Abstract
SIRT1, a NAD+ dependent class III deacetylase, takes part in many important biological processes. Previous studies show that SIRT1 is overexpressed in some cancers and plays an essential role in tumorigenesis. However, the association between SIRT1 and colorectal cancer (CRC) is still unclear. We found that many CRC specimens had strong SIRT1 expression, which had an obvious correlation with poor prognosis of CRC patients. Meanwhile, SIRT1 expression had a co-localization with CD133, a current universal marker to characterize colorectal cancer stem cells (CSCs). In vitro studies also revealed that SIRT1 was overexpressed in colorectal CSC-like cells. Moreover, SIRT1 deficiency decreased percentage of CD133+ cells, attenuated the abilities of colony and sphere formation and inhibited tumorigenicity in vivo in CRC cells. Further study demonstrated that the expressions of several stemness-associated genes, including Oct4, Nanog, Cripto, Tert and Lin28, were reduced by SIRT1 knockdown in CRC cells. Taken together, our findings suggest that SIRT1 plays a crucial role in keeping the characteristics of CSCs cells. SIRT1 is a potential independent prognostic factor of CRC patients after tumor resection with curative intent and will contribute to providing a promising new approach to target at CSCs in CRC treatment.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is a kind of cancer causing by uncontrolled cells growth in the colon or rectum. Although CRC has a fully-understood genetic risk1,2,3,4,5, it is still the third most common cancer in the world, with nearly 1.4 million new cases in 20126. In America, CRC is one of the four major cancers, with 9% of cancer deaths7. In Europe, the five-year survival rate of CRC is less than 60% and a third of patients die from it1. The principal treatment of CRC is operation combined with postoperative chemotherapy and radiation therapy8,9,10. However, radical cure for recurrent and metastasis CRC still is a major difficulty. Treatment status indicates that there is a subpopulation of cancer cells, in other words, cancer stem cells (CSCs), which cannot be eradicated by current therapies.
CSCs are a rare population of cancer cells which possess the ability of self-renewal and differentiation into multiple cell types. These cells can initiate and sustain tumor growth11. Meanwhile, CSCs have strong resistances towards chemotherapeutic agent and radiation therapy12,13,14. Owing to the ability of tumor formation and maintenance, CSCs are considered to be responsible for the poor prognosis. The CSCs subpopulation of CRC cells was also identified. These CSCs promoted the CRC progression and recurrence10,15. Increasing therapies targeted at CSCs have attracted tremendous attentions in recent years.
SIRT1, the human homolog of Sir2, is a member of sirtuins family. SIRT1 is a NAD+ dependent class III deacetylase (HDAC) which can deacetylate both histone and non-histone proteins. SIRT1 takes part in numerous cellular processes by the deacetylation of specific substrates. Previous evidence suggests that SIRT1 down regulates the activation of p53 as a transcription factor by deacetylating the C-terminal Lys120, Lys164 and Lys382 residues16,17,18. Moreover, SIRT1 influences cell survival by deacetylating Ku70, Bax16,19 and E2F120. SIRT1 also has an impact on senescence21, differentiation22,23 and oxidative stress resistance24. In addition, SIRT1 is considered as an essential role in tumorigenesis25,26. Recent studies demonstrate that SIRT1 is overexpressed in some cancers, such as prostate cancer27, breast cancer28,29 and leukemia lymphoblasts22. However, the function of SIRT1 in tumor initiation and progression is still under debate30. Meanwhile, SIRT1 also plays an essential role in maintaining the self-renewal ability and pluripotency of embryonic stem cells (ESCs)23,31,32. SIRT1 maintains the properties of ESCs by taking part in the Oct4-SIRT1-p53 axis32, and/or regulating the expression of Nanog23. These studies indicate that SIRT1 may have underlying association with CSCs. In the present work, we firstly detected whether the SIRT1 expression of cancer tissue had associations with prognosis and distribution of CSC-like cells in human CRC patients. Then we further explored the influences of SIRT1 on CSCs in tumorigenesis and their underlying mechanisms in CRC cell lines.
Results
Strong SIRT1 expression of tumor tissues had a correlation with poor prognosis in CRC patients
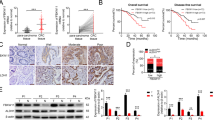
Data from a total of 102 patients with colorectal adenocarcinoma were evaluated. Demographics of 102 CRC patients are listed in Table 1. The expression of SIRT1 was detected by immunohistochemical analysis. Data revealed that SIRT1 had a nuclear localization in CRC tissues and CRC tissue had stronger SIRT1 expression compared with that of corresponding pericarcinomatous tissue (Figure 1A). Then SIRT1 expression was scored according to the SIRT1+ tumour glands. Tumours with more than 50% (including 50%) SIRT1+ glands were defined as SIRT1-Strong, whereas less than 50% stained glands were considered as SIRT1-Weak. Immunohistochemical staining of SIRT1 showed that CRC specimens with weak and strong expression were 58 and 44 samples, respectively (Figure 1B). Results suggested that SIRT1 had no significant correlation to clinicopathological features such as age, gender, location and T-category. On the other hand, the altered SIRT1 expression was significantly correlated with the number of cancer sites (P = 0.03), metastases (P = 0.02) and metastatic sites (P = 0.02). By using the Kaplan-Meier analysis, we found that strong SIRT1 expression of tumor tissue had an obvious association with poor prognosis in CRC patients (Log-rank test, P<0.01, Table 1, Figure 1C and D). Applying the multivariate Cox proportional hazards regression, SIRT1-Strong expression was significantly related to a lower survival rate, which displayed an independent relative risk of 2.459 compared with the weak group (P = 0.003, Table 2). While gender, age and metastatic had no significant association with the CRC patient outcome. TNM-classification of T4 was the significant variable in the multivariate analysis with a relative risk of 2.762 compared with the T2 and T3 group (P = 0.002). These data indicated that SIRT1 represented as an independent prognostic factor, which related to the prognosis of CRC patient.
