected to TUNEL staining using an In Situ Cell Death Detection Kit (Roche) in accordance with the manufacturer's instructions.
Coimmunoprecipitation
MIN6 cells were lysed in Pierce IP Lysis Buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40 and 5% glycerol) supplemented with protease and phosphatase inhibitors (Roche). The lysates were incubated with Protein G Sepharose Beads (GE Healthcare) for 10 min at 4°C. After centrifugation, the protein concentration of the supernatants was determined using a BCA Protein Assay Kit (Thermo Scientific). The supernatants were diluted to a protein concentration of 1 mg/ml with PBS and then incubated with mouse anti-poly (ADP-ribose) or mouse anti-GAPDH antibody overnight at 4°C. Subsequently the lysates were incubated with Protein G Sepharose Beads for 60 min at 4°C. The precipitated beads were washed with PBS and lysed in 2× sample buffer. The proteins in the lysates were analyzed by western blotting.
Purification of aggregates of ubiquitinated proteins at the LUb-ERGIC
We adapted a nuclear matrix protein isolation procedure30,31 to purify aggregates of ubiquitinated proteins at the LUb-ERGIC by extracting cytosolic and nuclear ubiquitinated material. MIN6 cells were washed with buffer A (20 mM Tris pH 7.4, 1.5 mM MgCl2 and 10 mM NaCl) and incubated with 1% Triton X-100 in buffer A for 30 min at 4°C, followed by 350 units/ml DNase I (Takara) in buffer A for 30 min at 37°C. Then nuclear material was extracted with 2 M NaCl in 20 mM Tris pH 7.4 and 1.5 mM MgCl2. The remaining material was subjected to immunofluorescence or western blotting to detect ubiquitinated aggregates or proteins that localized close to the aggregates.
GTP loading assay
MIN6 cells were washed with control KREBS buffer pH 7.4 (140 mM NaCl, 3.6 mM KCl, 0.5 mM NaH2PO4, 0.5 mM MgSO4, 1.5 mM CaCl2, 10 mM HEPES and 2 mM NaHCO3) and permeabilized with 50 μg/ml digitonin (Wako) in control KREBS buffer for 5 min at room temperature. The supernatant was removed and then the cells were incubated with control KREBS buffer that contained 20 μM desthiobiotin-GTP probe (GTPase Enrichment Kits, Thermo Scientific) and 20 mM MgCl2 (GTPase Enrichment Kits, Thermo Scientific) in the presence of an ATP generating system (40 IU/ml creatine phosphokinase and 1 mM phosphocreatine, Roche) for 20 min at 37°C. Then the supernatant was removed and the cells were lysed with Pierce IP Lysis Buffer supplemented with protease and phosphatase inhibitor (GTPase Enrichment Kits, Thermo Scientific). The lysates were diluted to a protein concentration of 2 mg/ml with Pierce IP Lysis Buffer. An equal volume of 8 M Urea (GTPase Enrichment Kits, Thermo Scientific) in Pierce IP Lysis Buffer was added to the lysates. The samples were mixed with Streptavidin Agarose Resin (50% slurry; GTPase Enrichment Kits, Thermo Scientific) for 60 min at room temperature and then centrifuged to precipitate the biotin-avidin complexes. The complexes were washed with 4 M Urea in Pierce IP Lysis Buffer, precipitated, lysed with 2× sample buffer and boiled for 5 min. The samples were analyzed by SDS-PAGE and western blotting.
GSIS
MIN6 cells were washed with pre-warmed control KREBS buffer and exposed to 3 mM glucose in control KREBS buffer for 60 min at 37°C. The cells were treated subsequently with 3 mM glucose or 25 mM glucose in pre-warmed control KREBS buffer for 60 min at 37°C to induce insulin secretion. The insulin secreted into the buffer was quantified using an AlphaLISA insulin kit (Perkin Elmer).
