-
PDF
- Split View
-
Views
-
Cite
Cite
N. Woodford, M. E. Ward, M. E. Kaufmann, J. Turton, E. J. Fagan, D. James, A. P. Johnson, R. Pike, M. Warner, T. Cheasty, A. Pearson, S. Harry, J. B. Leach, A. Loughrey, J. A. Lowes, R. E. Warren, D. M. Livermore, Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum β-lactamases in the UK, Journal of Antimicrobial Chemotherapy, Volume 54, Issue 4, October 2004, Pages 735–743, https://doi.org/10.1093/jac/dkh424
Close - Share Icon Share
Abstract
Objectives: During 2003, the Health Protection Agency's Antibiotic Resistance Monitoring and Reference Laboratory began to receive isolates of Escherichia coli for confirmation of extended-spectrum β-lactamase production with a phenotype implying a CTX-M-type β-lactamase, i.e. MICs of cefotaxime ≥8-fold higher than MICs of ceftazidime. Many were referred as being from community patients. We examined 291 CTX-M-producing isolates from the UK and investigated the genetic basis of their phenotype.
Methods: PCR was used to detect alleles encoding CTX-M enzymes and to assign these to their blaCTX-M phylogenetic groups. Selected alleles were sequenced. Producers were compared by analysis of banding patterns generated by pulsed-field gel electrophoresis of XbaI-digested genomic DNA. MICs were determined by an agar dilution method or by Etest.
Results: Of 291 CTX-M-producing E. coli isolates studied from 42 UK centres, 70 (24%) were reportedly from community patients, many of whom had only limited recent hospital contact. Community isolates were referred by 12 centres. Two hundred and seventy-nine (95.9%) producers contained genes encoding group 1 CTX-M enzymes and 12 contained blaCTX-M-9-like alleles. An epidemic CTX-M-15-producing strain was identified, with 110 community and inpatient isolates referred from six centres. Representatives of four other major strains also produced CTX-M-15, as did several sporadic isolates examined. Most producers were multi-resistant to fluoroquinolones, trimethoprim, tetracycline and aminoglycosides as well as to non-carbapenem β-lactams.
Conclusions: CTX-M-producing E. coli are a rapidly developing problem in the UK, with CTX-M-15 particularly common. The diversity of producers and geographical scatter of referring laboratories indicates wide dissemination of blaCTX-M genes. Because of the public health implications, including for the treatment of community-acquired urinary tract infections, the spread of these strains—and CTX-M-15 β-lactamase in particular—merits close monitoring.
Introduction
Extended-spectrum β-lactamases (ESBLs) are the major source of resistance to oxyimino-cephalosporins in Enterobacteriaceae.1 Most ESBLs are mutants of TEM and SHV enzymes, but CTX-M enzymes are increasingly important.2 The CTX-M types are diverse—with more than 30 alleles divided into five distinct phylogenetic groups—and have evolved via the genetic escape and mutation of the chromosomal β-lactamase genes of Kluyvera spp.2,3 CTX-M enzymes predominantly hydrolyse cefotaxime and most are only weakly active against ceftazidime, although some such as CTX-M-15 also have strong activity against ceftazidime.4 CTX-M enzymes have been the predominant ESBLs in Argentina for >10 years,5 but have a growing distribution and prevalence in many other parts of the world,2,6 including Europe.7,8 In the UK, CTX-M-type β-lactamases were first detected in 2000–2001, with a CTX-M-9-producing isolate of Klebsiella oxytoca in Leeds;9 a nosocomial cluster of Klebsiella pneumoniae isolates producing a CTX-M-25-like enzyme in Birmingham;10 and four unrelated CTX-M-15-producing isolates of Escherichia coli from three widely scattered hospitals.11
Most producers of TEM- and SHV-type ESBLs have been nosocomial isolates, predominantly klebsiellae, although recent data suggest a significant prevalence in Enterobacter spp.,12 where detection is complicated by the co-presence of AmpC. However, during 2003, the Health Protection Agency's Antibiotic Resistance Monitoring and Reference Laboratory (ARMRL) began to receive isolates of E. coli for confirmation of ESBL production, with a phenotype implying a CTX-M-type β-lactamase (see Materials and methods). Of particular concern was that many isolates were reported to be from patients attending general practice, with limited or no history of recent hospital contact. The emergence of ESBL-producing E. coli in the community is also being seen concurrently elsewhere in Europe and in Canada.2,13 ARMRL investigated the molecular nature of these isolates and their ESBLs in collaboration with the Laboratory of HealthCare-Associated Infections.
Materials and methods
Bacterial isolates and susceptibility testing
For this report, we analysed data for 287 CTX-M-producing E. coli isolates referred to ARMRL from 42 UK centres in the period January 2003–March 2004, and for four previously reported isolates from 2001.11 Detailed analysis of histories and outcomes for some patient groups has been undertaken14 and will be reported separately.
MICs were determined by agar dilution or by Etest, and were interpreted using BSAC breakpoints.15 A phenotype consistent with production of a CTX-M-type β-lactamase was defined by a cefotaxime MIC ≥8-fold higher than the ceftazidime MIC, with the MICs of both agents reduced significantly (again ≥8-fold) in the presence of 4 mg/L clavulanic acid.
Detection and characterization of blaCTX-M alleles
Isolates with a CTX-M phenotype were screened for blaCTX-M alleles by PCR, initially with universal primers MA1 and MA2 (amplicon size 554 bp),16 and then with primers specific for various blaCTX-M groups.16 Cycling conditions were: initial denaturation at 94°C for 3 min; 35 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 45 s; and a final elongation at 72°C for 5 min (G. Arlet, personal communication). Linkage of blaCTX-M alleles with ISEcp1-like elements, which have been implicated in their expression and mobilization,17–19 was sought with primers PROM+ and PRECTX-M-3B (expected amplicon size approx. 900 bp).4 Cycling conditions were: initial denaturation at 94°C for 5 min; 30 cycles of 94°C for 25 s, 52°C for 40 s and 72°C for 50 s; and a final elongation at 72°C for 6 min.
Both strands of selected amplicons were sequenced using dye-terminator chemistry on a CEQ8000 analyser (Beckman Coulter, High Wycombe, UK), either directly or after cloning into pCR2.1 (Invitrogen, Paisley, UK).
After determining the genetic context of blaCTX-M-15 in three representatives of epidemic strain A (see below), all isolates were screened with primers 5′-GCG GTA AAT CGT GGA GTG AT-3′ and 5′-ATT CGG CAA GTT TTT GCT GT-3′, which were designed to amplify a 400 bp fragment spanning the link between IS26 and blaCTX-M-15. Cycling conditions were the same as those for primers PROM+/PRECTX-M-3B.
Pulsed-field gel electrophoresis (PFGE) and serotyping
Isolates were compared by PFGE of XbaI-digested genomic DNA, as described previously,20 on a CHEF DRII apparatus (Bio-Rad Laboratories, Hemel Hempstead, UK). A linear ramp of 5–35 s was used and gels [1.2% (w/v) agarose in 0.5 × Tris/borate/EDTA buffer] were run for 30 h at 6 V/cm at 12°C. Banding patterns were analysed using BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium) and strains were defined as having PFGE profiles with ≥85% similarity (UPGMA, Dice coefficient). E. coli strain TN17, a CTX-M-15 producer from Paris that has an IS26 element linked to its blaCTX-M-15 gene,21 was included as a comparator.
