-
PDF
- Split View
-
Views
-
Cite
Cite
Donald J. Lehmann, Mario Cortina-Borja, Donald R. Warden, A. David Smith, Kristel Sleegers, Jonathan A. Prince, Cornelia M. van Duijn, Patrick G. Kehoe, Large Meta-Analysis Establishes the ACE Insertion-Deletion Polymorphism as a Marker of Alzheimer's Disease, American Journal of Epidemiology, Volume 162, Issue 4, 15 August 2005, Pages 305–317, https://doi.org/10.1093/aje/kwi202
Close - Share Icon Share
Abstract
Apolipoprotein E ε4 (APOE*4) is the only fully established susceptibility allele for Alzheimer's disease. One of the most studied candidates is the insertion (I)/deletion (D) polymorphism (indel) of the gene for angiotensin I-converting enzyme (ACE). This study aimed to clarify its association with Alzheimer's disease. The meta-analysis included 39 samples, comprising 6,037 cases of Alzheimer's disease and 12,099 controls, using mainly primary data. Potential interactions with gender, age, ethnic group, and carrier status of the apolipoprotein E ε4 allele were all examined. D homozygotes were at reduced risk of Alzheimer's disease (odds ratio = 0.81, 95% confidence interval: 0.72, 0.90; corrected p = 0.0004); I homozygotes showed no association with Alzheimer's disease, while heterozygotes were at increased risk. Although there were clear differences among the three ethnic groups examined (North Europeans, South Caucasians, and East Asians), in all groups D homozygotes were at reduced risk. These results confirm the association of the angiotensin I-converting enzyme indel with Alzheimer's disease across diverse populations, although this is probably due to linkage disequilibrium with the true risk factor. Further, in North Europeans, both association and Hardy-Weinberg analysis suggested partial heterosis, that is, an increased risk for heterozygotes, due to a hidden interaction with another, as yet unknown, risk factor. This interaction warrants further investigation.
In 1999, Kehoe et al. (1) first proposed an association with Alzheimer's disease of the insertion (I)/deletion (D) polymorphism (indel) in intron 16 of the gene for angiotensin I-converting enzyme (ACE) on chromosome 17q23. The enzyme converts angiotensin I to angiotensin II, which is potently hypertensive. The D allele is associated with raised plasma levels of the enzyme (2, 3). Kehoe et al. (1) found that I positives, that is, DI + II, were at increased risk of Alzheimer's disease. The study comprised three samples, from Cardiff and London in Britain and Belfast in Northern Ireland, with strikingly similar results.
Since then, this potential association has been examined in over 40 samples (4, 5) (table 1) from around the world. Several studies replicated the report of increased risk for I positives, just a few contradicted it, and many found no association with Alzheimer's disease. One study suggested an effect of age on the association (6), and another proposed an influence of gender (7). An association with onset age of Alzheimer's disease has also been proposed (8).
Cohorts used in the meta-analysis of the ACE* insertion-deletion polymorphism in Alzheimer's disease
Reference . | Location of samples . | Alzheimer's disease (no.) . | Controls (no.) . | Alzheimer's disease diagnosis† . | Stratified data supplied and used‡ . |
|---|---|---|---|---|---|
| Alvarez et al. (63) | Asturias, Spain | 351 | 536 | NINCDS-ADRDA probable | Yes |
| Buss et al. (40) | Hamburg, Germany; Basel, Switzerland; and Brescia, Italy | 261 | 306 | NINCDS-ADRDA | Yes |
| Camelo et al. (13) | Colombia | 98 | 72 | NINCDS-ADRDA probable | Yes |
| Carbonell et al. (64) | London, United Kingdom | 80 | 65 | NINCDS-ADRDA | |
| Chapman et al. (65) | Tel Aviv, Israel | 49 | 40 | NINCDS-ADRDA probable | |
| Cheng et al. (66) | Taipei, Taiwan | 173 | 116 | NINCDS-ADRDA probable | |
| Crawford et al. (7) | Florida, United States | 171 | 175 | NINCDS-ADRDA | |
| Farrer et al. (6) | Moscow, Russia | 151 | 206 | NINCDS-ADRDA | |
| Farrer et al. (6) | Toronto, Canada; Miami, Florida; and Florence, Italy | 236 | 169 | NINCDS-ADRDA | |
| Fernández-Novoa et al. (14) | Galicia, Spain | 185 | 117 | NINCDS-ADRDA and DSM-IV | Yes |
| Hu et al. (67) | Japan | 132 | 148 | NINCDS-ADRDA probable | |
| Isbir et al. (68) | Turkey | 35 | 29 | NINCDS-ADRDA probable | |
| Kehoe et al. (11) | Bristol and London, United Kingdom | 296 | 95 | CERAD or NINCDS- ADRDA probable | Yes |
| Kehoe et al. (1) | Cardiff, Wales | 198 | 77 | NINCDS-ADRDA probable | |
| Kehoe et al. (1) | London, United Kingdom | 201 | 205 | NINCDS-ADRDA | Yes |
| Kehoe et al. (1) | Belfast, Northern Ireland | 209 | 198 | NINCDS-ADRDA probable and DSM-IV | Yes |
| Keikhaee et al. (15) | Iran | 132 | 107 | Yes | |
| Lendon et al. (16) | Manchester, England | 206 | 33 | NINCDS-ADRDA probable or CERAD | Yes |
| Lendon et al. (16) | Scotland | 167 | 357 | NINCDS-ADRDA | Yes |
| Mattila et al. (69) | West Finland | 77 | 67 | CERAD or NINCDS-ADRDA probable | Yes |
| Monastero et al. (70) | Palermo, Italy | 149 | 149 | NINCDS-ADRDA probable | |
| Myllykangas et al. (71) | Vantaa, Finland | 121 | 75 | CERAD | |
| Narain et al. (10) | Cambridge, England | 107 | 163 | CERAD | Yes |
| Narain et al. (10) | Oxford, England | 26 | 179 | CERAD | Yes |
| Palumbo et al. (72) | Italy | 96 | 40 | NINCDS-ADRDA probable | |
| Panza et al. (17) | South Italy | 141 | 268 | NINCDS-ADRDA probable | Yes |
| Perry et al. (41) | Alabama, United States (African Americans) | 111 | 78 | NINCDS-ADRDA probable | Yes |
| Prince et al. (23) (cohort 1); Kehoe et al. (11) | Sweden | 201 | 161 | NINCDS-ADRDA or CERAD | Yes |
| Prince et al. (23) (cohort 2); Kehoe et al. (11) | Sweden | 168 | 322 | NINCDS-ADRDA | Yes |
| Richard et al. (cohort 1) (73) | North Europe | 421 | 474 | NINCDS-ADRDA probable | Yes |
| Richard et al. (73) (cohort 2) | North Europe | 56 | 221 | NINCDS-ADRDA probable | Yes |
| Scacchi et al. (74) | Rome, Italy | 80 | 153 | NINCDS-ADRDA probable | Yes |
| Seripa et al. (18) | Rome, Italy | 97 | 101 | NINCDS-ADRDA probable | Yes |
| Seripa et al. (18) | Wisconsin and Kentucky, United States | 102 | 66 | NINCDS-ADRDA probable and Khachaturian | Yes |
| Sleegers et al. (9) | Netherlands | 236 | 5,889 | NINCDS-ADRDA | Yes |
| Wu et al. (75) | Beijing, People's Republic of China | 96 | 96 | DSM-IV | Yes |
| Yang et al. (76) | Shanghai, People's Republic of China | 188 | 227 | NINCDS-ADRDA probable | |
| Zuliani et al. (77) | Northeast Italy | 35 | 51 | NINCDS-ADRDA probable | Yes |
| OPTIMA* study§ | Oxfordshire, England | 198 | 268 | CERAD or NINCDS-ADRDA probable | Yes |
| Totals | 6,037 | 12,099 |
Reference . | Location of samples . | Alzheimer's disease (no.) . | Controls (no.) . | Alzheimer's disease diagnosis† . | Stratified data supplied and used‡ . |
|---|---|---|---|---|---|
| Alvarez et al. (63) | Asturias, Spain | 351 | 536 | NINCDS-ADRDA probable | Yes |
| Buss et al. (40) | Hamburg, Germany; Basel, Switzerland; and Brescia, Italy | 261 | 306 | NINCDS-ADRDA | Yes |
| Camelo et al. (13) | Colombia | 98 | 72 | NINCDS-ADRDA probable | Yes |
| Carbonell et al. (64) | London, United Kingdom | 80 | 65 | NINCDS-ADRDA | |
| Chapman et al. (65) | Tel Aviv, Israel | 49 | 40 | NINCDS-ADRDA probable | |
| Cheng et al. (66) | Taipei, Taiwan | 173 | 116 | NINCDS-ADRDA probable | |
| Crawford et al. (7) | Florida, United States | 171 | 175 | NINCDS-ADRDA | |
| Farrer et al. (6) | Moscow, Russia | 151 | 206 | NINCDS-ADRDA | |
| Farrer et al. (6) | Toronto, Canada; Miami, Florida; and Florence, Italy | 236 | 169 | NINCDS-ADRDA | |
| Fernández-Novoa et al. (14) | Galicia, Spain | 185 | 117 | NINCDS-ADRDA and DSM-IV | Yes |
| Hu et al. (67) | Japan | 132 | 148 | NINCDS-ADRDA probable | |
| Isbir et al. (68) | Turkey | 35 | 29 | NINCDS-ADRDA probable | |
| Kehoe et al. (11) | Bristol and London, United Kingdom | 296 | 95 | CERAD or NINCDS- ADRDA probable | Yes |