Statistical analysis
The data were analyzed statistically by using the F-test to check the equality of variance and then Student's t-test alone or Student's t-test followed by the Bonferroni correction. Values were given as the mean ± standard deviation (s.d.) or standard error (s.e.).
References
Appenzeller-Herzog, C. & Hauri, H. P. The ER-Golgi intermediate compartment (ERGIC): in search of its identity and function. J. Cell. Sci. 119, 2173–2183 (2006).
Moyer, B. D., Allan, B. B. & Balch, W. E. Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis--Golgi tethering. Traffic 2, 268–276 (2001).
Tisdale, E. J., Kelly, C. & Artalejo, C. R. Glyceraldehyde-3-phosphate dehydrogenase interacts with Rab2 and plays an essential role in endoplasmic reticulum to Golgi transport exclusive of its glycolytic activity. J. Biol. Chem. 279, 54046–54052 (2004).
Tisdale, E. J. Rab2 interacts directly with atypical protein kinase C (aPKC) iota/lambda and inhibits aPKCiota/lambda-dependent glyceraldehyde-3-phosphate dehydrogenase phosphorylation. J. Biol. Chem. 278, 52524–52530 (2003).
Devalaraja-Narashimha, K. & Padanilam, B. J. PARP-1 inhibits glycolysis in ischemic kidneys. J. Am. Soc. Nephrol. 20, 95–103 (2009).
Du, X. et al. Inhibition of GAPDH activity by poly(ADP-ribose) polymerase activates three major pathways of hyperglycemic damage in endothelial cells. J. Clin. Invest. 112, 1049–1057 (2003).
Hammond, C. & Helenius, A. Quality control in the secretory pathway: retention of a misfolded viral membrane glycoprotein involves cycling between the ER, intermediate compartment and Golgi apparatus. J. Cell Biol. 126, 41–52 (1994).
Hsu, V. W. et al. A recycling pathway between the endoplasmic reticulum and the Golgi apparatus for retention of unassembled MHC class I molecules. Nature 352, 441–444 (1991).
Moolenaar, C. E. et al. A mutation in a highly conserved region in brush-border sucrase-isomaltase and lysosomal alpha-glucosidase results in Golgi retention. J. Cell. Sci. 110, 557–567 (1997).
Gilbert, A., Jadot, M., Leontieva, E., Wattiaux-De Coninck, S. & Wattiaux, R. Delta F508 CFTR localizes in the endoplasmic reticulum-Golgi intermediate compartment in cystic fibrosis cells. Exp. Cell Res. 242, 144–152 (1998).
VanSlyke, J. K., Deschenes, S. M. & Musil, L. S. Intracellular transport, assembly and degradation of wild-type and disease-linked mutant gap junction proteins. Mol. Biol. Cell 11, 1933–1946 (2000).
Ito, M., Amizuka, N., Ozawa, H. & Oda, K. Retention at the cis-Golgi and delayed degradation of tissue-non-specific alkaline phosphatase with an Asn153-->Asp substitution, a cause of perinatal hypophosphatasia. Biochem. J. 361, 473–480 (2002).
Elkabetz, Y. et al. Immunoglobulin light chains dictate vesicular transport-dependent and -independent routes for IgM degradation by the ubiquitin-proteasome pathway. J. Biol. Chem. 278, 18922–18929 (2003).
Zuber, C., Fan, J. Y., Guhl, B. & Roth, J. Misfolded proinsulin accumulates in expanded pre-Golgi intermediates and endoplasmic reticulum subdomains in pancreatic beta cells of Akita mice. FASEB J. 18, 917–919 (2004).
Hermosilla, R. et al. Disease-causing V(2) vasopressin receptors are retained in different compartments of the early secretory pathway. Traffic 5, 993–1005 (2004).
Ge, L., Melville, D., Zhang, M. & Schekman, R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife 2, e00947 (2013).