In addition, all isolates were confirmed biochemically as E. coli and serotyped using the internationally accepted serotyping scheme based on the agglutination reactions of heat-stable lipopolysaccharide somatic or ‘O’ antigen with polyclonal rabbit antisera.22 The scheme comprises somatic antigens O1–O173. Strains that could not be serogrouped using the scheme were classed as unidentifiable and designated ‘O ?’.
Transfer of blaCTX-M in vitro
Transfer of cefotaxime resistance by conjugation from 10 CTX-M-producing donor isolates was attempted in broth and on agar plates, using E. coli J62-2 (RifR) (NCTC 50170) as the recipient. The donors were the first 10 isolates subjected to blaCTX-M sequencing. Transconjugants were selected on nutrient agar containing cefotaxime (10 mg/L) and rifampicin (50 mg/L).
GenBank accession numbers
Sequences relevant to this work have been deposited in GenBank under accession numbers AY462238 (strain A), AY463958 and AY536259.
Results
Detection of blaCTX-M alleles
All 291 isolates, from 42 centres, with a CTX-M phenotype contained blaCTX-M alleles, as demonstrated by PCR amplicons with the universal primers used. Of these, 279 (95.9%) yielded products with group 1-specific primers and with primers PROM+ and PRECTX-M-3B, indicating linkage of an allele encoding a group 1 CTX-M enzyme with an ISEcp1-like element. One hundred and sixty-nine isolates yielded products of the expected size (approx. 900 bp) with PROM+/PRECTX-M-3B, but the remaining 110 isolates gave amplicons of approx. 1.9 kb.
Twelve of 291 (4.1%) CTX-M-producing isolates, referred from nine centres, did not yield products with primers PROM+/PRECTX-M-3B. Amplification with group-specific primers indicated the presence of blaCTX-M-9-like alleles.
PFGE analysis, serotyping and identification of an epidemic CTX-M-producing E. coli strain
PFGE banding patterns were obtained for 287 of the 291 CTX-M-producing E. coli isolates; DNA from four producers consistently autodigested and no banding patterns were obtained. The 287 isolates represented multiple strains (Figure 1), among which several clusters of related isolates (≥85% similarity) were observed.
Most notable among the clusters were 110 isolates, from six centres, that represented a single epidemic strain, designated A. Most of these isolates were referred by centres 1 (in the West Midlands), 2 (on the south coast of England) and 41 (in Northern Ireland), although single isolates were referred from two hospitals in London and two isolates from another Midlands centre (Table 1). Isolates of strain A were from urine (59), blood (15), faecal screens (13), sputa (2), wounds (1) and of unknown origin (20).
Four other substantial clusters of isolates were noted. These were designated: strain B, with 13 isolates from four centres; strain C (with subtypes C and C'), with 29 isolates from nine centres; strain D, with 21 isolates from a single centre; and strain E (with subtypes E and E'), with 15 isolates from seven centres (Table 1).
Overall, 188 of 291 CTX-M producers (64.6%) belonged to one of these five major strains, which were related at 78% similarity (Figure 1). Fifty-six tested isolates of the five major strains all serotyped as O25. The remaining isolates represented numerous PFGE patterns and serotypes, although some small groups of related isolates were observed (Figure 1 and Table 1). The four CTX-M-15-producing E. coli isolates reported previously from the UK11 were unrelated to the major strains described here.
Among those laboratories referring >10 isolates, laboratory 1 was affected primarily by isolates of strains A and D (91/114 isolates), laboratory 2 solely by strain A (26/26 isolates), laboratory 41 by strains A and C/C' (23/26 isolates) and laboratory 4 partly by strains C/C' and E/E' (12/31 isolates). In contrast, laboratory 43 referred diverse CTX-M-producers with 16 different PFGE-defined strains among 18 referred isolates (Table 1).
E. coli strain TN17, a CTX-M-15 producer from France,21 was distinct from all major and other UK CTX-M-producing strains (data not shown).
blaCTX-M PCR in relation to PFGE
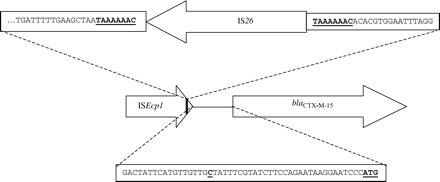
Twenty-five group 1 alleles were sequenced. Representatives of each of the five major strains (A–E) identified by PFGE produced CTX-M-15 β-lactamase. In 20 isolates from 17 centres, including representatives of strains B, C/C', D and E/E', blaCTX-M-15 was linked directly to an upstream ISEcp1-like element (e.g. GenBank AY463958). In contrast, all isolates belonging to epidemic strain A yielded approx. 1.9 kb amplicons with primers PROM+/PRECTX-M-3B. In the three isolates of strain A sequenced, from different centres, an IS26 element was found within the terminal inverted repeat of the ISEcpI-like element upstream of blaCTX-M-15 (Figure 2), thus separating the blaCTX-M-15 allele from its usual promoter. One of the three sequenced representatives of strain A possessed a T→C polymorphism in the spacer region between ISEcp1 and blaCTX-M-15 (GenBank AY462238; Figure 2).
The IS26 element in strain A was flanked by 8 bp direct repeats of ISEcp1 DNA, consistent with target site duplication following a transposition event (Figure 2). Based on this arrangement, a PCR assay was designed to amplify a 400 bp fragment spanning the link between IS26 and blaCTX-M-15. Members of strain A yielded amplicons of the expected size and most other producers failed to yield amplicons, but four isolates from centre 1, which were related by PFGE, gave products of approx. 800 bp. Sequencing of a representative isolate again revealed IS26 inserted within the ISEcp1-like element but, in this strain, the insertion was located upstream of the blaCTX-M promoter (and the PROM+ primer; GenBank AY536259).
A single PFGE-distinct isolate containing a group 1 allele and selected for sequencing produced CTX-M-3, which differs from CTX-M-15 by a single amino acid, Asp-240→Gly.4
All 12 producers of CTX-M-9-like enzymes had distinct PFGE banding patterns, including in those instances when more than one such isolate was received from a centre.
Antibiotic susceptibilities
In addition to extended-spectrum cephalosporins, most CTX-M producers were resistant to ciprofloxacin (MICs > 8 mg/L) and trimethoprim (MICs > 2 mg/L). All remained susceptible to carbapenems, and the MICs of mecillinam were also low (geometric mean MIC 1.5 mg/L). However, susceptibility to aminoglycosides and β-lactamase inhibitor combinations was variable between and within strains, but with resistance frequent (Tables 2 and 3).
When compared with other producers of group 1 CTX-M enzymes, isolates of epidemic strain A were less resistant to cefotaxime (geometric mean MIC 37.3 mg/L versus 81.3 mg/L), ceftazidime (geometric mean MIC 2.9 mg/L versus 30.4 mg/L), cefpodoxime (geometric mean MIC 49.7 mg/L versus 241.2 mg/L) and cefalexin (geometric mean MIC 49.7 mg/L versus 256 mg/L), and were markedly more susceptible to gentamicin (geometric mean MIC 1.1 mg/L versus 17.6 mg/L). Producers of CTX-M-9-like enzymes were also less resistant (Table 2). All tested isolates belonging to epidemic strain A were resistant to ciprofloxacin and trimethoprim, but remained susceptible to nitrofurantoin (MICs 8 mg/L) and fosfomycin (MICs ≤ 2 mg/L), as also did most isolates belonging to other strains (Table 3). Limited studies with co-trimoxazole and tetracycline suggested that resistance to these agents was frequent among CTX-M producers (data not shown).