SIRT1 expression in CRC tissue correlates with CRC patient survival and stem cell marker CD133 expression.
(A) Immunohistochemical results. Images showed that the levels of SIRT1 expression in colorectal pericarcinomatous and cancer tissues. (B) Levels of SIRT1 staining were classified as weak or strong according to the percent of SIRT1+ glands. (C) Time to progression curve of CRC patients. Patients with strong SIRT1 expression showed a higher proportion of progression compared with the weak SIRT1 expression (P<0.01). (D) The overall survival curve of CRC patients. Patients with strong SIRT1 expression exhibited a reduced lifespan after tumor resection compared with the weak SIRT1 expression (P<0.01). (E) Double-labeling immunofluorescence analysis demonstrated that SIRT1 expression had a correlation with CD133 expression. Scale bars were shown. Results are representatives of three independent experiments.
Meanwhile, we detected the expression of a common stem cell marker CD133 (Human prominin-1) in these specimens. CD133 was expressed by various stem and progenitor cells originating from diverse sources, which was used as a stem cell marker universally33. Double-labeling immunofluorescence analysis showed that CRC specimens with strong SIRT1 expression had more CD133-positive cells compared to those with weak SIRT1 expression (Figure 1E). This phenomenon demonstrated that SIRT1 had underlying association with CSCs in CRC.
SIRT1 is overexpressed in CSC-like cells in CRC
Then we attempted to confirm the association between SIRT1 and CSCs in CRC cell lines. Spheroid culture is a common method to enrich CSC-like cells34. After the stem cell conditioned culture, mammospheres were harvested. Immunofluorescence staining analysis showed that the SIRT1 was enriched in mammospheres in SW620 and SW480 cells compared to the controls (Figure 2A). Meanwhile, the mRNA and protein levels of SIRT1 were obviously increased in the mammospheres compared to the adherent cells in SW620 and SW480 cells (Figure 2B and C).
Increased SIRT1 expression in colorectal CSC-like cells.
(A) Immunofluorescence staining of SIRT1 in normal adherent cells and mammospheres of SW620 and SW480 cells. Scale bars (50 μm) were shown. (B) Quantitative real time PCR analysis of SIRT1 mRNA levels in normal adherent cells and mammospheres of SW620 and SW480 cells. (C) Western blotting and quantitative analysis of SIRT1 and GAPDH in normal adherent cells and mammospheres of SW620 and SW480 cells. (D) Representative flow cytometry plots and quantitative analysis showing the percentage of CD133+ cells in SW620 and SW480 cells. (E)Quantitative real time PCR analysis of SIRT1 mRNA levels in SW620 and SW480 cells. (F) Western blotting and quantitative analysis of SIRT1 and GAPDH in SW620 and SW480 cells. (G) Western blotting and quantitative analysis of SIRT1 and GAPDH in CD133- and CD133+ cells separated by CD133 microbeads or control cells of HCT116, SW620 and SW480 cells. All data are representative of three independent experiments. Significance: *P<0.05, **P<0.01, compared with the controls.
We further evaluated the relationship between the expression of SIRT1 and CD133 in CRC. SW620 and SW480 were derived from different stages of CRC of the same patient. Flow cytometry analysis that SW620 cells had higher percentage of CD133+ cell than SW480 cells (35.5% ± 2.0% versus 20.0% ± 3.0%) (Figure 2D). Then we detected SIRT1 expression in these two cell lines. Results showed that SIRT1 expression of SW620 cells was higher than SW480 cells at both mRNA and protein levels (Figure 2E and F). We further separated three kinds of CRC cells (HCT116, SW620 and SW480) into the CD133+ cells and CD133− cells by magnetic bead sorting. The identification of flow cytometry revealed that CD133+ cells had a purity >90% and CD133− cells had a purity >99%. Western blotting results demonstrated that SIRT1 had a higher expression in the CD133+ cells compared to the CD133− cells in these three cell lines (Figure 2G). These data demonstrated that SIRT1 had a high expression in CSC-like cells in CRC.
SIRT1 deficiency reduces percentage of CD133+ cells and the formation of colony and sphere in CRC cells
According to the positive-relationship between SIRT1 and CD133, we attempted to explore whether SIRT1 deficiency had an influence on the stemness of colorectal CSCs. Firstly, we used SIRT1 inhibitor nicotinamide (NAM, 20 mM) to pre-treat HCT116, SW620 and SW480 cells for 24 hours. Flow cytometry assay demonstrated that NAM significantly decreased the percentage of CD133+ cell in these three CRC cells (Figure 3A). Meanwhile, 20 mM NAM did not lead to the increase of apoptosis in these cells (figures were not shown). Then control cells and NAM pre-treated cells were seeded at 6-well plates in a density of 2000 cells/well. NAM pre-treated cells displayed lower ability of colony formation compared to the controls (Figure 3B). Then, we separated HCT116, SW620 and SW480 cells into the CD133+ cells and CD133− cells using the magnetic bead sorting. These cells were detected their abilities of colony and sphere formation in the absence or presence of 20 mM NAM. The results showed that CD133+ cells had stronger abilities of colony and sphere formation. Meanwhile, these abilities were apparently down-regulated when cells were treated by NAM. Moreover, there were no significant differences between CD133+ cells treated with NAM and CD133− cells treated with NAM in the abilities of colony and sphere formation (Figure 3C and D).