| Kehoe et al. (1) | Cardiff, Wales | 198 | 77 | NINCDS-ADRDA probable | |
| Kehoe et al. (1) | London, United Kingdom | 201 | 205 | NINCDS-ADRDA | Yes |
| Kehoe et al. (1) | Belfast, Northern Ireland | 209 | 198 | NINCDS-ADRDA probable and DSM-IV | Yes |
| Keikhaee et al. (15) | Iran | 132 | 107 | Yes | |
| Lendon et al. (16) | Manchester, England | 206 | 33 | NINCDS-ADRDA probable or CERAD | Yes |
| Lendon et al. (16) | Scotland | 167 | 357 | NINCDS-ADRDA | Yes |
| Mattila et al. (69) | West Finland | 77 | 67 | CERAD or NINCDS-ADRDA probable | Yes |
| Monastero et al. (70) | Palermo, Italy | 149 | 149 | NINCDS-ADRDA probable | |
| Myllykangas et al. (71) | Vantaa, Finland | 121 | 75 | CERAD | |
| Narain et al. (10) | Cambridge, England | 107 | 163 | CERAD | Yes |
| Narain et al. (10) | Oxford, England | 26 | 179 | CERAD | Yes |
| Palumbo et al. (72) | Italy | 96 | 40 | NINCDS-ADRDA probable | |
| Panza et al. (17) | South Italy | 141 | 268 | NINCDS-ADRDA probable | Yes |
| Perry et al. (41) | Alabama, United States (African Americans) | 111 | 78 | NINCDS-ADRDA probable | Yes |
| Prince et al. (23) (cohort 1); Kehoe et al. (11) | Sweden | 201 | 161 | NINCDS-ADRDA or CERAD | Yes |
| Prince et al. (23) (cohort 2); Kehoe et al. (11) | Sweden | 168 | 322 | NINCDS-ADRDA | Yes |
| Richard et al. (cohort 1) (73) | North Europe | 421 | 474 | NINCDS-ADRDA probable | Yes |
| Richard et al. (73) (cohort 2) | North Europe | 56 | 221 | NINCDS-ADRDA probable | Yes |
| Scacchi et al. (74) | Rome, Italy | 80 | 153 | NINCDS-ADRDA probable | Yes |
| Seripa et al. (18) | Rome, Italy | 97 | 101 | NINCDS-ADRDA probable | Yes |
| Seripa et al. (18) | Wisconsin and Kentucky, United States | 102 | 66 | NINCDS-ADRDA probable and Khachaturian | Yes |
| Sleegers et al. (9) | Netherlands | 236 | 5,889 | NINCDS-ADRDA | Yes |
| Wu et al. (75) | Beijing, People's Republic of China | 96 | 96 | DSM-IV | Yes |
| Yang et al. (76) | Shanghai, People's Republic of China | 188 | 227 | NINCDS-ADRDA probable | |
| Zuliani et al. (77) | Northeast Italy | 35 | 51 | NINCDS-ADRDA probable | Yes |
| OPTIMA* study§ | Oxfordshire, England | 198 | 268 | CERAD or NINCDS-ADRDA probable | Yes |
| Totals | 6,037 | 12,099 |
ACE, angiotensin I-converting enzyme gene; OPTIMA, Oxford Project to Investigate Memory and Ageing.
Based on criteria of the National Institute of Neurological and Communicative Disorders and Stroke–Alzheimer's Disease and Related Disorders Association (NINCDS-ADRDA) (26), Diagnostic and Statistical Manual of Mental Disorders: DSM-IV (DSM-IV) (78), Consortium to Establish a Registry for Alzheimer's Disease (CERAD) (25), or Khachaturian (79).
Full data were obtained on 84% of subjects.
Unpublished observations.
Cohorts used in the meta-analysis of the ACE* insertion-deletion polymorphism in Alzheimer's disease
Reference . | Location of samples . | Alzheimer's disease (no.) . | Controls (no.) . | Alzheimer's disease diagnosis† . | Stratified data supplied and used‡ . |
|---|---|---|---|---|---|
| Alvarez et al. (63) | Asturias, Spain | 351 | 536 | NINCDS-ADRDA probable | Yes |
| Buss et al. (40) | Hamburg, Germany; Basel, Switzerland; and Brescia, Italy | 261 | 306 | NINCDS-ADRDA | Yes |
| Camelo et al. (13) | Colombia | 98 | 72 | NINCDS-ADRDA probable | Yes |
| Carbonell et al. (64) | London, United Kingdom | 80 | 65 | NINCDS-ADRDA | |
| Chapman et al. (65) | Tel Aviv, Israel | 49 | 40 | NINCDS-ADRDA probable | |
| Cheng et al. (66) | Taipei, Taiwan | 173 | 116 | NINCDS-ADRDA probable | |
| Crawford et al. (7) | Florida, United States | 171 | 175 | NINCDS-ADRDA | |
| Farrer et al. (6) | Moscow, Russia | 151 | 206 | NINCDS-ADRDA | |
| Farrer et al. (6) | Toronto, Canada; Miami, Florida; and Florence, Italy | 236 | 169 | NINCDS-ADRDA | |
| Fernández-Novoa et al. (14) | Galicia, Spain | 185 | 117 | NINCDS-ADRDA and DSM-IV | Yes |
| Hu et al. (67) | Japan | 132 | 148 | NINCDS-ADRDA probable | |
| Isbir et al. (68) | Turkey | 35 | 29 | NINCDS-ADRDA probable | |
| Kehoe et al. (11) | Bristol and London, United Kingdom | 296 | 95 | CERAD or NINCDS- ADRDA probable | Yes |
| Kehoe et al. (1) | Cardiff, Wales | 198 | 77 | NINCDS-ADRDA probable | |
| Kehoe et al. (1) | London, United Kingdom | 201 | 205 | NINCDS-ADRDA | Yes |
| Kehoe et al. (1) | Belfast, Northern Ireland | 209 | 198 | NINCDS-ADRDA probable and DSM-IV | Yes |
| Keikhaee et al. (15) | Iran | 132 | 107 | Yes | |
| Lendon et al. (16) | Manchester, England | 206 | 33 | NINCDS-ADRDA probable or CERAD | Yes |
| Lendon et al. (16) | Scotland | 167 | 357 | NINCDS-ADRDA | Yes |
| Mattila et al. (69) | West Finland | 77 | 67 | CERAD or NINCDS-ADRDA probable | Yes |
| Monastero et al. (70) | Palermo, Italy | 149 | 149 | NINCDS-ADRDA probable | |
| Myllykangas et al. (71) | Vantaa, Finland | 121 | 75 | CERAD | |
| Narain et al. (10) | Cambridge, England | 107 | 163 | CERAD | Yes |
| Narain et al. (10) | Oxford, England | 26 | 179 | CERAD | Yes |
| Palumbo et al. (72) | Italy | 96 | 40 | NINCDS-ADRDA probable | |
| Panza et al. (17) | South Italy | 141 | 268 | NINCDS-ADRDA probable | Yes |
| Perry et al. (41) | Alabama, United States (African Americans) | 111 | 78 | NINCDS-ADRDA probable | Yes |
| Prince et al. (23) (cohort 1); Kehoe et al. (11) | Sweden | 201 | 161 | NINCDS-ADRDA or CERAD | Yes |
| Prince et al. (23) (cohort 2); Kehoe et al. (11) | Sweden | 168 | 322 | NINCDS-ADRDA | Yes |
| Richard et al. (cohort 1) (73) | North Europe | 421 | 474 | NINCDS-ADRDA probable | Yes |
| Richard et al. (73) (cohort 2) | North Europe | 56 | 221 | NINCDS-ADRDA probable | Yes |
| Scacchi et al. (74) | Rome, Italy | 80 | 153 | NINCDS-ADRDA probable | Yes |
| Seripa et al. (18) | Rome, Italy | 97 | 101 | NINCDS-ADRDA probable | Yes |
| Seripa et al. (18) | Wisconsin and Kentucky, United States | 102 | 66 | NINCDS-ADRDA probable and Khachaturian | Yes |
| Sleegers et al. (9) | Netherlands | 236 | 5,889 | NINCDS-ADRDA | Yes |
| Wu et al. (75) | Beijing, People's Republic of China | 96 | 96 | DSM-IV | Yes |
| Yang et al. (76) | Shanghai, People's Republic of China | 188 | 227 | NINCDS-ADRDA probable | |
| Zuliani et al. (77) | Northeast Italy | 35 | 51 | NINCDS-ADRDA probable | Yes |
| OPTIMA* study§ | Oxfordshire, England | 198 | 268 | CERAD or NINCDS-ADRDA probable | Yes |
| Totals | 6,037 | 12,099 |
Reference . | Location of samples . | Alzheimer's disease (no.) . | Controls (no.) . | Alzheimer's disease diagnosis† . | Stratified data supplied and used‡ . |
|---|---|---|---|---|---|
| Alvarez et al. (63) | Asturias, Spain | 351 | 536 | NINCDS-ADRDA probable | Yes |
| Buss et al. (40) | Hamburg, Germany; Basel, Switzerland; and Brescia, Italy | 261 | 306 | NINCDS-ADRDA | Yes |
| Camelo et al. (13) | Colombia | 98 | 72 | NINCDS-ADRDA probable | Yes |
| Carbonell et al. (64) | London, United Kingdom | 80 | 65 | NINCDS-ADRDA | |
| Chapman et al. (65) | Tel Aviv, Israel | 49 | 40 | NINCDS-ADRDA probable | |
| Cheng et al. (66) | Taipei, Taiwan | 173 | 116 | NINCDS-ADRDA probable | |
| Crawford et al. (7) | Florida, United States | 171 | 175 | NINCDS-ADRDA | |
| Farrer et al. (6) | Moscow, Russia | 151 | 206 | NINCDS-ADRDA | |
| Farrer et al. (6) | Toronto, Canada; Miami, Florida; and Florence, Italy | 236 | 169 | NINCDS-ADRDA | |
| Fernández-Novoa et al. (14) | Galicia, Spain | 185 | 117 | NINCDS-ADRDA and DSM-IV | Yes |
| Hu et al. (67) | Japan | 132 | 148 | NINCDS-ADRDA probable | |
| Isbir et al. (68) | Turkey | 35 | 29 | NINCDS-ADRDA probable | |
| Kehoe et al. (11) | Bristol and London, United Kingdom | 296 | 95 | CERAD or NINCDS- ADRDA probable | Yes |
| Kehoe et al. (1) | Cardiff, Wales | 198 | 77 | NINCDS-ADRDA probable | |
| Kehoe et al. (1) | London, United Kingdom | 201 | 205 | NINCDS-ADRDA | Yes |
| Kehoe et al. (1) | Belfast, Northern Ireland | 209 | 198 | NINCDS-ADRDA probable and DSM-IV | Yes |