Community-associated CTX-M-producing E. coli
Seventy (24%) CTX-M-producing isolates, mainly from urine specimens (52/65 with a stated origin), were considered by 12 referring centres to have been derived from samples obtained from patients in the community, although we cannot exclude the possibility that prior hospital contact had not been indicated to the laboratory that referred isolates to ARMRL. These isolates had diverse PFGE banding patterns, but representatives of each major strain were referred from community patients: 25 isolates belonged to epidemic strain A; one to strain B; 10 to strain C/C'; five to strain D; and four to strain E/E'. Among community-acquired CTX-M-producers, 67 produced group 1 enzymes and three produced CTX-M-9-like enzymes. The community isolates were as multi-resistant as hospital isolates.
Transfer of blaCTX-M in vitro
Transfer of CTX-M production was not achieved from 10 potential donor isolates tested, including representatives of strains A, B, D and E.
Discussion
ESBLs belonging to the CTX-M family were first reported in the UK in 2003, with the isolates having been collected in 2000–2002.9–11 Subsequently, CTX-M-producing strains of E. coli have emerged as a rapidly developing problem. As of July 2004, ARMRL had received almost 500 isolates from >70 centres located in all nine of the English Government Office Regions, as well as in Wales, Scotland and Northern Ireland. In many instances, referred isolates represent only a proportion of isolates from these centres, and were referred as examples of the strains. In this study, describing the first 291 E. coli producers referred to us, most isolates (95.9%) produced enzymes in phylogenetic group 1, particularly CTX-M-15, although a few isolates (4.1%) that produced CTX-M-9-like enzymes were also found. Since 2003, we have also detected CTX-M-15 in UK isolates of K. pneumoniae and Proteus mirabilis, and CTX-M-9 in an isolate of Enterobacter (N. Woodford, M. E. Ward, E. J. Fagan & D. M. Livermore, unpublished data).
CTX-M-15 was first reported in 2001 in E. coli, K. pneumoniae and Enterobacter aerogenes from India,17 but has now been widely reported, including from Bulgaria, Canada, France, Italy, Japan, Poland, Romania, Russia and Turkey.2,6 It is closely-related to CTX-M-3, differing by a single amino acid substitution (Asp-240→Gly). CTX-M-3 has been reported from many countries, including Poland, France, Japan, Taiwan, Greece and China.2 The same Asp-240→Gly substitution occurs in other CTX-M enzymes, such as CTX-M-16, -25, -27, -28, -29, -30 and -32, and, where kinetic data are available, has been associated with increased catalytic activity against ceftazidime.4,6,17,23,24 In this study, the expression of phenotypic ceftazidime resistance—previously recommended as a single screen for ESBLs—may have biased detection in diagnostic laboratories, and referral for reference testing, in favour of those CTX-M enzymes, such as CTX-M-15, that are most active against ceftazidime. Despite this, the MICs of cephalosporins for isolates of strain A, an epidemic CTX-M-15-producing E. coli strain with >100 individual patient isolates from six UK centres, were consistently lower than for other CTX-M-15 producers, and many isolates would be considered susceptible to ceftazidime by BSAC criteria (MICs ≤ 2 mg/L).15 As emphasized in several recent guidance updates,10,11,25 initial laboratory screens for ESBL-producing Enterobacteriaceae should be undertaken using both cefotaxime and ceftazidime or, if a single indicator is preferred, using cefpodoxime; resistance to cefotaxime and cefpodoxime was clear in all producers.
The blaCTX-M-15 allele is located on plasmids and is associated with an upstream copy of an ISEcp1-like element;17 similar genetic arrangements have been reported for several other blaCTX-M alleles.2,18,19 Indeed, the −35 and −10 promoter regions necessary for expression of these alleles are located within the 3′ end of the ISEcp1-like insertion sequences.16–18 We have not yet investigated comprehensively whether the blaCTX-M alleles in the numerous isolates reported here are plasmid-mediated (though transfer was not detected in the few representative isolates where conjugation was studied), but those encoding group 1 enzymes were all associated with an upstream ISEcp1-like element. Interestingly, isolates of strain A had an IS26 element within the terminal inverted repeat of their ISEcp1-like element. It seems likely that separation by IS26 of blaCTX-M-15 from its usual promoter was responsible for the lower cephalosporin MICs for strain A, as this organization may diminish transcription efficiency. However, it is unclear why the MICs of β-lactamase inhibitor combinations were not also lower for strain A as these should also reflect enzyme quantity. Further molecular studies are planned to identify the functional promoter of blaCTX-M-15 in this strain, to measure directly enzyme expression and to determine the reasons for the strain's susceptibility to gentamicin.
The presence of an IS26 element served, alongside the PFGE banding pattern, as a useful molecular marker for isolates belonging to epidemic strain A, and an ‘IS26-blaCTX-M link’ PCR assay was developed to allow rapid screening. Using this assay, we also identified four isolates, from a single centre, belonging to a second strain in which a copy of IS26 had inserted further upstream of blaCTX-M-15. This insertion sequence has also been associated with blaCTX-M alleles in E. coli isolates from Paris, France.16,21 Nevertheless, PFGE confirmed that a representative of these French strains, TN17, which also produces CTX-M-15,21 was unrelated to any CTX-M-producing strain from the UK. Thus, although the presence of the IS26-blaCTX-M link was a useful additional criterion for characterizing strain A, such linkage does not always indicate strain relatedness.
In addition to epidemic strain A, four other major CTX-M-15-producing strains were defined in this study on the basis of their PFGE banding patterns. Representative isolates of all major strains belonged to the same serotype, O25, and may have common ancestry. It is interesting that the five major strains appeared more closely related to each other (78% similarity) than to many of the other producers. Although strain D was only referred from one centre, the other major strains were referred from multiple locations. A cluster of cases involving an O25 ESBL-producing strain of E. coli was recently reported in the UK.26
Our data indicate that CTX-M-producing E. coli have become widely scattered throughout the UK, and that the underlying epidemiology is complex; it involves not only the spread of epidemic and other major strains between centres, but also instances of intra-hospital spread of minor strains and, potentially, horizontal transfer of plasmids or integrons carrying blaCTX-M alleles. Some of the centres that referred large numbers of isolates were affected largely by one or more of the major strains, whereas others, particularly around London, referred more diverse isolates.
The recognition of CTX-M-producing community strains is a cause of concern in many other countries besides the UK.2,13 In the current analysis, almost one quarter of UK isolates were derived from samples received from the community. Community-derived isolates included CTX-M-15 producers belonging to all of the major strains, including epidemic strain A, and producers of CTX-M-9-like enzymes. CTX-M-9-related enzymes have also been reported recently in non-hospitalized patients in Spain, although, here too, many patients had recent hospital contact.27 Clearly the epidemiology of CTX-M enzymes in the UK, as elsewhere, is very different from that of TEM- and SHV-derived ESBLs; specifically, CTX-M enzymes are not limited mainly to nosocomial klebsiellae, and their potential for spread beyond the hospital environment serves to exacerbate public health concerns.