Pharmacological inhibition of SIRT1 reduces percentage of CD133+ cells and the colony formation in CRC cells.
(A) Representative flow cytometry plots showing percentage of CD133+ cells in control cells and cells treated with NAM (20 mM) of HCT116, SW620 and SW480 cells. (B) The colony formation analysis of control cells and cells treated with NAM (20 mM) of HCT116, SW620 and SW480 cells. (C) The colony formation analysis of CD133+ cells and CD133- cells in the absence or presence of 20 mM NAM in HCT116, SW620 and SW480 cells. (D) The sphere formation analysis of CD133+ cells and CD133− cells in the absence or presence of 20 mM NAM in HCT116, SW620 and SW480 cells. All data are representative of three independent experiments. Significance: *P<0.05, **P<0.01, ***P<0.001, compared with the controls.
To further confirm above findings, we used ShRNA to suppress the expression of SIRT1. HCT116 and SW620 cells were transducted with anti-SIRT1 lentivirus vectors (ShSIRT1-1, ShSIRT1-2) or scrambled shRNA (Scr-ShRNA). The shRNAs effective inhibition of SIRT1 expression was confirmed by western blotting analysis (Figure 4A). The flow cytometry results illustrated that the cells transducted with SIRT1 shRNAs remarkably decreased percentages of CD133+ cells in both HCT116 and SW620 cells (Figure 4B). The colony formation assay outcome indicated that the ability of colony formation distinctly dropped following with the down-regulation of SIRT1 (Figure 4C). Simultaneously, sphere formation ability also had a positive-correlationship with the SIRT1 expression. SIRT1 knockdown obviously reduced the sphere formation in HCT116 and SW620 cells (Figure 4D). These data illustrated that SIRT1 inhibition significantly reduced the percentage of CD133+ cells and the abilities of colony and sphere formation in CRC cells.
SIRT1 knockdown reduces percentage of CD133+ cells and represses colony and sphere formations in CRC cells.
(A) Western blotting of SIRT1 and GAPDH in HCT116 and SW620 cells transduced with SIRT1 shRNAs (ShSIRT1-1 and ShSIRT1-2) or Scr-ShRNA. (B) Representative flow cytometry plots showing percentage of CD133+ cells in control cells and cells transduced with shRNAs or Scr-ShRNA of HCT116 and SW620 cells. (C) The colony formation analysis of control cells and cells transduced with shRNAs or Scr-ShRNA of HCT116 and SW620 cells. (D) The sphere formation analysis of control cells and cells transduced with shRNAs or Scr-ShRNA of HCT116 and SW620 cells. All data are representative of three independent experiments. Significance: **P<0.01, ***P<0.001, compared with the controls.
SIRT1 deficiency results in lower CRC tumorigenicity in vivo
To further explore the impact of SIRT1 knockdown on tumor forming in vivo, SW620 cells were transducted with SIRT1 ShRNAs or Scr-ShRNA. These cells were injected subcutaneously into the nude mice. Results demonstrated that 2.0 × 106 cells infected with ShSIRT1-1 or ShSIRT1-2 were able to initiate the tumor formation, while 2.5 × 105 control SW620 cells were enough to initiate the tumor growth (Supplementary Table 1). Then 5.0 × 106 infected SW620 cells were injected subcutaneously into nude mice. The 5.0 × 106 SW620 cells were operated as control. The period of observation was from the 1st day after injection to the 18th day. The growth of tumors was monitored with measurements of two diameters35. The observation of tumor growth illustrated that cells infected by Scr-ShRNA had a similar growth mode compared with the control SW620 cells, while the two SIRT1 ShRNA infected cells grew in another analogous manner obviously slower than controls (Figure 5A). Then tumors were harvested on the 18th day (Figure 5B). Tumor further examination displayed that SIRT1 knock-down cells exhibited obvious decrease of tumor volume and tumor weight compared with those of controls (Figure 5C and D). These results demonstrated that SIRT1 knockdown using SIRT1 ShRNA decreased the tumorigenicity of CRC in vivo.
SIRT1 knockdown using SIRT1 shRNA results in lower tumorigenicity in vivo.
(A) The BALB/c nude mice were divided into four groups randomly, with control group (n = 8) receiving 5 × 106 SW620 cells and other groups receiving 5 × 106 treated cells. Treated cells were SW620 cells which were transduced with SIRT1 ShRNAs (ShSIRT1-1 and ShSIRT1-2) or Scr-ShRNA. The tumor growth rate of the experimental mice measured by tumor volumes. Significance: **P<0.01, *P<0.05, means ShSIRT1-1 SW620 group vs. control SW620 group; ##P<0.01, #P<0.05, means ShSIRT1-2 SW620 group vs. control SW620 group. (B) Photographic illustration of tumor nodules excised from experimental mice on the harvesting time (18 days). (C) Tumor volumes of the experimental mice. The ShSIRT1-1 and ShSIRT1-2 groups were significantly decreased (*, P<0.05) compared to the controls. (D) Tumor weights of the experimental mice. The ShSIRT1-1 and ShSIRT1-2 groups were significantly decreased (*, P<0.05; **, P<0.01) compared to the control.