| Keikhaee et al. (15) | Iran | 132 | 107 | Yes | |
| Lendon et al. (16) | Manchester, England | 206 | 33 | NINCDS-ADRDA probable or CERAD | Yes |
| Lendon et al. (16) | Scotland | 167 | 357 | NINCDS-ADRDA | Yes |
| Mattila et al. (69) | West Finland | 77 | 67 | CERAD or NINCDS-ADRDA probable | Yes |
| Monastero et al. (70) | Palermo, Italy | 149 | 149 | NINCDS-ADRDA probable | |
| Myllykangas et al. (71) | Vantaa, Finland | 121 | 75 | CERAD | |
| Narain et al. (10) | Cambridge, England | 107 | 163 | CERAD | Yes |
| Narain et al. (10) | Oxford, England | 26 | 179 | CERAD | Yes |
| Palumbo et al. (72) | Italy | 96 | 40 | NINCDS-ADRDA probable | |
| Panza et al. (17) | South Italy | 141 | 268 | NINCDS-ADRDA probable | Yes |
| Perry et al. (41) | Alabama, United States (African Americans) | 111 | 78 | NINCDS-ADRDA probable | Yes |
| Prince et al. (23) (cohort 1); Kehoe et al. (11) | Sweden | 201 | 161 | NINCDS-ADRDA or CERAD | Yes |
| Prince et al. (23) (cohort 2); Kehoe et al. (11) | Sweden | 168 | 322 | NINCDS-ADRDA | Yes |
| Richard et al. (cohort 1) (73) | North Europe | 421 | 474 | NINCDS-ADRDA probable | Yes |
| Richard et al. (73) (cohort 2) | North Europe | 56 | 221 | NINCDS-ADRDA probable | Yes |
| Scacchi et al. (74) | Rome, Italy | 80 | 153 | NINCDS-ADRDA probable | Yes |
| Seripa et al. (18) | Rome, Italy | 97 | 101 | NINCDS-ADRDA probable | Yes |
| Seripa et al. (18) | Wisconsin and Kentucky, United States | 102 | 66 | NINCDS-ADRDA probable and Khachaturian | Yes |
| Sleegers et al. (9) | Netherlands | 236 | 5,889 | NINCDS-ADRDA | Yes |
| Wu et al. (75) | Beijing, People's Republic of China | 96 | 96 | DSM-IV | Yes |
| Yang et al. (76) | Shanghai, People's Republic of China | 188 | 227 | NINCDS-ADRDA probable | |
| Zuliani et al. (77) | Northeast Italy | 35 | 51 | NINCDS-ADRDA probable | Yes |
| OPTIMA* study§ | Oxfordshire, England | 198 | 268 | CERAD or NINCDS-ADRDA probable | Yes |
| Totals | 6,037 | 12,099 |
ACE, angiotensin I-converting enzyme gene; OPTIMA, Oxford Project to Investigate Memory and Ageing.
Based on criteria of the National Institute of Neurological and Communicative Disorders and Stroke–Alzheimer's Disease and Related Disorders Association (NINCDS-ADRDA) (26), Diagnostic and Statistical Manual of Mental Disorders: DSM-IV (DSM-IV) (78), Consortium to Establish a Registry for Alzheimer's Disease (CERAD) (25), or Khachaturian (79).
Full data were obtained on 84% of subjects.
Unpublished observations.
The association has recently been studied in the longitudinal, observational cohorts of the Oxford Project to Investigate Memory and Ageing (OPTIMA) and the Rotterdam Study (9). Both groups found a suggestion of a gender difference in their results, consistent with a previous report (7). We therefore undertook a meta-analysis of studies of the association of the ACE indel with Alzheimer's disease, stratified by gender, by age, by ethnic group, and by apolipoprotein E ε4 allele (APOE*4) carrier status, using mainly primary data, supplied by authors.
Although three previous meta-analyses of ACE studies had been reported (10–12), subsequent studies have now made further data available. Compared even with the latest meta-analysis (12), 10 more samples from eight studies were available to us (9, 13–18, and the above study of the Oxford Project to Investigate Memory and Ageing). In addition, all previous meta-analyses were based on published data, that is, largely unstratified, and were thus limited in the interpretations they allowed. Using primary data, we were able to explore more deeply, searching for sources of heterogeneity. Two previous stratified meta-analyses of Alzheimer's disease genes (19, 20), based on primary data, have both deepened our understanding.
Egger et al. (21) have stressed the need to examine sources of heterogeneity in meta-analyses of case-control studies. We therefore divided our analysis into two phases: first, a broad overall analysis and, second, the detailed examination of sources of heterogeneity. We aimed to discover whether the ACE indel is associated with Alzheimer's disease and, if so, the nature of that association.
MATERIALS AND METHODS
Phase 1: broad analysis
In April 2003, following PubMed searches, the study of references and of abstracts, and communication with experts, we wrote to the authors of all association studies of the ACE indel and Alzheimer's disease known to us, 33 studies in all, involving 41 samples (4, 5) (table 1), including seven samples from unpublished studies. The searches were updated in November 2004, without further data arising. For the stratified analyses, we used the raw data or stratified summaries received from authors on 26 samples. For the overall analyses, we also used published data, making 39 samples altogether (table 1). Very early onset cases (<50 years) and young controls (<50 years) were excluded, where identifiable, as were subjects with incomplete data. Distinct populations were treated separately, where possible. The large number of controls in the cohort of Sleegers et al. (9) had all been fully assessed (22). All samples had been genotyped for the ACE indel, except for the two of Prince et al. (23, 24), who had used the ACE rs4343 marker, which is in 85–90 percent linkage disequilibrium with the indel in North Europeans.
Phase 2: examination of heterogeneity
We examined six possible sources of heterogeneity. We performed analyses stratified by age, gender, and APOE*4 status. For the age stratifications (<65 years, 65–75 years, and >75 years), we divided the samples into two groups, according to how the ages of cases had been defined, for example, at onset (first group) or at examination or death (second group). We also conducted analyses restricted to controls and to diagnoses of probable or definite Alzheimer's disease according to criteria of the Consortium to Establish a Registry for Alzheimer's Disease (CERAD) (25) or of the National Institute of Neurological and Communicative Disorders and Stroke–Alzheimer's Disease and Related Disorders Association (NINCDS-ADRDA) (26) (26 full samples and two part samples). In separate analyses, we excluded eight samples where Hardy-Weinberg analysis and genotyping methods were compatible with mistyping. Underreporting of heterozygotes may occur unless either 5 percent dimethyl sulfoxide is included in the polymerase chain reaction (27) or the DD genotype is confirmed by an insertion-specific second amplification (28). In further analyses, samples were placed where possible (34 of 39 samples) in one of three ethnic groups: North European (19 samples); South Caucasian (11 samples), defined here as from the Mediterranean or from the Middle East; and East Asian (four samples), from China or from Japan.
Data analysis and statistical methods
Three methods are commonly used to produce odds ratios in genetic association studies: method 1, allelic (i.e., D vs. I in this case); method 2, comparing each genotype with one other in turn (e.g., DD vs. DI); method 3, comparing each genotype in turn with the other two combined. All three methods are subject to bias, the degree of bias depending on how imperfectly the model fits reality. For instance, method 1 is best suited to testing a codominant model, where there is an allelic dose effect (e.g., DD > DI > II). Method 3, on the other hand, may be used to test either an I-dominant/D-recessive or a D-dominant/I-recessive model. We chose to follow the hypothesis-generating study (1) in using method 3, partly because preliminary examination of subsequent studies had shown no evidence of codominance. We also used both methods 2 and 3 to examine heterosis (heterozygotes at greater or lesser risk than either homozygote). Our main approach, method 3, gives an estimate of the risk associated with each genotype when compared with the rest of the population.