The occurrence of CTX-M enzymes in the community presents treatment problems. Among oral agents licensed for treatment of urinary tract infection (UTI), only nitrofurantoin and fosfomycin were generally active and neither is ideal. Nitrofurantoin is unsuitable for empirical use, being inactive against Proteeae; while fosfomycin is not marketed in the UK. Other oral treatment options that may deserve investigation—at least in UTI—include mecillinam, which was surprisingly active in vitro, and simultaneous administration of co-amoxiclav and a third-generation cephalosporin. For more severe infections, including bacteraemia, necessitating intravenous therapy, carbapenems appear to be the only rational choice, with their empirical use being considered in settings and locales where CTX-M producers appear likely, e.g. in a patient with a community-acquired bacteraemia following a poorly-responsive UTI.
In summary, because of the significant public health implications, including for the treatment of community-acquired urinary tract infections, the spread of CTX-M producers in general, and of epidemic and non-epidemic strains with CTX-M-15 in particular, merits close monitoring.
Present address. Department of Healthcare-Associated Infection and Antimicrobial Resistance, CDSC, Health Protection Agency, London, UK
Dendrogram to illustrate the relatedness of 287 CTX-M-producing E. coli isolates from UK centres. Major strains (n=188 isolates), defined by PFGE profiles with ≥85% similarity (UPGMA, Dice; black vertical line), are indicated (see text for details). Each gradation on the scale represents a 5% difference in similarity. Banding patterns were not obtained from four isolates due to DNA autodigestion.
Details of CTX-M-producing E. coli isolates from 42 UK centres, including instances of possible community origin
| Referring laboratorya . | Government Office Region . | Number of isolates (n=291) . | Number of PFGE-defined strainsb . | Isolates of major strains (n=188) . | Possible community origin (n=70)c . |
|---|---|---|---|---|---|
| 1 | W Mids | 114 | 17 | A (70), B (3), C/C' (1), D (21), E/E' (3) | 36 |
| 4 | London | 31 | 16 | C/C' (9), E/E' (3) | 18 |
| 2 | S East | 26 | 1 | A (26) | 1 |
| 41 | Northern Ireland | 26 | 3 | A (10), C/C' (13) | 5 |
| 43 | London | 18 | 16 | A (1), B (1) | 0 |
| 18 | W Mids | 6 | 1 | B (6) | 0 |
| 3 | W Mids | 5 | 4 | A (2) | 2 |
| 24 | E Mids | 5 | 2 | – | 0 |
| 27 | S East | 5 | 2 | E/E' (4) | 0 |
| 36 | W Mids | 4 | 2 | B (3) | 2 |
| 38 | London | 4 | 3 | A (1) | 1 |
| 40 | W Mids | 4 | 2 | – | 0 |
| 9 | S East | 3 | 2 | C/C' (1), E/E' (1) | 1 |
| 16 | East | 3 | 1 | – | 0 |
| 23 | East | 3 | 3 | – | 0 |
| 13 | S East | 2 | 2 | E/E' (1) | 0 |
| 14 | London | 2 | 2 | – | 0 |
| 19 | Scotland | 2 | 2 | – | 0 |
| 25 | East | 2 | 1 | C/C' (1), E/E' (1) | 0 |
| 34 | London | 2 | 2 | – | 0 |
| 39 | W Mids | 2 | 2 | – | 0 |
| 53 | W Mids | 2 | 1 | – | 0 |
| Others (n=20) | – | 20 | – | C/C' (4), E/E' (2) | 4 |
| Referring laboratorya . | Government Office Region . | Number of isolates (n=291) . | Number of PFGE-defined strainsb . | Isolates of major strains (n=188) . | Possible community origin (n=70)c . |
|---|---|---|---|---|---|
| 1 | W Mids | 114 | 17 | A (70), B (3), C/C' (1), D (21), E/E' (3) | 36 |
| 4 | London | 31 | 16 | C/C' (9), E/E' (3) | 18 |
| 2 | S East | 26 | 1 | A (26) | 1 |
| 41 | Northern Ireland | 26 | 3 | A (10), C/C' (13) | 5 |
| 43 | London | 18 | 16 | A (1), B (1) | 0 |
| 18 | W Mids | 6 | 1 | B (6) | 0 |
| 3 | W Mids | 5 | 4 | A (2) | 2 |
| 24 | E Mids | 5 | 2 | – | 0 |
| 27 | S East | 5 | 2 | E/E' (4) | 0 |
| 36 | W Mids | 4 | 2 | B (3) | 2 |
| 38 | London | 4 | 3 | A (1) | 1 |
| 40 | W Mids | 4 | 2 | – | 0 |
| 9 | S East | 3 | 2 | C/C' (1), E/E' (1) | 1 |
| 16 | East | 3 | 1 | – | 0 |
| 23 | East | 3 | 3 | – | 0 |
| 13 | S East | 2 | 2 | E/E' (1) | 0 |
| 14 | London | 2 | 2 | – | 0 |
| 19 | Scotland | 2 | 2 | – | 0 |
| 25 | East | 2 | 1 | C/C' (1), E/E' (1) | 0 |
| 34 | London | 2 | 2 | – | 0 |
| 39 | W Mids | 2 | 2 | – | 0 |
| 53 | W Mids | 2 | 1 | – | 0 |
| Others (n=20) | – | 20 | – | C/C' (4), E/E' (2) | 4 |
Referring laboratory code numbers exceed 42 because the presented data were extracted from a larger database.
Strains were defined as isolates with PFGE banding patterns showing ≥85% similarity. No PFGE profiles were obtained for 4/291 isolates due to DNA autodigestion.
Isolates considered by the referring laboratory to have been community-derived.