Wahlman, J. et al. Real-time fluorescence detection of ERAD substrate retrotranslocation in a mammalian in vitro system. Cell 129, 943–955 (2007).
Ye, Y., Shibata, Y., Yun, C., Ron, D. & Rapoport, T. A. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429, 841–847 (2004).
Bukau, B., Weissman, J. & Horwich, A. Molecular chaperones and protein quality control. Cell 125, 443–451 (2006).
Vernace, V. A., Schmidt-Glenewinkel, T. & Figueiredo-Pereira, M. E. Aging and regulated protein degradation: who has the UPPer hand? Aging Cell 6, 599–606 (2007).
Kaniuk, N. A. et al. Ubiquitinated-protein aggregates form in pancreatic beta-cells during diabetes-induced oxidative stress and are regulated by autophagy. Diabetes 56, 930–939 (2007).
Costes, S. et al. β-cell dysfunctional ERAD/ubiquitin/proteasome system in type 2 diabetes mediated by islet amyloid polypeptide-induced UCH-L1 deficiency. Diabetes 60, 227–238 (2011).
Bensellam, M., Laybutt, D. R. & Jonas, J. C. The molecular mechanisms of pancreatic β-cell glucotoxicity: recent findings and future research directions. Mol. Cell. Endocrinol. 364, 1–27 (2012).
Leibowitz, G. et al. Glucose regulation of β-cell stress in type 2 diabetes. Diabetes Obes Metab 12, 66–75 (2010).
Tisdale, E. J., Bourne, J. R., Khosravi-Far, R., Der, C. J. & Balch, W. E. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J. Cell Biol. 119, 749–761 (1992).
Tisdale, E. J. A Rab2 mutant with impaired GTPase activity stimulates vesicle formation from pre-Golgi intermediates. Mol. Biol. Cell 10, 1837–1849 (1999).
Hou, J. C., Min, L. & Pessin, J. E. Insulin granule biogenesis, trafficking and exocytosis. Vitam. Horm. 80, 473–506 (2009).
Rubenstein, E. M., Kreft, S. G., Greenblatt, W., Swanson, R. & Hochstrasser, M. Aberrant substrate engagement of the ER translocon triggers degradation by the Hrd1 ubiquitin ligase. J. Cell Biol. 197, 761–773 (2012).
Tiwari, A. et al. SDF2L1 interacts with the ER-associated degradation machinery and retards the degradation of mutant proinsulin in pancreatic β-cells. J. Cell. Sci. 126, 1962–1968 (2013).
Staufenbiel, M. & Deppert, W. Preparation of nuclear matrices from cultured cells: subfractionation of nuclei in situ. J. Cell Biol. 98, 1886–1894 (1984).
Lelouard, H. et al. Transient aggregation of ubiquitinated proteins during dendritic cell maturation. Nature 417, 177–182 (2002).
Kano, F. et al. Yip1A regulates the COPI-independent retrograde transport from the Golgi complex to the ER. J. Cell. Sci. 122, 2218–2227 (2009).
Ben-Tekaya, H., Miura, K., Pepperkok, R. & Hauri, H. P. Live imaging of bidirectional traffic from the ERGIC. J. Cell. Sci. 118, 357–367 (2005).
Lippincott-Schwartz, J. et al. Microtubule-dependent retrograde transport of proteins into the ER in the presence of brefeldin A suggests an ER recycling pathway. Cell 60, 821–836 (1990).
Long, C. A. et al. Poly-ADP-ribose-polymerase inhibition ameliorates hind limb ischemia reperfusion injury in a murine model of type 2 diabetes. Ann. Surg. 258, 1087–1095 (2013).
Hou, Z. Q. et al. Involvement of chronic stresses in rat islet and INS-1 cell glucotoxicity induced by intermittent high glucose. Mol. Cell. Endocrinol. 291, 71–78 (2008).
Cavener, D. R., Gupta, S. & McGrath, B. C. PERK in beta cell biology and insulin biogenesis. Trends Endocrinol. Metab. 21, 714–721 (2010).