Pooled odds ratios were calculated three times, by the fixed-effects method of Mantel and Haenszel (29), by the random-effects method of DerSimonian and Laird (30), and by the Bayesian random-effects method of Warn et al. (31). However, as the results of all three methods were very similar, only those of the first two are shown (table 2). The methods of meta-analysis permit the accumulation of results across independent studies, even where the number of subjects or the ratio of cases to controls varies considerably, as here in the cohort of Sleegers et al. (9), for example. The heterogeneity test was by the method of Armitage (32). Logistic regression analysis was used to compare odds ratios among ethnic groups. Funnel plots (33) and cumulative meta-analysis (34) were used to investigate bias. Analyses were performed using “R” (35) and the package “rmeta” by Thomas Lumley (http://www.cran.r-project.org/src/contrib/Descriptions/rmeta.html). All tests of significance were two sided. A Bonferroni correction factor of 3 was applied to all p values by genotype.
Comparison‡ . | Fixed effects§ . | . | Random effects§ . | . | Heterogeneity (p value)§ . | ||
|---|---|---|---|---|---|---|---|
| . | Odds ratio . | 95% confidence interval . | Odds ratio . | 95% confidence interval . | . | ||
| DD versus (DI + II) | 0.82 | 0.76, 0.89 | 0.81 | 0.72, 0.90 | 0.01 | ||
| DI versus (DD + II) | 1.12 | 1.04, 1.20 | 1.12 | 1.04, 1.22 | 0.08¶ | ||
| II versus (DD + DI) | 1.07 | 0.98, 1.17 | 1.07 | 0.95, 1.21 | 0.02 | ||
Comparison‡ . | Fixed effects§ . | . | Random effects§ . | . | Heterogeneity (p value)§ . | ||
|---|---|---|---|---|---|---|---|
| . | Odds ratio . | 95% confidence interval . | Odds ratio . | 95% confidence interval . | . | ||
| DD versus (DI + II) | 0.82 | 0.76, 0.89 | 0.81 | 0.72, 0.90 | 0.01 | ||
| DI versus (DD + II) | 1.12 | 1.04, 1.20 | 1.12 | 1.04, 1.22 | 0.08¶ | ||
| II versus (DD + DI) | 1.07 | 0.98, 1.17 | 1.07 | 0.95, 1.21 | 0.02 | ||
ACE, angiotensin I-converting enzyme gene.
All analyses were based on 39 samples, comprising 6,037 cases of Alzheimer's disease and 12,099 controls.
D represents the deletion allele; I represents the insertion allele.
Fixed-effects odds ratios were by the method of Mantel and Haenszel (29), random-effects odds ratios by that of DerSimonian and Laird (30), and the heterogeneity test by that of Armitage (32).
Not significant at 0.05.
Comparison‡ . | Fixed effects§ . | . | Random effects§ . | . | Heterogeneity (p value)§ . | ||
|---|---|---|---|---|---|---|---|
| . | Odds ratio . | 95% confidence interval . | Odds ratio . | 95% confidence interval . | . | ||
| DD versus (DI + II) | 0.82 | 0.76, 0.89 | 0.81 | 0.72, 0.90 | 0.01 | ||
| DI versus (DD + II) | 1.12 | 1.04, 1.20 | 1.12 | 1.04, 1.22 | 0.08¶ | ||
| II versus (DD + DI) | 1.07 | 0.98, 1.17 | 1.07 | 0.95, 1.21 | 0.02 | ||
Comparison‡ . | Fixed effects§ . | . | Random effects§ . | . | Heterogeneity (p value)§ . | ||
|---|---|---|---|---|---|---|---|
| . | Odds ratio . | 95% confidence interval . | Odds ratio . | 95% confidence interval . | . | ||
| DD versus (DI + II) | 0.82 | 0.76, 0.89 | 0.81 | 0.72, 0.90 | 0.01 | ||
| DI versus (DD + II) | 1.12 | 1.04, 1.20 | 1.12 | 1.04, 1.22 | 0.08¶ | ||
| II versus (DD + DI) | 1.07 | 0.98, 1.17 | 1.07 | 0.95, 1.21 | 0.02 | ||
ACE, angiotensin I-converting enzyme gene.
All analyses were based on 39 samples, comprising 6,037 cases of Alzheimer's disease and 12,099 controls.
D represents the deletion allele; I represents the insertion allele.
Fixed-effects odds ratios were by the method of Mantel and Haenszel (29), random-effects odds ratios by that of DerSimonian and Laird (30), and the heterogeneity test by that of Armitage (32).
Not significant at 0.05.
RESULTS
Phase 1: broad analysis
Examination of bias.
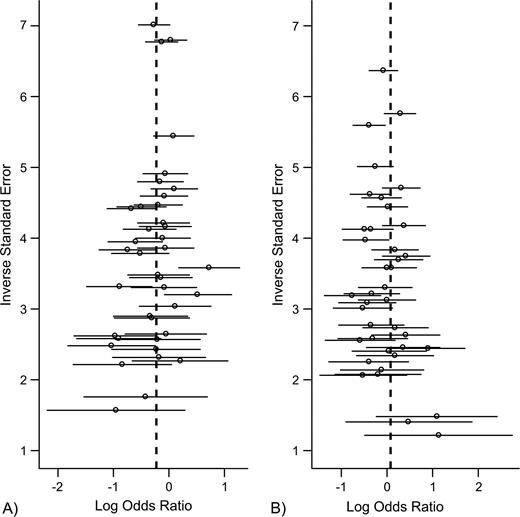
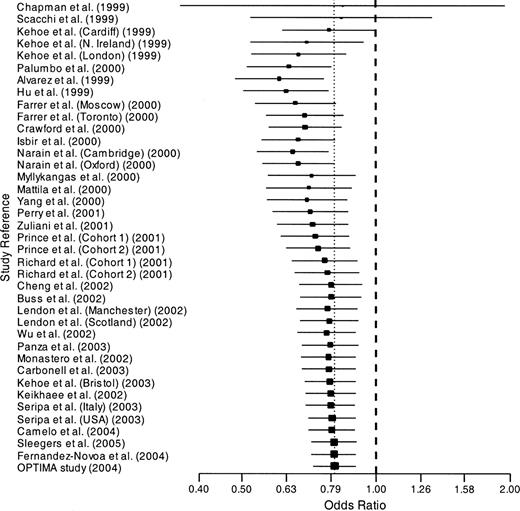
We found no evidence of publication bias. Funnel plots for the comparisons most commonly made, that is, for D homozygotes versus I positives (i.e., DI + II) and for I homozygotes versus D positives, are shown in figure 1. These plots gave p = 0.19 and p = 0.21, respectively. Figure 2 shows the cumulative meta-analysis after each study from July 1998 to November 2004, for D homozygotes versus I positives, that is, the comparison made in the hypothesis-generating study of Kehoe et al. (1). After only four of 39 cohorts had been studied, the odds ratio by DerSimonian and Laird (30) became significantly less than 1 and remained so thereafter. It changed very little from around 0.8 after 2001.
Funnel plots of angiotensin I-converting enzyme gene (ACE) studies: A, D homozygotes versus (DI + II) (p = 0.21); B, I homozygotes versus (DI + DD) (p = 0.19). D represents the deletion allele, and I represents the insertion allele. Horizontal lines are 95% confidence intervals; dashed vertical lines are summary log odds ratios.
Cumulative meta-analysis of the odds ratio of Alzheimer's disease for ACE*D homozygotes versus I positives. D represents the deletion allele, and I represents the insertion allele. Horizontal lines are 95% confidence intervals; the vertical dotted line is the summary odds ratio. OPTIMA, Oxford Project to Investigate Memory and Ageing; ACE, angiotensin I-converting enzyme gene. Additional study information is given in table 1.
Risk of Alzheimer's disease, by genotype.
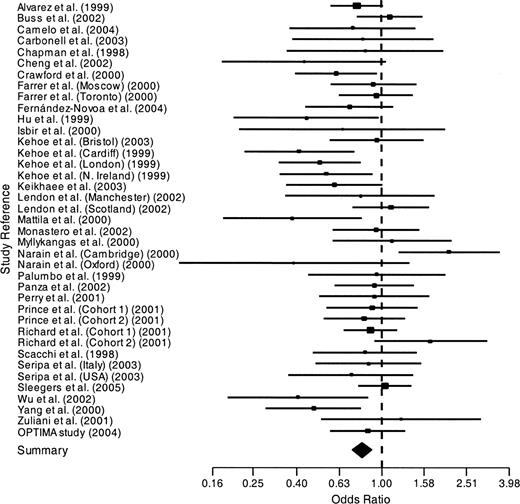
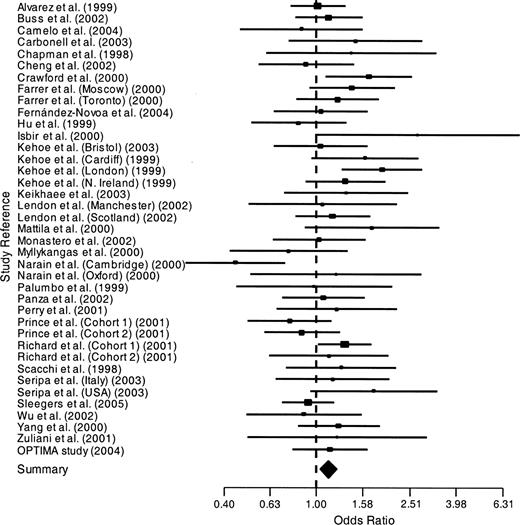
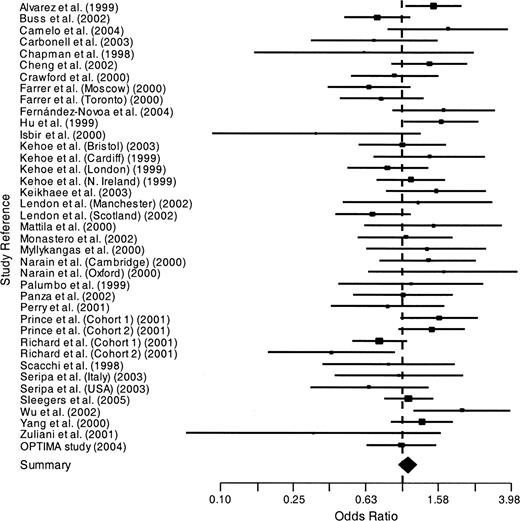
The odds ratios for each genotype versus the other two are shown in figures 3, 4, and 5 and are summarized in table 2. D homozygotes were at reduced risk of Alzheimer's disease, with an overall odds ratio of 0.81 (95 percent confidence interval: 0.72, 0.90; corrected p = 0.0004) by the method of DerSimonian and Laird (30). Heterozygotes were at increased risk of Alzheimer's disease, with an overall odds ratio of 1.12 (95 percent confidence interval: 1.04, 1.22; corrected p = 0.006). The odds ratio of Alzheimer's disease for I homozygotes versus D positives did not differ significantly from 1. Two of these three results were heterogeneous.