SIRT1 deficiency affects the expressions of p53 and stemness-associated genes in CRC
It was reported that p53 was involved in the formation of induced pluripotent stem (iPS) cell. The suppression of p53 increased the efficiency of iPS cell generation in both mouse and human36. Meanwhile, SIRT1 down regulated p53 activation in CML37. Therefore, we detected the expression of p53 when SIRT1 was knocked down in CRC cells. The result showed that p53 expression had an increase in SW620 when SIRT1 was knocked down (Figure 6A, B and C). Then we further explore the mechanism of SIRT1 in keeping the stemness by detecting the expression of several associated genes, including Oct4, Nanog, Sox2, Bmi-1, Cripto, Tert, Smo and Lin28. Quantitative real time PCR results showed that the mRNA levels of Oct4, Nanog, Cripto, Tert and Lin28 all had apparent reductions when SIRT1 was inhibited. However, SIRT1 inhibition had no significant impact on the expressions of Sox2, Smo and Bmi-1 (Figure 6D). These results revealed that SIRT1 inhibition led to the increase of p53 expression and the decrease of several stemness-associated genes expressions in CRC cells.
SIRT1 deficiency affects the expression of p53 and genes involved in self-renewal of SW620 cells.
(A) Quantitative real time PCR analysis of p53 mRNA levels in control SW620 cells and cells transduced with shRNAs or Scr-ShRNA. (B) Western blotting of p53 and GAPDH of control SW620 cells and cells transduced with shRNAs or Scr-ShRNA. (C) Quantitative western blotting analysis of p53 protein levels in control SW620 cells and cells transduced with shRNAs or Scr-ShRNA. (D) Quantitative real time PCR analysis of the mRNA levels of stemness-associated genes (Oct4, Nanog, Cripto, Tert, Lin28, Sox2, Smo and Bmi-1) in control SW620 cells and cells transduced with shRNAs or Scr-ShRNA. All data are representative of three independent experiments. Significance: *P<0.05, **P<0.01, compared with the controls.
Discussion
It has been well accepted that SIRT1 played a considerable role in many important biological processes such as apoptosis, senescence and metabolism. Here we reported that SIRT1 plays a crucial role in tumorigenesis of CRC by keeping the stemness of CSC-like cells. Immunochemical analysis demonstrated that SIRT1 was overexpressed in CRC tissues compared with their pericarcinomatous tissues. Moreover, CRC patients with high SIRT1 expression tended to exhibit poor prognosis. Meanwhile, we observed co-localization of SIRT1 with a stem marker CD133 in CRC tissues, indicating that SIRT1 had an underlying association with CSC-like cells of CRC. In vitro study showed that CSC-like cells had higher SIRT1 expression in CRC cell lines. More importantly, the stemness of CSC-like cells of CRC attenuated when SIRT1 was inhibited. Further studies revealed that inhibition of SIRT1 in CRC cells resulted in the increase of p53 expression and the decrease of several stemness-associated genes, including Oct4, Nanog, Cripto, Tert and Lin28.
The role of SIRT1 in cancer is under debate30,38,39. Many previous studies have reported that high SIRT1 expression in some cancers, including prostate cancer27, colorectal cancer28,39, breast cancer28,29, leukemia lymphoblasts22, glioblastoma40 and skin carcinogenesis41. In breast cancer, SIRT1 was reported to contribute to the tumorigenesis by inactivating tumor suppressor genes, such as cyclin G229. Inhibition of SIRT1 also caused gene re-expression of tumor suppressor genes with full retention of DNA hypermethylation42. At the same time, the balance between SIRT1 and DBC1 was lost in various cancers, such as breast cancer43 and acute myeloid leukemia44. On the other hand, RH Wang, et al. reported that activation of SIRT1 reduced tumorigenesis of various cancers via enhancing DNA damage response and maintain genome stability in mice model. Meanwhile, SIRT1 was lower expressed in HCC tissues compared to the corresponding pericarcinomatous tissues in the clinical samples45. These researches indicate that SIRT1 may play different roles in the different kinds of cancers. Menssen A, et al. reported that in colorectal cancer, SIRT1 was activated by oncoprotein c-MYC and could promote c-MYC function in turn46. The positive feedback loop formed by c-MYC, SIRT1 and DBC1 contributed to the tumor development46. In our study, we also found that SIRT1 was overexpressed in colorectal tumor tissues compared to their pericarcinomatous tissues. More importantly, clinical data analysis showed that high SIRT1 expression of tumor tissue had a significant correlation with poor prognosis in CRC patients. Cox's proportional hazards regression model further indicated that SIRT1 is an independent factor of prognosis of CRC patients. And the influence of SIRT1 on CSCs plays a crucial role on this phenomenon.