Details of CTX-M-producing E. coli isolates from 42 UK centres, including instances of possible community origin
| Referring laboratorya . | Government Office Region . | Number of isolates (n=291) . | Number of PFGE-defined strainsb . | Isolates of major strains (n=188) . | Possible community origin (n=70)c . |
|---|---|---|---|---|---|
| 1 | W Mids | 114 | 17 | A (70), B (3), C/C' (1), D (21), E/E' (3) | 36 |
| 4 | London | 31 | 16 | C/C' (9), E/E' (3) | 18 |
| 2 | S East | 26 | 1 | A (26) | 1 |
| 41 | Northern Ireland | 26 | 3 | A (10), C/C' (13) | 5 |
| 43 | London | 18 | 16 | A (1), B (1) | 0 |
| 18 | W Mids | 6 | 1 | B (6) | 0 |
| 3 | W Mids | 5 | 4 | A (2) | 2 |
| 24 | E Mids | 5 | 2 | – | 0 |
| 27 | S East | 5 | 2 | E/E' (4) | 0 |
| 36 | W Mids | 4 | 2 | B (3) | 2 |
| 38 | London | 4 | 3 | A (1) | 1 |
| 40 | W Mids | 4 | 2 | – | 0 |
| 9 | S East | 3 | 2 | C/C' (1), E/E' (1) | 1 |
| 16 | East | 3 | 1 | – | 0 |
| 23 | East | 3 | 3 | – | 0 |
| 13 | S East | 2 | 2 | E/E' (1) | 0 |
| 14 | London | 2 | 2 | – | 0 |
| 19 | Scotland | 2 | 2 | – | 0 |
| 25 | East | 2 | 1 | C/C' (1), E/E' (1) | 0 |
| 34 | London | 2 | 2 | – | 0 |
| 39 | W Mids | 2 | 2 | – | 0 |
| 53 | W Mids | 2 | 1 | – | 0 |
| Others (n=20) | – | 20 | – | C/C' (4), E/E' (2) | 4 |
| Referring laboratorya . | Government Office Region . | Number of isolates (n=291) . | Number of PFGE-defined strainsb . | Isolates of major strains (n=188) . | Possible community origin (n=70)c . |
|---|---|---|---|---|---|
| 1 | W Mids | 114 | 17 | A (70), B (3), C/C' (1), D (21), E/E' (3) | 36 |
| 4 | London | 31 | 16 | C/C' (9), E/E' (3) | 18 |
| 2 | S East | 26 | 1 | A (26) | 1 |
| 41 | Northern Ireland | 26 | 3 | A (10), C/C' (13) | 5 |
| 43 | London | 18 | 16 | A (1), B (1) | 0 |
| 18 | W Mids | 6 | 1 | B (6) | 0 |
| 3 | W Mids | 5 | 4 | A (2) | 2 |
| 24 | E Mids | 5 | 2 | – | 0 |
| 27 | S East | 5 | 2 | E/E' (4) | 0 |
| 36 | W Mids | 4 | 2 | B (3) | 2 |
| 38 | London | 4 | 3 | A (1) | 1 |
| 40 | W Mids | 4 | 2 | – | 0 |
| 9 | S East | 3 | 2 | C/C' (1), E/E' (1) | 1 |
| 16 | East | 3 | 1 | – | 0 |
| 23 | East | 3 | 3 | – | 0 |
| 13 | S East | 2 | 2 | E/E' (1) | 0 |
| 14 | London | 2 | 2 | – | 0 |
| 19 | Scotland | 2 | 2 | – | 0 |
| 25 | East | 2 | 1 | C/C' (1), E/E' (1) | 0 |
| 34 | London | 2 | 2 | – | 0 |
| 39 | W Mids | 2 | 2 | – | 0 |
| 53 | W Mids | 2 | 1 | – | 0 |
| Others (n=20) | – | 20 | – | C/C' (4), E/E' (2) | 4 |
Referring laboratory code numbers exceed 42 because the presented data were extracted from a larger database.
Strains were defined as isolates with PFGE banding patterns showing ≥85% similarity. No PFGE profiles were obtained for 4/291 isolates due to DNA autodigestion.
Isolates considered by the referring laboratory to have been community-derived.
Susceptibilities of CTX-M-producing E. coli from the UK to β-lactams, alone and in combination with β-lactamase inhibitors
| . | MIC (mg/L) . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme produced/ strain . | AMP . | AMC . | CTX . | CTX . | CAZ . | CAZ . | CPD . | CPD . | RAD . | LEX . | MEC . | PIP . | TZP . | IPM . | MEM . | |||||||||||||||
| . | . | . | . | +CLA . | . | +CLA . | . | +CLA . | . | . | . | . | . | . | . | |||||||||||||||
| Group 1 | ||||||||||||||||||||||||||||||
| strain Aa | ||||||||||||||||||||||||||||||
| range | 64–256 | 8–64 | 8–256 | 0.06–1 | 0.5–32 | 0.125–0.5 | 32–256 | 0.5–2 | 32–256 | 16–256 | 1–8 | 64–256 | 8–256 | 0.06–0.25 | 0.016–0.125 | |||||||||||||||
| geometric mean | 94.2 | 18.1 | 37.3 | 0.1 | 2.9 | 0.2 | 49.7 | 0.7 | 53.0 | 49.7 | 1.4 | 94.2 | 20.1 | 0.2 | 0.05 | |||||||||||||||
| no. isolates tested | 61 | 61 | 63 | 58 | 63 | 58 | 22 | 22 | 22 | 22 | 22 | 61 | 61 | 60 | 61 | |||||||||||||||
| other major strainsa | ||||||||||||||||||||||||||||||
| range | 64–256 | 8–128 | 8–256 | 0.06–32 | 1–128 | 0.06–32 | 32–256 | 1–2 | 256.0 | 256.0 | 0.25–8 | 64–256 | 2–64 | 0.125–1 | 0.016–2 | |||||||||||||||
| geometric mean | 108.7 | 20.1 | 93.2 | 0.2 | 23.0 | 0.4 | 233.9 | 1.6 | 256.0 | 256.0 | 1.5 | 107.3 | 13.2 | 0.2 | 0.1 | |||||||||||||||
| no. isolates tested | 55 | 55 | 59 | 49 | 59 | 49 | 23 | 23 | 23 | 23 | 23 | 55 | 55 | 54 | 55 | |||||||||||||||
| non-major strains | ||||||||||||||||||||||||||||||
| range | 64–256 | 8–64 | 1–256 | 0.06–0.5 | 1–128 | 0.125–2 | 256.0 | 1–2 | 256.0 | 256.0 | 0.5–64 | 64–256 | 2–256 | 0.06–0.5 | 0.016–0.25 | |||||||||||||||
| geometric mean | 92.2 | 17.0 | 73.0 | 0.2 | 37.89 | 0.4 | 256.0 | 1.7 | 256.0 | 256.0 | 2.0 | 92.2 | 14.7 | 0.2 | 0.06 | |||||||||||||||
| no. isolates tested | 57 | 56 | 74 | 59 | 74 | 59 | 12 | 12 | 12 | 12 | 12 | 57 | 59 | 54 | 57 | |||||||||||||||
| CTX-M-9-like | ||||||||||||||||||||||||||||||
| Range | 64–256 | 8–16 | 16–256 | 0.06–0.25 | 1–32 | 0.125–0.5 | 128 | 2 | 128 | 128 | 0.5 | 64–256 | 2–16 | 0.125–0.5 | 0.06 | |||||||||||||||