Odds ratios of Alzheimer's disease for ACE*D homozygotes versus I positives. D represents the deletion allele, and I represents the insertion allele. Horizontal lines are 95% confidence intervals. OPTIMA, Oxford Project to Investigate Memory and Ageing; ACE, angiotensin I-converting enzyme gene. Additional study information is given in table 1.
Odds ratios of Alzheimer's disease for ACE heterozygotes versus all homozygotes. Horizontal lines are 95% confidence intervals. OPTIMA, Oxford Project to Investigate Memory and Ageing; ACE, angiotensin I-converting enzyme gene. Additional study information is given in table 1.
Odds ratios of Alzheimer's disease for ACE*I homozygotes versus D positives. D represents the deletion allele, and I represents the insertion allele. Horizontal lines are 95% confidence intervals. OPTIMA, Oxford Project to Investigate Memory and Ageing; ACE, angiotensin I-converting enzyme gene. Additional study information is given in table 1.
Onset age of Alzheimer's disease.
Onset age data were available from 13 samples, comprising 2,309 cases of Alzheimer's disease. Both stratified analyses and linear regression analysis were performed on these data and also on a subset of 10 North European samples, comprising 1,822 cases of Alzheimer's disease. No effect of ACE*I carrier status on onset age of Alzheimer's disease and, surprisingly, only a modest effect of APOE*4 carrier status on onset age were seen (data not shown).
Phase 2: examination of heterogeneity
We examined six potential sources of heterogeneity: age, gender, race, APOE*4 status, misdiagnosis, and mistyping.
Stratification by gender and by APOE*4 status.
No significant effects were seen of gender or of APOE*4 status on the associations with Alzheimer's disease, nor did stratification by either of these two factors remove the heterogeneity (data not shown).
Stratification by age.
We calculated the odds ratios of Alzheimer's disease for ACE*D homozygotes versus I positives when stratified by age (<65 years, 65–75 years, and >75 years). Rather little effect of age was seen on the association of D homozygotes with Alzheimer's disease (data not shown), nor was there any reduction in heterogeneity on stratification by age.
Examination of diagnosis.
We repeated the overall analyses of the three genotypes, restricting the data to the 28 samples that could be limited to the more rigorous diagnoses of probable or definite Alzheimer's disease according to criteria of the Consortium to Establish a Registry for Alzheimer's Disease (25) or the National Institute of Neurological and Communicative Disorders and Stroke–Alzheimer's Disease and Related Disorders Association (26). The odds ratios were similar to those shown in table 2, that is, significant odds ratios by the method of DerSimonian and Laird (30) of 0.77 for D homozygotes and of 1.15 for heterozygotes and a nonsignificant odds ratio of 1.10 for I homozygotes. Heterogeneity remained for D homozygotes (p = 0.005) but was no longer significant (p = 0.16) for I homozygotes.
Examination of genotyping.
There were 12 instances of Hardy-Weinberg disequilibrium in individual studies and four in the six groups of pooled cases and controls analyzed by race (table 3). We therefore excluded the eight samples where mistyping was considered possible, that is, underreporting of heterozygotes, taking into account both genotyping methods and Hardy-Weinberg analysis. We then repeated the overall analyses with the remaining 31 samples. The odds ratios were similar to those in table 2, that is, significant odds ratios of 0.81 for D homozygotes and of 1.12 for heterozygotes and a nonsignificant odds ratio of 1.09 for I homozygotes. Heterogeneity remained for I homozygotes (p = 0.02) but was no longer significant for D homozygotes (p = 0.19).
. | North Europeans . | . | South Caucasians (Mediterranean and Middle East) . | . | East Asians (China and Japan) . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| Alzheimer's disease cases (no.)/controls (no.) | 3,380/9,361 | 1,350/1,591 | 589/587 | ||||||
| D frequencies in controls (% (95% CI*)) | 53.0 | 52.2, 53.7 | 63.4 | 61.7, 65.0 | 41.3 | 38.5, 44.1 | |||
| DD versus (DI + II) (OR*,‡ (95% CI)) | 0.86 (a)§ | 0.71, 1.05 | 0.81 | 0.70, 0.95 | 0.45 | 0.32, 0.63 | |||
| 0.81 (b)§ | 0.70, 0.95 | ||||||||
| DI versus (DD + II) (OR‡ (95% CI)) | 1.11 (a)§ | 0.97, 1.28 | 1.11 | 0.95, 1.30 | 0.98 | 0.78, 1.25 | |||
| 1.15 (b)§ | 1.04, 1.28 | ||||||||
| II versus (DD + DI) (OR‡ (95% CI)) | 0.99 (a)§ | 0.85, 1.16 | 1.12 | 0.86, 1.46 | 1.51 | 1.19, 1.91 | |||
| 0.99 (b)§ | 0.88, 1.12 | ||||||||
| HW* excess (p value) | |||||||||
| Alzheimer's disease | Heterozygotes (0.0009) | Homozygotes (0.01) | Equilibrium | ||||||
| Controls | Equilibrium | Homozygotes (0.02) | Homozygotes (0.003) | ||||||
. | North Europeans . | . | South Caucasians (Mediterranean and Middle East) . | . | East Asians (China and Japan) . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| Alzheimer's disease cases (no.)/controls (no.) | 3,380/9,361 | 1,350/1,591 | 589/587 | ||||||
| D frequencies in controls (% (95% CI*)) | 53.0 | 52.2, 53.7 | 63.4 | 61.7, 65.0 | 41.3 | 38.5, 44.1 | |||
| DD versus (DI + II) (OR*,‡ (95% CI)) | 0.86 (a)§ | 0.71, 1.05 | 0.81 | 0.70, 0.95 | 0.45 | 0.32, 0.63 | |||
| 0.81 (b)§ | 0.70, 0.95 | ||||||||
| DI versus (DD + II) (OR‡ (95% CI)) | 1.11 (a)§ | 0.97, 1.28 | 1.11 | 0.95, 1.30 | 0.98 | 0.78, 1.25 | |||
| 1.15 (b)§ | 1.04, 1.28 | ||||||||
| II versus (DD + DI) (OR‡ (95% CI)) | 0.99 (a)§ | 0.85, 1.16 | 1.12 | 0.86, 1.46 | 1.51 | 1.19, 1.91 | |||
| 0.99 (b)§ | 0.88, 1.12 | ||||||||
| HW* excess (p value) | |||||||||
| Alzheimer's disease | Heterozygotes (0.0009) | Homozygotes (0.01) | Equilibrium | ||||||
| Controls | Equilibrium | Homozygotes (0.02) | Homozygotes (0.003) | ||||||
ACE, angiotensin I-converting enzyme gene; CI, confidence interval; OR, odds ratio; HW, Hardy-Weinberg.
D represents the deletion allele; I represents the insertion allele.
All odds ratios shown were by the random-effects method of DerSimonian and Laird (30), which is generally more conservative than the fixed-effects method; all significant odds ratios were very closely replicated by the Bayesian random-effects method of Warn et al. (31), except for the II odds ratio for East Asians, whose result was 1.53 (95% CI: 0.96, 2.54) by the Bayesian method.
Heterogeneity was seen only in North European cohorts; it was removed upon exclusion of two cohorts with possible genotyping problems. Results for North Europeans are shown for all 19 cohorts (a) and after the removal of those two cohorts (b). North Europeans versus East Asians: OR for DD versus (DI + II), p = 0.0008; OR for II versus (DD + DI), p = 0.0005. South Caucasians versus East Asians: OR for DD versus (DI + II), p = 0.0003.