A lot of work has been done about the maintenance of characteristics of CSCs. Marhold M, et al. reported that HIF-1 alpha deregulated the PI3K/AKT/mTOR pathway of CSC to attenuate stem cells metabolism and growth in prostate cancer47. Meanwhile, both Oct4 and Nanog overexpression enhanced the expression of EMT-related genes in CSC in breast cancer. The overexpression of Oct4 and Nanog accelerated the invasiveness of CSC48. Our above results indicated that SIRT1 played an important role in keeping the properties of CSC-like cells in CRC. Further researches showed that the inhibition of SIRT1 in CRC cells resulted in the increase of p53 expression and reduction of various stemness-associated genes, including Oct4, Nanog, Cripto, Tert and Lin28.
p53, a critical tumor suppressor, plays an important role in tumorigenesis by regulating the expressions of several apoptosis-, cell cycle-, senescence-associated genes49. Recent studies demonstrated that p53 had a strong impact on stem cells. p53 was considered as a crucial barrier of iPS cell generation36. Akita H, et al. also found that c-MYC enhanced the self-renewal capacity of liver CSCs in a p53-dependent manner50. SIRT1 could inhibit p53 activation induced by genotoxic stress44. WY Chen, et al. also reported that tumor suppressor HIC1 regulated p53-dependent DNA-damage responses by the modulation of SIRT151. Previous study suggested that SIRT1 could regulate p53 activation by various pathways. As a class III histone deacetylase, SIRT1 can deacetylate some lysine residues of the tumor p53 protein, which leads to the instability and inactivation of p5317,18,25,52,53. Han, M. K. et al also reported that SIRT1 could upregulate Nanog expression in mouse ESCs by controlling ROS-related p53 subcellular localization23. Our research found that silencing SIRT1 led to the increases of mRNA and protein levels of p53 and the decrease of Nanog mRNA level in CRC cells. However, the clear mechanism remains to be confirmed.
SIRT1 has a complicated association with Oct4, which as an important transcription factor is frequently used as a marker for undifferentiated cells54. Low expression of Oct4 caused the differentiation of cells55. It has been reported that Oct4 can directly bind to the promoter region of SIRT1 to activate the SIRT1 expression32. Moreover, Oct4 could form Oct4-SIRT1-p53 axis to regulate pluripotency and DNA damage pathways to maintain the pluripotency and genomic stability of hESCs32. Our data showed that the inhibition of SIRT1 had a down-regulation on the expression of Oct4, which indicated there was a reciprocal regulation between SIRT1 and Oct4 through a feedback loop. By examining the published protein sequence analysis data (InterPro) of the function domain of SIRT1 protein, we found that SIRT1 has not DNA binding domain. It revealed that SIRT1 did not regulate the Oct4 expression by directly binding to Oct4 promoter. Additionally, Lingxia Wang, et al. reported that PCAF/SIRT1 balance played an important role in the regulation of Lin28 activity. Lin28 was acetylated by PCAF, which can be reversed by SIRT1. SIRT1 inhibitor NAM led to apparent decrease of Lin28 protein level56. Our result also demonstrated that Lin28 mRNA levels were reduced obviously when SIRT1 was knocked down in CRC cells. However, the underlying mechanisms by which SIRT1 regulates the mRNA expressions of these stemness-associated genes need to be further explored.
In conclusion, clinical samples analysis revealed that high expression of SIRT1 was associated with poor prognosis in CRC patients. Further study suggested that SIRT1 was overexpressed in CSC-like cells of CRC and played a critical role in the tumorigenesis of CRC by maintaining stemness of CSC-like cells. All the results indicate that SIRT1 is a potential independent prognostic factor of CRC patients after tumor resection with curative intent and reveals a promising treatment targeting CSCs in CRC.
Methods
Ethics statement
The procedure of human sample acquisition and experiments were approved by the Hospital Research Ethics Committees of Ren Ji Hospital. All animal studies were approved by the Animal Care Committee of Shanghai Jiao Tong University. And all methods were carried out in accordance with the approved guidelines.
Clinical samples
Colorectal cancer samples were obtained from patients in Department of Gastrointestinal Surgery, Ren Ji Hospital. From January 2006 to December 2013, patients who were confirmed as CRC were recruited for prospective follow-up. All subjects signed an informed consent form. All experimental protocols were conducted in accordance with the Declaration of Helsniki of the World Medical Association.
Cell lines and culture
Human colorectal carcinoma cell lines HCT116, SW620 and SW480 were obtained from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), where they were authenticated. These procedures were based on cross species checks, DNA authentication and quarantine. Cells were maintained in RPMI1640 with 10% serum, penicillin (100 IU/ml), streptomycin (100 μg/ml) (Invitrogen). Cells were grown at 37°C in an atmosphere containing 5% CO2. Nicotinamide (NAM) was purchased from Sigma.
Mice
Male BALB/c nude mice (6–8 weeks, 18–22 g) were purchased from Shanghai SLAC Laboratory Animals Co.,Ltd (Shanghai, China). The mice were housed under specific pathogen-free conditions. All animal experiments were carried out in accordance with the guidelines of China Animal Welfare Legislation. All efforts were made to minimize suffering.
Cell transfection and selection
HCT116 and SW620 cells were transduced with lentivirus vectors expressing SIRT1 ShRNA. These cells were cultured for 24 hours, followed by the exposures to virus-containing supernatants (MOI = 20) via polybrene. Cells were selected by puromycin (2 μg/ml) (Sigma) 48 hours later.
ShRNA1-1 and ShRNA1-2 were the anti-SIRT1 ShRNA sequences. To turn off the effect of ShRNA, a scrambled sequence was used as a control. ShRNA sequences were as follows:
ShSIRT1-1: GAAGTGCCTCAGATATTAA
ShSIRT1-2: GTTGACCTCCTCATTGTTA
ShScramble: GCGCGCTTTGTAGGATTCG.