| Geometric mean | 87.1 | 12.7 | 56.4 | 0.1 | 2.1 | 0.3 | 128.0 | 2.0 | 128.0 | 128.0 | 0.5 | 74.7 | 4.3 | 0.3 | 0.1 | |||||||||||||||
| No. isolates tested | 9 | 9 | 11 | 8 | 11 | 8 | 1 | 1 | 1 | 1 | 1 | 9 | 9 | 9 | 9 | |||||||||||||||
| . | MIC (mg/L) . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme produced/ strain . | AMP . | AMC . | CTX . | CTX . | CAZ . | CAZ . | CPD . | CPD . | RAD . | LEX . | MEC . | PIP . | TZP . | IPM . | MEM . | |||||||||||||||
| . | . | . | . | +CLA . | . | +CLA . | . | +CLA . | . | . | . | . | . | . | . | |||||||||||||||
| Group 1 | ||||||||||||||||||||||||||||||
| strain Aa | ||||||||||||||||||||||||||||||
| range | 64–256 | 8–64 | 8–256 | 0.06–1 | 0.5–32 | 0.125–0.5 | 32–256 | 0.5–2 | 32–256 | 16–256 | 1–8 | 64–256 | 8–256 | 0.06–0.25 | 0.016–0.125 | |||||||||||||||
| geometric mean | 94.2 | 18.1 | 37.3 | 0.1 | 2.9 | 0.2 | 49.7 | 0.7 | 53.0 | 49.7 | 1.4 | 94.2 | 20.1 | 0.2 | 0.05 | |||||||||||||||
| no. isolates tested | 61 | 61 | 63 | 58 | 63 | 58 | 22 | 22 | 22 | 22 | 22 | 61 | 61 | 60 | 61 | |||||||||||||||
| other major strainsa | ||||||||||||||||||||||||||||||
| range | 64–256 | 8–128 | 8–256 | 0.06–32 | 1–128 | 0.06–32 | 32–256 | 1–2 | 256.0 | 256.0 | 0.25–8 | 64–256 | 2–64 | 0.125–1 | 0.016–2 | |||||||||||||||
| geometric mean | 108.7 | 20.1 | 93.2 | 0.2 | 23.0 | 0.4 | 233.9 | 1.6 | 256.0 | 256.0 | 1.5 | 107.3 | 13.2 | 0.2 | 0.1 | |||||||||||||||
| no. isolates tested | 55 | 55 | 59 | 49 | 59 | 49 | 23 | 23 | 23 | 23 | 23 | 55 | 55 | 54 | 55 | |||||||||||||||
| non-major strains | ||||||||||||||||||||||||||||||
| range | 64–256 | 8–64 | 1–256 | 0.06–0.5 | 1–128 | 0.125–2 | 256.0 | 1–2 | 256.0 | 256.0 | 0.5–64 | 64–256 | 2–256 | 0.06–0.5 | 0.016–0.25 | |||||||||||||||
| geometric mean | 92.2 | 17.0 | 73.0 | 0.2 | 37.89 | 0.4 | 256.0 | 1.7 | 256.0 | 256.0 | 2.0 | 92.2 | 14.7 | 0.2 | 0.06 | |||||||||||||||
| no. isolates tested | 57 | 56 | 74 | 59 | 74 | 59 | 12 | 12 | 12 | 12 | 12 | 57 | 59 | 54 | 57 | |||||||||||||||
| CTX-M-9-like | ||||||||||||||||||||||||||||||
| Range | 64–256 | 8–16 | 16–256 | 0.06–0.25 | 1–32 | 0.125–0.5 | 128 | 2 | 128 | 128 | 0.5 | 64–256 | 2–16 | 0.125–0.5 | 0.06 | |||||||||||||||
| Geometric mean | 87.1 | 12.7 | 56.4 | 0.1 | 2.1 | 0.3 | 128.0 | 2.0 | 128.0 | 128.0 | 0.5 | 74.7 | 4.3 | 0.3 | 0.1 | |||||||||||||||
| No. isolates tested | 9 | 9 | 11 | 8 | 11 | 8 | 1 | 1 | 1 | 1 | 1 | 9 | 9 | 9 | 9 | |||||||||||||||
AMP, ampicillin; AMC, co-amoxiclav; CTX, cefotaxime; CAZ, ceftazidime; CLA, clavulanic acid; CPD, cefpodoxime; RAD, cefradine; LEX, cefalexin; MEC, mecillinam; PIP, piperacillin; TZP, piperacillin/tazobactam; IPM, imipenem; MEM, meropenem.
Defined by PFGE (see text and Figure 1)
Susceptibilities of CTX-M-producing E. coli from the UK to β-lactams, alone and in combination with β-lactamase inhibitors
| . | MIC (mg/L) . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme produced/ strain . | AMP . | AMC . | CTX . | CTX . | CAZ . | CAZ . | CPD . | CPD . | RAD . | LEX . | MEC . | PIP . | TZP . | IPM . | MEM . | |||||||||||||||
| . | . | . | . | +CLA . | . | +CLA . | . | +CLA . | . | . | . | . | . | . | . | |||||||||||||||
| Group 1 | ||||||||||||||||||||||||||||||
| strain Aa | ||||||||||||||||||||||||||||||
| range | 64–256 | 8–64 | 8–256 | 0.06–1 | 0.5–32 | 0.125–0.5 | 32–256 | 0.5–2 | 32–256 | 16–256 | 1–8 | 64–256 | 8–256 | 0.06–0.25 | 0.016–0.125 | |||||||||||||||
| geometric mean | 94.2 | 18.1 | 37.3 | 0.1 | 2.9 | 0.2 | 49.7 | 0.7 | 53.0 | 49.7 | 1.4 | 94.2 | 20.1 | 0.2 | 0.05 | |||||||||||||||
| no. isolates tested | 61 | 61 | 63 | 58 | 63 | 58 | 22 | 22 | 22 | 22 | 22 | 61 | 61 | 60 | 61 | |||||||||||||||
| other major strainsa | ||||||||||||||||||||||||||||||
| range | 64–256 | 8–128 | 8–256 | 0.06–32 | 1–128 | 0.06–32 | 32–256 | 1–2 | 256.0 | 256.0 | 0.25–8 | 64–256 | 2–64 | 0.125–1 | 0.016–2 | |||||||||||||||
| geometric mean | 108.7 | 20.1 | 93.2 | 0.2 | 23.0 | 0.4 | 233.9 | 1.6 | 256.0 | 256.0 | 1.5 | 107.3 | 13.2 | 0.2 | 0.1 | |||||||||||||||
| no. isolates tested | 55 | 55 | 59 | 49 | 59 | 49 | 23 | 23 | 23 | 23 | 23 | 55 | 55 | 54 | 55 | |||||||||||||||
| non-major strains | ||||||||||||||||||||||||||||||
| range | 64–256 | 8–64 | 1–256 | 0.06–0.5 | 1–128 | 0.125–2 | 256.0 | 1–2 | 256.0 | 256.0 | 0.5–64 | 64–256 | 2–256 | 0.06–0.5 | 0.016–0.25 | |||||||||||||||
| geometric mean | 92.2 | 17.0 | 73.0 | 0.2 | 37.89 | 0.4 | 256.0 | 1.7 | 256.0 | 256.0 | 2.0 | 92.2 | 14.7 | 0.2 | 0.06 | |||||||||||||||
| no. isolates tested | 57 | 56 | 74 | 59 | 74 | 59 | 12 | 12 | 12 | 12 | 12 | 57 | 59 | 54 | 57 | |||||||||||||||
| CTX-M-9-like | ||||||||||||||||||||||||||||||
| Range | 64–256 | 8–16 | 16–256 | 0.06–0.25 | 1–32 | 0.125–0.5 | 128 | 2 | 128 | 128 | 0.5 | 64–256 | 2–16 | 0.125–0.5 | 0.06 | |||||||||||||||
| Geometric mean | 87.1 | 12.7 | 56.4 | 0.1 | 2.1 | 0.3 | 128.0 | 2.0 | 128.0 | 128.0 | 0.5 | 74.7 | 4.3 | 0.3 | 0.1 | |||||||||||||||