. | North Europeans . | . | South Caucasians (Mediterranean and Middle East) . | . | East Asians (China and Japan) . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| Alzheimer's disease cases (no.)/controls (no.) | 3,380/9,361 | 1,350/1,591 | 589/587 | ||||||
| D frequencies in controls (% (95% CI*)) | 53.0 | 52.2, 53.7 | 63.4 | 61.7, 65.0 | 41.3 | 38.5, 44.1 | |||
| DD versus (DI + II) (OR*,‡ (95% CI)) | 0.86 (a)§ | 0.71, 1.05 | 0.81 | 0.70, 0.95 | 0.45 | 0.32, 0.63 | |||
| 0.81 (b)§ | 0.70, 0.95 | ||||||||
| DI versus (DD + II) (OR‡ (95% CI)) | 1.11 (a)§ | 0.97, 1.28 | 1.11 | 0.95, 1.30 | 0.98 | 0.78, 1.25 | |||
| 1.15 (b)§ | 1.04, 1.28 | ||||||||
| II versus (DD + DI) (OR‡ (95% CI)) | 0.99 (a)§ | 0.85, 1.16 | 1.12 | 0.86, 1.46 | 1.51 | 1.19, 1.91 | |||
| 0.99 (b)§ | 0.88, 1.12 | ||||||||
| HW* excess (p value) | |||||||||
| Alzheimer's disease | Heterozygotes (0.0009) | Homozygotes (0.01) | Equilibrium | ||||||
| Controls | Equilibrium | Homozygotes (0.02) | Homozygotes (0.003) | ||||||
. | North Europeans . | . | South Caucasians (Mediterranean and Middle East) . | . | East Asians (China and Japan) . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| Alzheimer's disease cases (no.)/controls (no.) | 3,380/9,361 | 1,350/1,591 | 589/587 | ||||||
| D frequencies in controls (% (95% CI*)) | 53.0 | 52.2, 53.7 | 63.4 | 61.7, 65.0 | 41.3 | 38.5, 44.1 | |||
| DD versus (DI + II) (OR*,‡ (95% CI)) | 0.86 (a)§ | 0.71, 1.05 | 0.81 | 0.70, 0.95 | 0.45 | 0.32, 0.63 | |||
| 0.81 (b)§ | 0.70, 0.95 | ||||||||
| DI versus (DD + II) (OR‡ (95% CI)) | 1.11 (a)§ | 0.97, 1.28 | 1.11 | 0.95, 1.30 | 0.98 | 0.78, 1.25 | |||
| 1.15 (b)§ | 1.04, 1.28 | ||||||||
| II versus (DD + DI) (OR‡ (95% CI)) | 0.99 (a)§ | 0.85, 1.16 | 1.12 | 0.86, 1.46 | 1.51 | 1.19, 1.91 | |||
| 0.99 (b)§ | 0.88, 1.12 | ||||||||
| HW* excess (p value) | |||||||||
| Alzheimer's disease | Heterozygotes (0.0009) | Homozygotes (0.01) | Equilibrium | ||||||
| Controls | Equilibrium | Homozygotes (0.02) | Homozygotes (0.003) | ||||||
ACE, angiotensin I-converting enzyme gene; CI, confidence interval; OR, odds ratio; HW, Hardy-Weinberg.
D represents the deletion allele; I represents the insertion allele.
All odds ratios shown were by the random-effects method of DerSimonian and Laird (30), which is generally more conservative than the fixed-effects method; all significant odds ratios were very closely replicated by the Bayesian random-effects method of Warn et al. (31), except for the II odds ratio for East Asians, whose result was 1.53 (95% CI: 0.96, 2.54) by the Bayesian method.
Heterogeneity was seen only in North European cohorts; it was removed upon exclusion of two cohorts with possible genotyping problems. Results for North Europeans are shown for all 19 cohorts (a) and after the removal of those two cohorts (b). North Europeans versus East Asians: OR for DD versus (DI + II), p = 0.0008; OR for II versus (DD + DI), p = 0.0005. South Caucasians versus East Asians: OR for DD versus (DI + II), p = 0.0003.
Ethnic stratification.
The samples were divided into three ethnic groups, where possible: North European (19 samples), South Caucasian (Mediterranean and Middle Eastern, 11 samples), and East Asian (Chinese and Japanese, four samples). This removed nearly all heterogeneity (table 3). Table 3 shows clear differences among the three ethnic groups. There was no overlap whatsoever in allelic frequencies among the three groups. North Europeans and East Asians differed in the odds ratios for D homozygotes versus I positives (p = 0.0008) and for I homozygotes versus D positives (p = 0.0005). South Caucasians also differed from East Asians in the odds ratio for D homozygotes versus I positives (p = 0.0003). D homozygotes were at reduced risk in all groups, but particularly in East Asians. Heterozygotes were at increased risk in North Europeans. I homozygotes were at higher risk in East Asians, except by Bayesian analysis. We further examined heterosis (heterozygotes at greater risk) in North Europeans, by comparing DI with DD and with II in turn. We found that heterozygotes were at higher risk than D homozygotes (odds ratio = 1.25, 95 percent confidence interval: 1.08, 1.45) but not significantly higher than I homozygotes (odds ratio = 1.08, 95 percent confidence interval: 0.95, 1.22).
DISCUSSION
This was a large meta-analysis, by the standards of Alzheimer's disease genetics, with 39 samples, comprising 6,037 cases of Alzheimer's disease and 12,099 elderly controls (table 1). We found no evidence of publication bias.
ACE*D homozygotes were at reduced risk of Alzheimer's disease (table 2) in each of the three main ethnic groups examined (table 3). I homozygotes were neutral as regards Alzheimer's disease risk (table 2), except in East Asians, in whom I homozygotes may be at higher risk (table 3). Heterozygotes were at increased risk of Alzheimer's disease overall (table 2) and mainly in North Europeans (table 3). This example of heterosis will be discussed below. Very similar results were achieved by three methods: fixed effects, random effects, and Bayesian random effects (refer to Materials and Methods and table 2). Only genotypic comparisons were made (refer to Materials and Methods), which showed that there was no allelic dose effect, except in East Asians (table 3), and that thus allelic comparisons would have proved less informative.
These findings of odds ratios close to 1, even though significantly different from 1 (tables 2 and 3), suggest either linkage disequilibrium with the true risk factor or confounding by a hidden interaction or both. The second possibility is discussed below. Regarding the former, Kehoe et al. (1) in their study of ACE haplotypes gave evidence that the indel may not be the actual risk factor but rather in linkage disequilibrium with a nearby functional polymorphism that is the true risk factor. ACE haplotype analysis has also proved of value with other phenotypes, such as enzyme levels (36), insulin levels, myocardial infarction, and obesity (37). Future Alzheimer's disease studies may do better to examine polymorphisms in the regulatory regions, rather than the indel.
The overall results were consistent with those of Kehoe et al. (1) from 1999 and also with those of the recent meta-analyses of Kehoe et al. (11) from 2003 and of Elkins et al. (12) from 2004. The latter report (12) also suggested an age difference, however, although actual ages were not available to them, only mean cohort ages, and they did not take into account the different ways of defining case ages (refer to Materials and Methods), a strongly confounding factor. Because of the latter problem, we cannot exclude the possibility that the association might be stronger in younger cases, for example, in those aged less than 65 years at onset or less than 75 years at death.
Heterogeneity
Our overall results showed heterogeneity (table 2). Heterogeneity may be due to differences in sample selection (e.g., in age, gender, or diagnosis) or in methods (e.g., of genotyping), or it may be due to real differences in populations (e.g., in race) or in interactions with other risk factors (e.g., APOE*4). We examined all of these potential sources of heterogeneity. We found little effect of age, gender, or APOE*4 status, thus confounding our initial hypothesis. Both diagnostic and genotyping differences contributed some heterogeneity. The former was mainly due to the diagnosis of “possible Alzheimer's disease,” reported to have a poor record of confirmation at autopsy (38). The latter was largely due to certain genotyping methods that may lead to underreporting of heterozygotes (27, 28), as shown by Hardy-Weinberg analysis.
However, the greatest source of heterogeneity was ethnic differences. Ethnic stratification removed nearly all heterogeneity (table 3). There were clear contrasts among the three ethnic groups examined (North Europeans, South Caucasians, and East Asians) in allelic frequencies and in odds ratios (table 3). The latter might be due to differences in linkage disequilibrium with the true risk polymorphism (see above). Some of these ethnic differences had previously been pointed out by Panza et al. (17, 39), by Buss et al. (40), and by Elkins et al. (12). These contrasts highlight the dangers of combining ethnic groups within a single cohort, even North and South Europeans, as indicated by Panza et al. (17, 39). Unfortunately, there was only one cohort of African origin, from the United States (41). Thus, no conclusions could be drawn about Africans.
Heterosis
Deviations from Hardy-Weinberg equilibrium are common in studies of the ACE indel, as previously pointed out by Buss et al. (40). This may be due to either mistyping or heterosis. Several cases of such disequilibrium were indeed compatible with mistyping. Most examples of disequilibrium in North Europeans, however, were of an excess of heterozygotes in Alzheimer's disease, which requires another explanation. We believe that explanation is heterosis. Heterosis occurs when heterozygotes show either a greater or lesser association with a given trait than does either group of homozygotes. In this case, in North Europeans, both an excess of heterozygotes in Alzheimer's disease and an association of heterozygotes with Alzheimer's disease were found. Comings and MacMurray (42) have suggested three explanations for heterosis, of which their second can be applied here. This proposes that heterosis may follow from the inadvertent combination of two unlike subsets due to a hidden interaction with another, unknown risk factor. To test this proposal, we constructed a mathematical model with two equal subsets based on a median split of an unknown factor X (table 4). The model illustrates how, although heterozygotes may have intermediate odds ratios in each subset, they can emerge with the highest odds ratio when the subsets are combined (i.e., heterosis). The overall results of this illustrative model (table 4) were very close to those of the present meta-analysis (table 2). We should stress, however, that heterosis was found only in North Europeans. Moreover, although heterozygotes were at greater risk than D homozygotes (odds ratio = 1.25, 95 percent confidence interval: 1.08, 1.45), they were not at significantly higher risk than I homozygotes (odds ratio = 1.08, 95 percent confidence interval: 0.95, 1.22). Thus, partial heterosis better describes this case.