Real time PCR analysis
Total RNA was isolated from CRC cells using Trizol reagent method. 1 μg RNA was reversely transcribed into cDNA. The mRNA level was measured by real time PCR. Primers used in this work were as follows:
SIRT1 forward primer: TGCTGGCCTAATAGAGTGGCA
SIRT1 reverse primer: CTCAGCGCCATGGAAAATGT
Oct4 forward primer: GAGCAAAACCCGGAGGAGT
Oct4 reverse primer: TTCTCTTTCGGGCCTGCAC
Sox2 forward primer: GGCAGCTACAGCATGATGCAGGAGC
Sox2 reverse primer: CTGGTCATGGAGTTGTACTGCAGG
Nanog forward primer: GCTTGCCTTGCTTTGAAGCA
Nanog reverse primer: TTCTTGACTGGGACCTTGTC
Cripto forward primer: TACCTGGCCTTCAGAGAT
Cripto reverse primer: CCAGCATTTACACAGGGAACAC
Tert forward primer: TGTGCACCAACATCTACAAG
Tert reverse primer: GCGTTCTTGGCTTTCAGGAT
Lin28 forward primer: GGAGGCCAAGAAAGGGAATA
Lin28 reverse primer: CCGCCCCATAAATTCAAGAT
Smo forward primer: ATCTCCACAGGAGAGACTGGTTCGG
Smo reverse primer: AAAGTGGGGCCTTGGGAACATG
Bmi-1 forward primer: GGAGACCAGCAAGTATTGTCCTTTTG
Bmi-1 reverse primer: CATTGCTGCTGGGCATCGTAAG
p53 forward primer: AAGTCTGTGACTTGCACGTACTCC
p53 reverse primer: GTCATGTGCTGTGACTGCTTGRTAG
GAPDH forward primer: AATGGGCAGCCGTTAGGAAA
GAPDH reverse primer: GCGCCCAATACGACCAAATC
Western blotting assay
Cells were lysed in the cell lysates (Thermo) supplemented with protease inhibitors PMSF and Cocktail (Roche). Proteins were separated in 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose NC membranes (0.22 μm, Whatman). Membranes were blocked with blocking buffer (Li-COR), sequentially incubated in primary antibodies and secondary antibody. The primary antibodies included rabbit anti-human SIRT1 (Abcam) and rabbit anti-human GAPDH (CST). The secondary antibody was Goat Anti-Rabbit IgG (Invitrogen). Protein levels were measured by gray value with Quantity One software.
Immunofluorescence staining
Cells were grown on sterile glass coverslips. After 24 hours, these cells were fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, blocked with 5% goat serum and sequentially incubated in primary antibody against SIRT1 (Rabbit monoclonal to SIRT1, Abcam) and Secondary Antibody FITC-conjugated Goat Anti-Rabbit IgG (CST). Cells were washed in PBS twice and stained with DAPI (Sigma). Finally, examined under a TCS_SP5 (Leica) confocal microscope.
Colony formation assay
The cells which more than 95% cells with viability were seeded in RPMI1640 with 10% FBS at a density of 2000 cells/well on 6-well plates (Corning). After 14 days, the plates were stained with crystal violet (Sigma) and counted the number of the colonies. The number of colonies was used to assess the ability of holoclone formation.
Sphere formation assay
The cells were seeded at a density of 5000 cells/well on ultra low attachment 6-well plates (Corning). Cells were cultured in serum-free DMEM/F12 supplemented with 100 IU/ml penicillin, 100 μg/ml streptomycin, 10 ng/ml basic FGF, 20 ng/ml EGF, 2% B27 (Invitrogen). Cells with a three dimensional spherical structure (spheres) were collected 7 to 10 days later by gentle centrifugation (300 g/min, 3 min). The spheres were dissociated into single cells with trypsin-EDTA. The resulting single cells were re-suspended in the above medium to re-form spheres.
Magnetic separation
CD133 positive cells were isolated by magnetic bead sorting using the MidiMACS system (Miltenyi). The cells were prepared in a single-cell suspension in 300 μl of running buffer per 108 total cells. Then cells were incubated with CD133 MicroBeads for 30 minutes in the refrigerator (2–8°C). The CD133+ cells were enriched by LS MACS Column and MACS Separator as the manufacturer's instructions. The purity of the cells was detected by flow cytometry (BD) using antibody against human CD133/255 (Miltenyi).
Tumorigenicity experiments in vivo
The SW620 cells and SW620 cells transducted with SIRT1 ShRNAs or Scr-ShRNA were used in tumorigenicity experiments. The cells were prepared in suspensions of single-cell type using 100 μl PBS. 5.0 × 106 cells were injected into male BALB/c nude mice subcutaneously. Mice were examined once every 6 days. Tumor growth was evaluated by measuring the two diameters of the tumor nodules. Tumor volumes were calculated by the formula: Volume = XY2/2, where ‘X’ was the maximum diameter and ‘Y’ was the minimal diameter. Experimental mice were killed at 18 days after injection. Tumors were harvested for the further experiments.