| No. isolates tested | 9 | 9 | 11 | 8 | 11 | 8 | 1 | 1 | 1 | 1 | 1 | 9 | 9 | 9 | 9 | |||||||||||||||
| . | MIC (mg/L) . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme produced/ strain . | AMP . | AMC . | CTX . | CTX . | CAZ . | CAZ . | CPD . | CPD . | RAD . | LEX . | MEC . | PIP . | TZP . | IPM . | MEM . | |||||||||||||||
| . | . | . | . | +CLA . | . | +CLA . | . | +CLA . | . | . | . | . | . | . | . | |||||||||||||||
| Group 1 | ||||||||||||||||||||||||||||||
| strain Aa | ||||||||||||||||||||||||||||||
| range | 64–256 | 8–64 | 8–256 | 0.06–1 | 0.5–32 | 0.125–0.5 | 32–256 | 0.5–2 | 32–256 | 16–256 | 1–8 | 64–256 | 8–256 | 0.06–0.25 | 0.016–0.125 | |||||||||||||||
| geometric mean | 94.2 | 18.1 | 37.3 | 0.1 | 2.9 | 0.2 | 49.7 | 0.7 | 53.0 | 49.7 | 1.4 | 94.2 | 20.1 | 0.2 | 0.05 | |||||||||||||||
| no. isolates tested | 61 | 61 | 63 | 58 | 63 | 58 | 22 | 22 | 22 | 22 | 22 | 61 | 61 | 60 | 61 | |||||||||||||||
| other major strainsa | ||||||||||||||||||||||||||||||
| range | 64–256 | 8–128 | 8–256 | 0.06–32 | 1–128 | 0.06–32 | 32–256 | 1–2 | 256.0 | 256.0 | 0.25–8 | 64–256 | 2–64 | 0.125–1 | 0.016–2 | |||||||||||||||
| geometric mean | 108.7 | 20.1 | 93.2 | 0.2 | 23.0 | 0.4 | 233.9 | 1.6 | 256.0 | 256.0 | 1.5 | 107.3 | 13.2 | 0.2 | 0.1 | |||||||||||||||
| no. isolates tested | 55 | 55 | 59 | 49 | 59 | 49 | 23 | 23 | 23 | 23 | 23 | 55 | 55 | 54 | 55 | |||||||||||||||
| non-major strains | ||||||||||||||||||||||||||||||
| range | 64–256 | 8–64 | 1–256 | 0.06–0.5 | 1–128 | 0.125–2 | 256.0 | 1–2 | 256.0 | 256.0 | 0.5–64 | 64–256 | 2–256 | 0.06–0.5 | 0.016–0.25 | |||||||||||||||
| geometric mean | 92.2 | 17.0 | 73.0 | 0.2 | 37.89 | 0.4 | 256.0 | 1.7 | 256.0 | 256.0 | 2.0 | 92.2 | 14.7 | 0.2 | 0.06 | |||||||||||||||
| no. isolates tested | 57 | 56 | 74 | 59 | 74 | 59 | 12 | 12 | 12 | 12 | 12 | 57 | 59 | 54 | 57 | |||||||||||||||
| CTX-M-9-like | ||||||||||||||||||||||||||||||
| Range | 64–256 | 8–16 | 16–256 | 0.06–0.25 | 1–32 | 0.125–0.5 | 128 | 2 | 128 | 128 | 0.5 | 64–256 | 2–16 | 0.125–0.5 | 0.06 | |||||||||||||||
| Geometric mean | 87.1 | 12.7 | 56.4 | 0.1 | 2.1 | 0.3 | 128.0 | 2.0 | 128.0 | 128.0 | 0.5 | 74.7 | 4.3 | 0.3 | 0.1 | |||||||||||||||
| No. isolates tested | 9 | 9 | 11 | 8 | 11 | 8 | 1 | 1 | 1 | 1 | 1 | 9 | 9 | 9 | 9 | |||||||||||||||
AMP, ampicillin; AMC, co-amoxiclav; CTX, cefotaxime; CAZ, ceftazidime; CLA, clavulanic acid; CPD, cefpodoxime; RAD, cefradine; LEX, cefalexin; MEC, mecillinam; PIP, piperacillin; TZP, piperacillin/tazobactam; IPM, imipenem; MEM, meropenem.
Defined by PFGE (see text and Figure 1)
Susceptibilities of CTX-M-producing E. coli from the UK to non-β-lactam antibiotics
| . | MIC (mg/L) . | . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme produced/strain . | CIP . | AMK . | GEN . | TMP . | NIT . | FOF . | ||||||
| Group 1 | ||||||||||||
| strain Aa | ||||||||||||
| range | 8–256 | 2–32 | 0.5–4 | 256 | 8 | 0.5–2 | ||||||
| geometric mean | 17.5 | 9.0 | 1.1 | 256.0 | 8.0 | 0.9 | ||||||
| no. isolates tested | 61 | 61 | 62 | 15 | 15 | 22 | ||||||
| other major strainsa | ||||||||||||
| range | 2–256 | 8–256 | 0.5–256 | 0.25–256 | 4–16 | 0.5–1 | ||||||
| geometric mean | 6.7 | 18.2 | 28.6 | 9.6 | 7.3 | 0.6 | ||||||
| no. isolates tested | 55 | 55 | 56 | 15 | 15 | 23 | ||||||
| non-major strains | ||||||||||||
| range | 2–64 | 0.03–256 | 0.25–128 | 0.25–256 | 4–256 | 0.5–256 | ||||||
| geometric mean | 6.1 | 9.3 | 12.2 | 45.3 | 22.6 | 1.9 | ||||||
| no. isolates tested | 57 | 59 | 73 | 10 | 10 | 12 | ||||||
| CTX-M-9-like | ||||||||||||
| Range | 0.125–32 | 1–16 | 0.25–32 | – | – | 0.5 | ||||||
| Geometric mean | 1.6 | 2.3 | 2.3 | – | – | 0.5 | ||||||
| No. isolates tested | 9 | 9 | 11 | 0 | 0 | 1 | ||||||
| . | MIC (mg/L) . | . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme produced/strain . | CIP . | AMK . | GEN . | TMP . | NIT . | FOF . | ||||||
| Group 1 | ||||||||||||
| strain Aa | ||||||||||||
| range | 8–256 | 2–32 | 0.5–4 | 256 | 8 | 0.5–2 | ||||||
| geometric mean | 17.5 | 9.0 | 1.1 | 256.0 | 8.0 | 0.9 | ||||||
| no. isolates tested | 61 | 61 | 62 | 15 | 15 | 22 | ||||||
| other major strainsa | ||||||||||||
| range | 2–256 | 8–256 | 0.5–256 | 0.25–256 | 4–16 | 0.5–1 | ||||||
| geometric mean | 6.7 | 18.2 | 28.6 | 9.6 | 7.3 | 0.6 | ||||||
| no. isolates tested | 55 | 55 | 56 | 15 | 15 | 23 | ||||||
| non-major strains | ||||||||||||
| range | 2–64 | 0.03–256 | 0.25–128 | 0.25–256 | 4–256 | 0.5–256 | ||||||
| geometric mean | 6.1 | 9.3 | 12.2 | 45.3 | 22.6 | 1.9 | ||||||
| no. isolates tested | 57 | 59 | 73 | 10 | 10 | 12 | ||||||
| CTX-M-9-like | ||||||||||||
| Range | 0.125–32 | 1–16 | 0.25–32 | – | – | 0.5 | ||||||
| Geometric mean | 1.6 | 2.3 | 2.3 | – | – | 0.5 | ||||||
| No. isolates tested | 9 | 9 | 11 | 0 | 0 | 1 | ||||||
CIP, ciprofloxacin; AMK, amikacin; GEN, gentamicin; TMP, trimethoprim; NIT, nitrofurantoin; FOF, fosfomycin.