Model to show how heterosis may follow from the combination of two unlike subsets
Model . | Odds ratios for each ACE* genotype versus both others . | . | . | ||
|---|---|---|---|---|---|
| . | DD . | DI . | II . | ||
| Low factor X† | 0.50 | 1.17 | 1.59 | ||
| High factor X† | 1.24 | 1.09 | 0.63 | ||
| All | 0.82 | 1.13 | 1.05 | ||
| Present meta-analysis‡ | 0.81 | 1.12 | 1.07 | ||
Model . | Odds ratios for each ACE* genotype versus both others . | . | . | ||
|---|---|---|---|---|---|
| . | DD . | DI . | II . | ||
| Low factor X† | 0.50 | 1.17 | 1.59 | ||
| High factor X† | 1.24 | 1.09 | 0.63 | ||
| All | 0.82 | 1.13 | 1.05 | ||
| Present meta-analysis‡ | 0.81 | 1.12 | 1.07 | ||
ACE, angiotensin I-converting enzyme gene.
Factor X is hypothesized to interact with the ACE insertion-deletion polymorphism in Alzheimer's disease risk; the two subsets (“low” and “high”) are based on a median split of values of factor X. The model was devised to give opposing results in each subset, while maintaining Hardy-Weinberg equilibrium, but to produce overall results similar to those of this meta-analysis.
See table 2.
Model to show how heterosis may follow from the combination of two unlike subsets
Model . | Odds ratios for each ACE* genotype versus both others . | . | . | ||
|---|---|---|---|---|---|
| . | DD . | DI . | II . | ||
| Low factor X† | 0.50 | 1.17 | 1.59 | ||
| High factor X† | 1.24 | 1.09 | 0.63 | ||
| All | 0.82 | 1.13 | 1.05 | ||
| Present meta-analysis‡ | 0.81 | 1.12 | 1.07 | ||
Model . | Odds ratios for each ACE* genotype versus both others . | . | . | ||
|---|---|---|---|---|---|
| . | DD . | DI . | II . | ||
| Low factor X† | 0.50 | 1.17 | 1.59 | ||
| High factor X† | 1.24 | 1.09 | 0.63 | ||
| All | 0.82 | 1.13 | 1.05 | ||
| Present meta-analysis‡ | 0.81 | 1.12 | 1.07 | ||
ACE, angiotensin I-converting enzyme gene.
Factor X is hypothesized to interact with the ACE insertion-deletion polymorphism in Alzheimer's disease risk; the two subsets (“low” and “high”) are based on a median split of values of factor X. The model was devised to give opposing results in each subset, while maintaining Hardy-Weinberg equilibrium, but to produce overall results similar to those of this meta-analysis.
See table 2.
A hidden interaction
Partial heterosis in North Europeans suggests an interaction with another risk factor for Alzheimer's disease, another potential source of heterogeneity. Candidates include cardiovascular risk factors, since many also contribute to Alzheimer's disease risk and since the ACE indel has been associated with cardiovascular risk (43–47).
Biologic background
Full discussion of the biologic basis of these associations is outside the scope of this mainly analytical study. However, we note the associations of the ACE indel and of nearby polymorphisms with various phenotypes, including cardiovascular risk (37, 43–47). In addition, ACE protein levels are raised in certain brain regions with Alzheimer's disease (48–50), the enzyme has been reported to modify β-amyloid aggregation (51), ACE inhibitors have been reported to maintain memory in aged rats (52) and in human patients (53), and brain-penetrating ACE inhibitors have been associated with both reduced incidence of Alzheimer's disease in elderly hypertensive patients (54) and less cognitive decline in cases with mild-to-moderate Alzheimer's disease (55). Sleegers et al. (9) have recently provided evidence from magnetic resonance imaging of increased atrophy in both the hippocampus and amygdala of nondemented women homozygous for the ACE*I allele, together with a modest but significant increase in risk of Alzheimer's disease, independent of vascular factors. The D allele is associated with raised plasma levels of ACE (2), but that association is thought to be due to linkage disequilibrium with other nearby polymorphisms (3, 36). There is an apparent conflict between the benefits of ACE inhibitor therapy and the associations of the indel with Alzheimer's disease risk and pathology and with plasma levels of ACE. However, the influence of ACE polymorphisms on brain levels of the enzyme is not yet known and cannot be assumed to reflect the levels in plasma. This subject warrants fuller discussion than is possible here. A more detailed discussion is given by Kehoe (56), although it will need to be reconsidered and perhaps revised in the light of any interacting factor or factors that may emerge in future studies.
Limitations of this meta-analysis
The lack of information from some authors was a limitation, but we obtained full data on 84 percent of subjects. The quality of diagnosis and of genotyping varied among studies, but our overall results were not changed when these questions were taken into account. There was considerable heterogeneity in our initial results. However, we found the main sources of that heterogeneity and were able to remove it. Since cardiovascular factors are strong candidates for a potential interaction with ACE variants, the lack of data available to us on those factors was a limitation, which we hope will be overcome by future studies. Cardiovascular factors might also have influenced our results through selective mortality. However, D homozygotes have only a very slightly increased risk of cardiovascular complications (43–47, 57) and little evidence of any association with reduced life span (57–62). Thus, the influence of selective mortality, if any, is likely to be very small and not enough to provide an alternative explanation for our findings.
Conclusions
We suggest that this large study has established the ACE indel as a marker of Alzheimer's disease risk, since we found no evidence of bias, we found and removed the main sources of heterogeneity, we excluded various potential interactions, and significant results remained in each of the three ethnic groups examined. The indel, however, is probably in linkage disequilibrium with the true risk polymorphism (11). There were marked contrasts among the three ethnic groups studied (table 3). We also found evidence of an interaction with another risk factor in North Europeans. Potential interactions with other factors than age, gender, and APOE*4 status should be examined, particularly interactions with cardiovascular risk factors.
Financial support was received from Bristol Myers Squibb, Phytopharm plc, the Medical Research Council, the Norman Collisson Foundation, and the Takayama Foundation. Dr. P. G. Kehoe is supported by a Gestetner Foundation Fellowship.
The authors thank all those groups who have undertaken ACE/Alzheimer's disease association studies, thereby providing the data for these analyses. They especially thank those authors who generously responded to requests for primary data: Dr. V. Alvarez, Dr. H. Arboleda, Dr. A. Capurso, Dr. L. Cook, Dr. M. M. Esiri, Dr. R. Fellin, Dr. L. Fernández-Novoa, Dr. U. Finkh, Dr. R. C. P. Go, Dr. H. Gurling, Dr. N. Helbecque, Dr. M. R. Keikhaee, Dr. C. L. Lendon, Dr. S. Lovestone, Dr. D. M. Mann, Dr. K. M. Mattila, Dr. S. P. McIlroy, Dr. L. Myllykangas, Dr. F. Panza, Dr. R. T. Perry, Dr. D. C. Rubinsztein, Dr. R. Scacchi, Dr. D. Seripa, Dr. H. Weiner, Dr. C. Wu, and Dr. G. Zuliani. The authors are particularly grateful to Dr. U. Finkh and colleagues for highlighting the issue of Hardy-Weinberg disequilibrium in ACE studies (40) and to them and to Dr. A. Capurso and colleagues (17, 39) for pointing out the ethnic differences in ACE indel frequencies. The authors also warmly thank Dr. M. Pembrey for advice on genetics, Dr. J. Marchini for advice on statistics, C. A. Aldridge for administrative support, M. G. Lehmann for help with mathematical calculations, and Dr. J. Wu for Chinese translations.
Conflict of interest: none declared.
Note added in proof:Two further association studies have been published since submission of this article: Kölsch et al. (Neurosci Lett 2005;377:37–9) and Zhang et al. (Dement Geriatr Cogn Disord 2005;20:52–6).
References
Kehoe PG, Russ C, McIlroy S, et al. Variation in DCP1, encoding ACE, is associated with susceptibility to Alzheimer disease.
Rigat B, Hubert C, Alhenc-Gelas F, et al. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels.
Tiret L, Rigat B, Visvikis S, et al. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels.
Nacmias B, Tedde A, Forleo P, et al. Angiotensin converting enzyme polymorphism in sporadic and presenilin-linked Alzheimer's disease families. (Abstract).
Wakutani Y, Kowa H, Kusumi M, et al. Genetic analysis of vascular factors in Alzheimer's disease.
Farrer LA, Sherbatich T, Keryanov SA, et al. Association between angiotensin-converting enzyme and Alzheimer disease.
Crawford F, Abdullah L, Schinka J, et al. Gender-specific association of the angiotensin converting enzyme gene with Alzheimer's disease.
Kehoe PG, Katzov H, Andreasen N, et al. Common variants of ACE contribute to variable age-at-onset of Alzheimer's disease.
Sleegers K, den Heijer T, van Dijk EJ, et al. ACE gene is associated with Alzheimer's disease and atrophy of hippocampus and amygdala.
Narain Y, Yip A, Murphy T, et al. The ACE gene and Alzheimer's disease susceptibility.
Kehoe PG, Katzov H, Feuk L, et al. Haplotypes extending across ACE are associated with Alzheimer's disease.
Elkins JS, Douglas VC, Johnston SC. Alzheimer disease risk and genetic variation in ACE. A meta-analysis.
Camelo D, Arboleda G, Yunis JJ, et al. Angiotensin-converting enzyme and alpha-2-macroglobulin gene polymorphisms are not associated with Alzheimer's disease in Colombian patients.