Lipson, K. L. et al. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 4, 245–254 (2006).
Breuza, L. et al. Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J. Biol. Chem. 279, 47242–47253 (2004).
Zuber, C. et al. Immunolocalization of UDP-glucose:glycoprotein glucosyltransferase indicates involvement of pre-Golgi intermediates in protein quality control. Proc. Natl. Acad. Sci. U.S.A. 98, 10710–10715 (2001).
Cosson, P. & Letourneur, F. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science 263, 1629–1631 (1994).
Greenfield, J. J. & High, S. The Sec61 complex is located in both the ER and the ER-Golgi intermediate compartment. J. Cell. Sci. 112, 1477–1486 (1999).
Kamhi-Nesher, S. et al. A novel quality control compartment derived from the endoplasmic reticulum. Mol. Biol. Cell 12, 1711–1723 (2001).
Frenkel, Z., Shenkman, M., Kondratyev, M. & Lederkremer, G. Z. Separate roles and different routing of calnexin and ERp57 in endoplasmic reticulum quality control revealed by interactions with asialoglycoprotein receptor chains. Mol. Biol. Cell 15, 2133–2142 (2004).
Kondratyev, M., Avezov, E., Shenkman, M., Groisman, B. & Lederkremer, G. Z. PERK-dependent compartmentalization of ERAD and unfolded protein response machineries during ER stress. Exp. Cell Res. 13, 3395–3407 (2007).
Leitman, J. et al. Herp coordinates compartmentalization and recruitment of HRD1 and misfolded proteins for ERAD. Mol. Biol. Cell 25, 1050–1060 (2014).
Tisdale, E. J. & Artalejo, C. R. A GAPDH mutant defective in Src-dependent tyrosine phosphorylation impedes Rab2-mediated events. Traffic 8, 733–741 (2007).
Cnop, M. et al. Progressive loss of beta-cell function leads to worsening glucose tolerance in first-degree relatives of subjects with type 2 diabetes. Diabetes Care 30, 677–682 (2007).
Steele-Mortimer, O., Méresse, S., Gorvel, J. P., Toh, B. H. & Finlay, B. B. Biogenesis of Salmonella typhimurium-containing vacuoles in epithelial cells involves interactions with the early endocytic pathway. Cell. Microbiol. 1, 33–49 (1999).
Acknowledgements
The authors thank Ms. Kishiko Osaka and Ms. Naomi Okamoto for the experimental assistance. This work was supported by grants from PRESTO, Japan Science and Technology Agency (to F.K.) and from Technology Development Program for Advanced Measurement and Analysis, JST (to M.M.).
Author information
Authors and Affiliations
Contributions
T.S. designed and performed all of the experiments, analyzed the data and wrote the paper; F.K. contributed to the design of the study and the revision of the manuscript; M.M. contributed to the conception and design of the study and the preparation of the manuscript. All authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Movie S1
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Sugawara, T., Kano, F. & Murata, M. Rab2A is a pivotal switch protein that promotes either secretion or ER-associated degradation of (pro)insulin in insulin-secreting cells. Sci Rep 4, 6952 (2014). https://doi.org/10.1038/srep06952
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep06952
This article is cited by
-
The Role of Rab GTPases in the development of genetic and malignant diseases
Molecular and Cellular Biochemistry (2024)
-
Potential molecular mechanism of ACE gene at different time points in STEMI patients based on genome-wide microarray dataset
Lipids in Health and Disease (2019)
-
Establishment and phenotyping of disease model cells created by cell-resealing technique
Scientific Reports (2017)
-
The impact of erdosteine on cisplatin-induced ototoxicity: a proteomics approach
European Archives of Oto-Rhino-Laryngology (2017)
-
Rab1A mediates proinsulin to insulin conversion in β-cells by maintaining Golgi stability through interactions with golgin-84
Protein & Cell (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.