Defined by PFGE (see text and Figure 1).
Susceptibilities of CTX-M-producing E. coli from the UK to non-β-lactam antibiotics
| . | MIC (mg/L) . | . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme produced/strain . | CIP . | AMK . | GEN . | TMP . | NIT . | FOF . | ||||||
| Group 1 | ||||||||||||
| strain Aa | ||||||||||||
| range | 8–256 | 2–32 | 0.5–4 | 256 | 8 | 0.5–2 | ||||||
| geometric mean | 17.5 | 9.0 | 1.1 | 256.0 | 8.0 | 0.9 | ||||||
| no. isolates tested | 61 | 61 | 62 | 15 | 15 | 22 | ||||||
| other major strainsa | ||||||||||||
| range | 2–256 | 8–256 | 0.5–256 | 0.25–256 | 4–16 | 0.5–1 | ||||||
| geometric mean | 6.7 | 18.2 | 28.6 | 9.6 | 7.3 | 0.6 | ||||||
| no. isolates tested | 55 | 55 | 56 | 15 | 15 | 23 | ||||||
| non-major strains | ||||||||||||
| range | 2–64 | 0.03–256 | 0.25–128 | 0.25–256 | 4–256 | 0.5–256 | ||||||
| geometric mean | 6.1 | 9.3 | 12.2 | 45.3 | 22.6 | 1.9 | ||||||
| no. isolates tested | 57 | 59 | 73 | 10 | 10 | 12 | ||||||
| CTX-M-9-like | ||||||||||||
| Range | 0.125–32 | 1–16 | 0.25–32 | – | – | 0.5 | ||||||
| Geometric mean | 1.6 | 2.3 | 2.3 | – | – | 0.5 | ||||||
| No. isolates tested | 9 | 9 | 11 | 0 | 0 | 1 | ||||||
| . | MIC (mg/L) . | . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme produced/strain . | CIP . | AMK . | GEN . | TMP . | NIT . | FOF . | ||||||
| Group 1 | ||||||||||||
| strain Aa | ||||||||||||
| range | 8–256 | 2–32 | 0.5–4 | 256 | 8 | 0.5–2 | ||||||
| geometric mean | 17.5 | 9.0 | 1.1 | 256.0 | 8.0 | 0.9 | ||||||
| no. isolates tested | 61 | 61 | 62 | 15 | 15 | 22 | ||||||
| other major strainsa | ||||||||||||
| range | 2–256 | 8–256 | 0.5–256 | 0.25–256 | 4–16 | 0.5–1 | ||||||
| geometric mean | 6.7 | 18.2 | 28.6 | 9.6 | 7.3 | 0.6 | ||||||
| no. isolates tested | 55 | 55 | 56 | 15 | 15 | 23 | ||||||
| non-major strains | ||||||||||||
| range | 2–64 | 0.03–256 | 0.25–128 | 0.25–256 | 4–256 | 0.5–256 | ||||||
| geometric mean | 6.1 | 9.3 | 12.2 | 45.3 | 22.6 | 1.9 | ||||||
| no. isolates tested | 57 | 59 | 73 | 10 | 10 | 12 | ||||||
| CTX-M-9-like | ||||||||||||
| Range | 0.125–32 | 1–16 | 0.25–32 | – | – | 0.5 | ||||||
| Geometric mean | 1.6 | 2.3 | 2.3 | – | – | 0.5 | ||||||
| No. isolates tested | 9 | 9 | 11 | 0 | 0 | 1 | ||||||
CIP, ciprofloxacin; AMK, amikacin; GEN, gentamicin; TMP, trimethoprim; NIT, nitrofurantoin; FOF, fosfomycin.
Defined by PFGE (see text and Figure 1).
We are grateful to our many colleagues in clinical microbiology laboratories throughout the UK who refer E. coli isolates for confirmation of ESBL production. Thanks also to Dr Guillaume Arlet (Laboratoire de Bactériologie, Université Paris) for providing strain TN17. Work by D. M. L. and N. W. on emerging β-lactamases is supported, in part, by the EU/FP6-funded COBRA project (6-PCRD LSHM-CT-2003–503–335). Part of this work was presented at the 14th ECCMID (Prague, May 2004; poster P 758).
References
Bradford, P. A. (
Bonnet, R. (
Poirel, L., Kampfer, P. & Nordmann, P. (
Poirel, L., Gniadkowski, M. & Nordmann, P. (
Radice, M., Power, P., Di Conza, J. et al. (
Walther-Rasmussen, J. & Hoiby, N. (
Baraniak, A., Fiett, J., Sulikowska, A. et al. (
Bou, G., Cartelle, M., Tomas, M. et al. (
Alobwede, I., M'Zali, F. H., Livermore, D. M. et al. (
Brenwald, N. P., Jevons, G., Andrews, J. M. et al. (
Mushtaq, S., Woodford, N., Potz, N. et al. (
Reynolds, R., Potz, N., Colman, M. et al. (
Pitout, J. D., Hanson, N. D., Church, D. L. et al. (
Warren, R., Doroshenko, A., Carr, A. et al. (
Andrews, J. M. (
Saladin, M., Cao, V. T., Lambert, T. et al. (
Karim, A., Poirel, L., Nagarajan, S. et al. (
Poirel, L., Decousser, J. W. & Nordmann, P. (
Lartigue, M. F., Poirel, L. & Nordmann, P. (
Kaufmann, M. E. (
Eckert, C., Gautier, V., Saladin-Allard, M. et al. (
Gross, R. J. & Rowe, B. (
Bonnet, R., Recule, C., Baraduc, R. et al. (
Cartelle, M., Del Mar, T. M., Molina, F. et al. (
Livermore, D. M. & Brown, D. F. (
Author notes
1Antibiotic Resistance Monitoring and Reference Laboratory, 2Laboratory of HealthCare-Associated Infections, and 3Laboratory of Enteric Pathogens, Specialist and Reference Microbiology Division–Colindale, Health Protection Agency, London; 4Communicable Disease Surveillance Centre, Health Protection Agency, London; 5Local and Regional Services, Health Protection Agency, London; 6Kingston Hospital NHS Trust, Kingston, Surrey; 7Belfast City Hospital, Belfast; 8HPA South-East, Southampton Laboratory, Southampton; 9Shrewsbury and Telford Hospital NHS Trust, Shrewsbury, UK
- alleles
- phenotype
- polymerase chain reaction
- patient referral
- antibiotic resistance, bacterial
- carbapenem
- cefotaxime
- urinary tract infections
- agar
- ceftazidime
- dna
- electrophoresis, gel, pulsed-field
- fluoroquinolones
- genes
- genome
- inpatients
- laboratory
- lactams
- trimethoprim
- enzymes
- genetics
- public health medicine
- tetracycline
- aminoglycosides
- epidemics
- escherichia coli
- multi-antibiotic resistance
- community
- dilution technique
- dilute (action)
- malnutrition-inflammation-cachexia syndrome