Fernández-Novoa L, Lombardi VRM, Seoane S, et al. The ACE gene in Alzheimer's disease. In: Hanin I, Fisher A, Cacabelos R, eds. New trends in Alzheimer and Parkinson related disorders. Bologna, Italy: Monduzzi Editore,
Keikhaee MR, Najmabadi H, Pasalar P. Angiotensin-converting enzyme and apolipoprotein E genotype in Alzheimer's disease in Iranian population. (Abstract).
Lendon CL, Thaker U, Harris JM, et al. The angiotensin 1-converting enzyme insertion (I)/deletion (D) polymorphism does not influence the extent of amyloid or tau pathology in patients with sporadic Alzheimer's disease.
Panza F, Solfrizzi V, D'Introno A, et al. Lack of association between ACE polymorphism and Alzheimer's disease in southern Italy.
Seripa D, Dal Forno G, Matera MG, et al. Methylenetetrahydrofolate reductase and angiotensin converting enzyme gene polymorphisms in two genetically and diagnostically distinct cohorts of Alzheimer patients.
Farrer LA, Cupples LA, Haines JL, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease: a meta-analysis.
Lehmann DJ, Williams J, McBroom J, et al. Using meta-analysis to explain the diversity of results in genetic studies of late-onset Alzheimer's disease and to identify high-risk subgroups.
Egger M, Davey Smith G, Altman DGE. Systematic reviews in health care: meta-analysis in context. London, United Kingdom: BMJ Books,
Ott A, Breteler MM, van Harskamp F, et al. Incidence and risk of dementia. The Rotterdam Study.
Prince JA, Feuk L, Sawyer SL, et al. Lack of replication of association findings in complex disease: an analysis of 15 polymorphisms in prior candidate genes for sporadic Alzheimer's disease.
Prince J, Feuk L, Brookes A, et al. Genetic variation in ACE influences the severity of Alzheimer's disease. (Abstract).
Mirra SS, Heyman A, McKeel D, et al. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease.
McKhann G, Drachman D, Folstein M, et al. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA work group under the auspices of Department of Health and Human Services task force on Alzheimer's disease.
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease.
Warn DE, Thompson SG, Spiegelhalter DJ. Bayesian random effects meta-analysis of trials with binary outcomes: methods for the absolute risk difference and relative risk scales.
Armitage P. Statistical methods in medical research. Oxford, United Kingdom: Blackwell Scientific Publications,
Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test.
Lau J, Antman EM, Jimenez-Silva J, et al. Cumulative meta-analysis of therapeutic trials for myocardial infarction.
Ihaka R, Gentleman R. R: a language for data analysis and graphics.
Cox R, Bouzekri N, Martin S, et al. Angiotensin-1-converting enzyme (ACE) plasma concentration is influenced by multiple ACE-linked quantitative trait nucleotides.
Katzov H, Bennet AM, Kehoe P, et al. A cladistic model of ACE sequence variation with implications for myocardial infarction, Alzheimer disease and obesity.
Nagy Z, Esiri MM, Hindley NJ, et al. Accuracy of clinical operational diagnostic criteria for Alzheimer's disease in relation to different pathological diagnostic protocols.
Panza F, Solfrizzi V, D'Introno A, et al. Shifts in angiotensin 1 converting enzyme insertion allele frequency across Europe: implications for Alzheimer's disease risk.
Buss S, Müller-Thomsen T, Hock C, et al. No association between DCP1 genotype and late-onset Alzheimer disease.
Perry RT, Collins JS, Harrell LE, et al. Investigation of association of 13 polymorphisms in eight genes in southeastern African American Alzheimer disease patients as compared to age-matched controls.
Samani NJ, Thompson JR, O'Toole L, et al. A meta-analysis of the association of the deletion allele of the angiotensin-converting enzyme gene with myocardial infarction.
Staessen JA, Wang JG, Ginocchio G, et al. The deletion/insertion polymorphism of the angiotensin converting enzyme gene and cardiovascular-renal risk.
Sharma P. Meta-analysis of the ACE gene in ischaemic stroke.
Keavney B, McKenzie C, Parish S, et al. Large-scale test of hypothesised associations between the angiotensin-converting-enzyme insertion/deletion polymorphism and myocardial infarction in about 5000 cases and 6000 controls.
Sayed-Tabatabaei FA, Houwing-Duistermaat JJ, van Duijn CM, et al. Angiotensin-converting enzyme gene polymorphism and carotid artery wall thickness. A meta-analysis.
Arregui A, Perry EK, Rossor M, et al. Angiotensin converting enzyme in Alzheimer's disease increased activity in caudate nucleus and cortical areas.
Barnes NM, Cheng CH, Costall B, et al. Angiotensin converting enzyme density is increased in temporal cortex from patients with Alzheimer's disease.
Savaskan E, Hock C, Olivieri G, et al. Cortical alterations of angiotensin converting enzyme, angiotensin II and AT1 receptor in Alzheimer's dementia.
Hu J, Igarashi A, Kamata M, et al. Angiotensin-converting enzyme degrades Alzheimer amyloid beta-peptide (Ab); retards Ab aggregation, deposition, fibril formation; and inhibits cytotoxicity.
Hirawa N, Uehara Y, Kawabata Y, et al. Long-term inhibition of renin-angiotensin system sustains memory function in aged Dahl rats.
Sudilovsky A, Turnbull B, Croog SH, et al. Angiotensin converting enzyme and memory: preclinical and clinical data.
Ohrui T, Matsui T, Yamaya M, et al. Angiotensin-converting enzyme inhibitors and incidence of Alzheimer's disease in Japan.
Ohrui T, Tomita N, Sato-Nakagawa T, et al. Effects of brain-penetrating ACE inhibitors on Alzheimer disease progression.
Kehoe PG. The renin-angiotensin-aldosterone system and Alzheimer's disease?
Agerholm-Larsen B, Nordestgaard BG, Steffensen R, et al. ACE gene polymorphism: ischemic heart disease and longevity in 10,150 individuals. A case-referent and retrospective cohort study based on the Copenhagen City Heart Study.
Galinsky D, Tysoe C, Brayne CE, et al. Analysis of the apoe E/apo C-I, angiotensin converting enzyme and methylenetetrahydrofolate reductase genes as candidates affecting human longevity.
Shimokata H, Yamada Y, Nakagawa M, et al. Distribution of geriatric disease-related genotypes in the National Institute for Longevity Sciences, Longitudinal Study of Aging (NILS-LSA).
Blanché H, Cabanne L, Sahbatou M, et al. A study of French centenarians: are ACE and APOE associated with longevity?
Panza F, Solfrizzi V, D'Introno A, et al. Angiotensin I converting enzyme (ACE) gene polymorphism in centenarians: different allele frequencies between the North and South of Europe.
Da Cruz IB, Oliveira G, Taufer M, et al. Angiotensin I-converting enzyme gene polymorphism in two ethnic groups living in Brazil's southern region: association with age.
Alvarez R, Alvarez V, Lahoz CH, et al. Angiotensin converting enzyme and endothelial nitric oxide synthase DNA polymorphisms and late onset Alzheimer's disease.
Carbonell J, Allen R, Kalsi G, et al. Variation in the DCP1 gene, encoding the angiotensin converting enzyme ACE, is not associated with increased susceptibility to Alzheimer's disease.
Chapman J, Wang N, Treves TA, et al. ACE, MTHFR, factor V Leiden, and APOE polymorphisms in patients with vascular and Alzheimer's dementia.
Cheng CY, Hong CJ, Liu HC, et al. Study of the association between Alzheimer's disease and angiotensin-converting enzyme gene polymorphism using DNA from lymphocytes.
Hu J, Miyatake F, Aizu Y, et al. Angiotensin-converting enzyme genotype is associated with Alzheimer disease in the Japanese population.
Isbir T, Agaçhan B, Yilmaz H, et al. Angiotensin converting enzyme gene polymorphism in Alzheimer's disease.
Mattila KM, Rinne JO, Röyttä M, et al. Dipeptidyl carboxypeptidase 1 (DCP1) and butyrylcholinesterase (BCHE) gene interactions with the apolipoprotein E ε4 allele as risk factors in Alzheimer's disease and in Parkinson's disease with coexisting Alzheimer pathology.
Monastero R, Caldarella R, Mannino M, et al. Lack of association between angiotensin converting enzyme polymorphism and sporadic Alzheimer's disease.
Myllykangas L, Polvikoski T, Sulkava R, et al. Cardiovascular risk factors and Alzheimer's disease: a genetic association study in a population aged 85 or over.
Palumbo B, Cadini D, Nocentini G, et al. Angiotensin converting enzyme deletion allele in different kinds of dementia disorders.
Richard F, Fromentin-David I, Ricolfi F, et al. The angiotensin I converting enzyme gene as a susceptibility factor for dementia.
Scacchi R, De Bernardini L, Mantuano E, et al. DNA polymorphisms of apolipoprotein B and angiotensin 1-converting enzyme genes and relationships with lipid levels in Italian patients with vascular dementia or Alzheimer's disease.
Wu C, Zhou D, Guan Z, et al. The association between angiotensin I converting enzyme gene polymorphism and Chinese late onset Alzheimer disease. (In Chinese).
Yang JD, Feng G, Zhang J, et al. Association between angiotensin-converting enzyme gene and late onset Alzheimer's disease in Han Chinese.
Zuliani G, Ble' A, Zanca R, et al. Genetic polymorphisms in older subjects with vascular or Alzheimer's dementia.
American Psychiatric Association. Diagnostic and statistical manual of mental disorders: DSM-IV. 4th ed. Washington, DC: American Psychiatric Association,