Immunohistochemistry analysis
The clinical samples were deparaffinized with xylene and rehydrated through graded alcohol series. For SIRT1 immunohistochemistry, antigen retrieval was conducted. Tissue sections were treated with microwave in 15 min in Citra Solution. 3% H2O2 (20 min) were used to block endogenous peroxidase. Tissue sections were sequentially blocked and incubated in 5% goat serum and primary antibodies. Primary antibodies against SIRT1 (Rabbit monoclonal to SIRT1, Sigma) (dilution 1:300) were applied and maintained overnight at 4°C. Secondary antibodies anti-mouse IgG and anti-rabbit IgG (Invitrogen) were applied for 30 min at 37°C. The slides were visualized by DAB (5 min) and hematoxylin counterstain (Invitrogen).
Evaluation of SIRT1 immunohistochemistry
SIRT1 expression was evaluated on entire tissue sections of colorectal cancer specimens. Five fields of every section were viewed. Every specimen was given a score of 0, < 50%, or ≥ 50% SIRT1+ expression. Strong was defined as a score of ≥ 50% SIRT1+ expression. Scores of 0 and < 50% were considered to be weak. Immunohistochemistry results were evaluated independently by two skilled pathologists blinded from other data.
Statistics
All values from independent experiments were reported as the mean ± SEM. Student's t-test analysis and One-way ANOVA were performed to determine statistical significance using GraphPad Prism software (Version 6.0). Log-rank test was performed for categorical data of survival rates analysis using GraphPad Prism. Multivariate analysis was conducted on the Cox regression model. Statistical procedures were performed using SPSS. Statistical significance was set at p < 0.05.
References
Cunningham, D. et al. Colorectal cancer. Lancet 375, 1030–47 (2010).
Fearon, E. R. & Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 61, 759–67 (1990).
Fearon, E. R. & Jones, P. A. Progressing toward a molecular description of colorectal cancer development. FASEB J 6, 2783–90 (1992).
Radtke, F. & Clevers, H. Self-renewal and cancer of the gut: two sides of a coin. Science 307, 1904–9 (2005).
Watson, A. J. & Collins, P. D. Colon cancer: a civilization disorder. Dig Dis 29, 222–8 (2011).
Siegel, R., Desantis, C. & Jemal, A. Colorectal cancer statistics, 2014. CA Cancer J Clin 64, 104–17 (2014).
Siegel, R., Naishadham, D. & Jemal, A. Cancer statistics, 2013. CA Cancer J Clin 63, 11–30 (2013).
Nelson, H. et al. Guidelines 2000 for colon and rectal cancer surgery. J Natl Cancer Inst 93, 583–96 (2001).
O'Connell, J. B., Maggard, M. A. & Ko, C. Y. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst 96, 1420–5 (2004).
O'Brien, C. A., Pollett, A., Gallinger, S. & Dick, J. E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 445, 106–10 (2007).
Reya, T., Morrison, S. J., Clarke, M. F. & Weissman, I. L. Stem cells, cancer and cancer stem cells. Nature 414, 105–11 (2001).
Dean, M., Fojo, T. & Bates, S. Tumour stem cells and drug resistance. Nat Rev Cancer 5, 275–84 (2005).
Bao, S. et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444, 756–60 (2006).
Diehn, M. et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 458, 780–3 (2009).
Ricci-Vitiani, L. et al. Identification and expansion of human colon-cancer-initiating cells. Nature 445, 111–5 (2007).
Cohen, H. Y. et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305, 390–2 (2004).
Vaziri, H. et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107, 149–59 (2001).
Luo, J. et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107, 137–48 (2001).
Li, Y. et al. Bax-inhibiting peptide protects cells from polyglutamine toxicity caused by Ku70 acetylation. Cell Death Differ 14, 2058–67 (2007).
Wang, C. et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol 8, 1025–31 (2006).
Langley, E. et al. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J 21, 2383–96 (2002).
Audrito, V. et al. Nicotinamide blocks proliferation and induces apoptosis of chronic lymphocytic leukemia cells through activation of the p53/miR-34a/SIRT1 tumor suppressor network. Cancer Res 71, 4473–83 (2011).
Han, M. K. et al. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell 2, 241–51 (2008).
Brunet, A. et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011–5 (2004).
Brooks, C. L. & Gu, W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer 9, 123–8 (2009).
Liu, T., Liu, P. Y. & Marshall, G. M. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res 69, 1702–5 (2009).
Huffman, D. M. et al. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res 67, 6612–8 (2007).
Kuzmichev, A. et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci U S A 102, 1859–64 (2005).
Elangovan, S. et al. SIRT1 is essential for oncogenic signaling by estrogen/estrogen receptor alpha in breast cancer. Cancer Res 71, 6654–64 (2011).
Song, N. Y. & Surh, Y. J. Janus-faced role of SIRT1 in tumorigenesis. Ann N Y Acad Sci 1271, 10–9 (2012).
Ou, X. et al. SIRT1 deficiency compromises mouse embryonic stem cell hematopoietic differentiation and embryonic and adult hematopoiesis in the mouse. Blood 117, 440–50 (2011).
Zhang, Z. N., Chung, S. K., Xu, Z. & Xu, Y. Oct4 maintains the pluripotency of human embryonic stem cells by inactivating p53 through Sirt1-mediated deacetylation. Stem Cells 32, 157–65 (2014).
Fargeas, C. A., Corbeil, D. & Huttner, W. B. AC133 antigen, CD133, prominin-1, prominin-2, etc.: prominin family gene products in need of a rational nomenclature. Stem Cells 21, 506–8 (2003).
Vermeulen, L. et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci U S A 105, 13427–32 (2008).
Price, J. E., Polyzos, A., Zhang, R. D. & Daniels, L. M. Tumorigenicity and metastasis of human breast carcinoma cell lines in nude mice. Cancer Res 50, 717–21 (1990).
Hong, H. et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 460, 1132–5 (2009).
Li, L. et al. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell 21, 266–81 (2012).
Deng, C. X. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci 5, 147–52 (2009).
Bosch-Presegue, L. & Vaquero, A. The dual role of sirtuins in cancer. Genes Cancer 2, 648–62 (2011).
Liu, G. et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer 5, 67 (2006).
Hida, Y., Kubo, Y., Murao, K. & Arase, S. Strong expression of a longevity-related protein, SIRT1, in Bowen's disease. Arch Dermatol Res 299, 103–6 (2007).
Pruitt, K. et al. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet 2, e40 (2006).
Yu, E. J. et al. Reciprocal roles of DBC1 and SIRT1 in regulating estrogen receptor alpha activity and co-activator synergy. Nucleic Acids Res 39, 6932–43 (2011).
Sasca, D. et al. SIRT1 prevents genotoxic stress-induced p53 activation in acute myeloid leukemia. Blood 124, 121–33 (2014).
Wang, R. H. et al. Impaired DNA damage response, genome instability and tumorigenesis in SIRT1 mutant mice. Cancer Cell 14, 312–23 (2008).
Menssen, A. et al. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1 and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci U S A 109, E187–96 (2012).
Marhold, M. et al. HIF-1alpha Regulates mTOR Signaling and Viability of Prostate Cancer Stem Cells. Mol Cancer Res Oct 27, early online (2014).
Wang, D. et al. Oct-4 and nanog promote the epithelial-mesenchymal transition of breast cancer stem cells and are associated with poor prognosis in breast cancer patients. Oncotarget Oct 4, early online (2014).
Vazquez, A., Bond, E. E., Levine, A. J. & Bond, G. L. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov 7, 979–87 (2008).
Akita, H. et al. MYC Activates Stem-like Cell Potential in Hepatocarcinoma by a p53-Dependent Mechanism. Cancer Res 74, 5903–13 (2014).
Chen, W. Y. et al. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell 123, 437–48 (2005).
Prives, C. & Manley, J. L. Why is p53 acetylated? Cell 107, 815–8 (2001).
Tang, Y., Zhao, W., Chen, Y., Zhao, Y. & Gu, W. Acetylation is indispensable for p53 activation. Cell 133, 612–26 (2008).
Boyer, L. A. et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–56 (2005).
Niwa, H., Miyazaki, J. & Smith, A. G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 24, 372–6 (2000).
Wang, L. X., Wang, J., Qu, T. T., Zhang, Y. & Shen, Y. F. Reversible acetylation of Lin28 mediated by PCAF and SIRT1. Biochim Biophys Acta 1843, 1188–95 (2014).
Acknowledgements
This project was supported by the Key Basic Research Project of China (Grant NO. 2011CB966203); Major State Scientific Research Program of China (Grant NO. 2012CBA01303); National Natural Science Foundation of China (Grant NO. 31171321, 81201584, 81372312, 81301715); Special Funds for National key Sci-Tech Sepcial Project of China (Grant NO.2012ZX10002-016, 2012ZX10002011-011); Shanghai Science and Technology Committee (Grant NO. 12431900802, 12ZR1454200); Innovation Program of Shanghai Municipal Education Commission(14YZ041) and Science Fund for Creative Research Groups, NSFC, China (Grant NO. 81221061).
Author information
Authors and Affiliations
Contributions
X.J.C. and K.S. contributed equally to this manuscript. X.J.C., K.S., M.Z. and L.X.W. conceived and designed the study. X.J.C., S.F.J. and N.C. performed animal experiments. X.J.C. and X.Z. performed cell experiments. H.B.Z. and K.S. performed flow cytometry experiments. Z.S.W. collected the colorectal cancer samples. Y.X.X. and K.S. performed the statistical analysis of the clinical samples. X.J.C. wrote the draft manuscript. Z.M. and L.X.W. finalized the manuscript. All authors read and approve the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
supplemental material
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Chen, X., Sun, K., Jiao, S. et al. High levels of SIRT1 expression enhance tumorigenesis and associate with a poor prognosis of colorectal carcinoma patients. Sci Rep 4, 7481 (2014). https://doi.org/10.1038/srep07481
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07481
This article is cited by
-
Histone modifications in drug-resistant cancers: From a cancer stem cell and immune evasion perspective
Experimental & Molecular Medicine (2023)
-
Expression Changes of SIRT1 and FOXO3a Significantly Correlate with Oxidative Stress Resistance Genes in AML Patients
Indian Journal of Hematology and Blood Transfusion (2023)
-
Propofol inhibits colon cancer cell stemness and epithelial-mesenchymal transition by regulating SIRT1, Wnt/β-catenin and PI3K/AKT/mTOR signaling pathways
Discover Oncology (2023)
-
m6A methyltransferase KIAA1429 acts as an oncogenic factor in colorectal cancer by regulating SIRT1 in an m6A-dependent manner
Cell Death Discovery (2022)
-
Association of sirtuins (SIRT1-7) with lung and intestinal diseases
Molecular and Cellular Biochemistry (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.