-
PDF
- Split View
-
Views
-
Cite
Cite
George A. Pankey, Tigecycline, Journal of Antimicrobial Chemotherapy, Volume 56, Issue 3, September 2005, Pages 470–480, https://doi.org/10.1093/jac/dki248
Close - Share Icon Share
Abstract
New antimicrobial agents are urgently needed for clinical use due to the increasing prevalence and spread of multidrug-resistant bacteria that are commonly responsible for serious and life-threatening diseases. The need to develop new agents that effectively overcome existing mechanisms of resistance displayed by bacteria resistant to currently available drugs has become paramount. Tigecycline, the first in a new class of antimicrobials, the glycylcyclines, is an analogue of minocycline with additional properties that negate most mechanisms mediating resistance to the tetracyclines. In vitro testing has revealed that tigecycline has activity against vancomycin-resistant enterococci, methicillin-resistant Staphylococcus aureus, penicillin-resistant Streptococcus pneumoniae and many species of multidrug-resistant Gram-negative bacteria, although resistance to tigecycline by Pseudomonas aeruginosa and reduced susceptibility among Proteus species do occur. Tigecycline is being evaluated in multicentre Phase III clinical trials for therapy of many serious and life-threatening infections in which multidrug-resistant bacterial organisms may be found. Tigecycline appears to hold promise as a novel expanded spectrum antibiotic.
Introduction
Resistance to currently available antibiotics is increasing at an alarming rate. At the same time, the development of new antimicrobial agents to treat serious bacterial infections is decreasing.1,2 As a result of the emergence and spread of multidrug resistance in many pathogenic bacterial species, the recognition of new bacterial pathogens, and the fear of bioterrorism with multidrug-resistant microbes, the need for new antimicrobial agents is more urgent and greater than ever. Many pharmaceutical companies have abandoned or diminished their programmes for developing new antimicrobial agents. It has been recently reported that only six of 506 drugs under development by the largest biotechnology and pharmaceutical companies are new antibacterial agents.3 In addition, FDA approval for new antibacterial drugs has decreased by over 50% in the past 20 years.3 A recent position paper published by the Infectious Diseases Society of America has identified emerging and worsening resistance among common bacteria as significantly contributing to the need for expanded antibacterial drug development.4
In some settings, the magnitude of antibiotic resistance found in bacterial pathogens isolated from the community has become strikingly similar to that previously reported in hospital-based studies.5 Community-associated methicillin-resistant Staphylococcus aureus (MRSA) is increasing in prevalence and complicates empirical antimicrobial therapy as previously-active β-lactams no longer have activity against these resistant bacteria.6,7 It is of great concern that the loss of activity of previously efficacious antimicrobial agents occurs both in the hospital setting and the community.
When patients are diagnosed as having a serious infection, tests are performed and empirical antibiotics are started. The use of appropriate empirical therapy in bacteraemia and other serious infections can be life-saving, and it is critical to choose an antimicrobial regimen to which the bacterial organisms are susceptible. Additionally, the accelerated emergence of resistance to previously active antibiotics has created limitations in the choice of appropriate antibiotics available to physicians.4,8–10 Moreover, recently published data suggest that resistance to antimicrobial agents among pathogenic bacteria is causing worse outcomes for patients, including increased mortality rates in both Gram-negative and Gram-positive bacterial infections.11,12
Specifically, MRSA, vancomycin-resistant enterococci, penicillin-resistant Streptococcus pneumoniae and multidrug-resistant Gram-negative bacteria containing expanded spectrum β-lactamases (ESBLs), commonly found in hospitals, have severely limited the number of appropriate antimicrobial agents available for use.10,13 Also of great concern has been the isolation of vancomycin-resistant S. aureus in the hospital setting and the increasing identification of MRSA in the community.14,15
An important class of antibiotics is the tetracyclines. When they were first introduced into clinical practice after their discovery in the 1940s, the tetracyclines exhibited a wide range of activity against Gram-positive and Gram-negative bacteria, chlamydiae, mycoplasmas, rickettsiae and protozoan parasites. Initially, due to their broad spectrum of activity, tetracyclines were used widely in the prophylaxis and therapy of human and animal infections. By 1953, the first tetracycline-resistant bacterium was isolated, since then tetracycline resistance has increased substantially and now occurs in a significant number of bacteria. The dissemination of tetracycline-resistant microbes currently limits the use of tetracyclines in the treatment of many infections.16
As a result of the spread of tetracycline resistance, a new class of antibiotics, the glycylcyclines, is currently being developed. These new agents exhibit potent in vitro activity when tested against a broad spectrum of both tetracycline-susceptible and tetracycline-resistant Gram-positive and Gram-negative bacteria. Glycylcycline activity is maintained even in the presence of drug efflux pumps and ribosomal protection mechanisms that otherwise confer tetracycline resistance.17
Tigecycline (9-t-butylglycylamido-minocycline; GAR-936, Wyeth Laboratories, Collegeville, PA, USA) is the glycylcycline that is farthest along in clinical trials. Tigecycline has potent in vitro activity against most Gram-positive and Gram-negative aerobic and anaerobic bacteria including S. aureus, Enterococcus spp., S. pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, Neisseria gonorrhoeae, peptostreptococci, Clostridium spp., Enterobacteriaceae and Bacteroides spp.18,19 Tigecycline has limited activity against Pseudomonas aeruginosa, and reduced activity against indole-positive Proteae and Proteus mirabilis has been noted.20–22 Tigecycline is very active against bacteria resistant to other classes of antibiotics, including the quinolones and β-lactams. Tigecycline, additionally, resists deactivation by most known tetracycline resistance mechanisms.23 Because of this promising profile against clinically important bacteria, as well as promising pharmacodynamic and pharmacokinetic data, tigecycline is being studied in clinical trials.24,25 The suitability of tigecycline for serious infections is being assessed in patients with complicated skin and skin-structure infections, intra-abdominal infections and pneumonia.26,27
Mechanism of action, chemistry, and ability of tigecycline to overcome resistance mechanisms
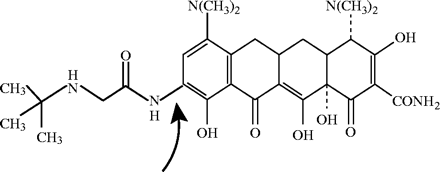
Glycylcyclines, a novel group of antibacterial agents, have the central four-ring carbocyclic skeleton present in the tetracyclines that is necessary for antibacterial activity. In the glycylcyclines, substitution of an N-alkyl-glycylamido group on the D ring at the 9th position facilitates the broader spectrum of activity, and additionally, this creates the ability to overcome most tetracycline resistance mechanisms. Tigecycline has a 9-t-butyl-glycylamido side chain on the central skeleton, and it is an analogue of minocycline (Figure 1). Tigecycline's molecular weight is 585.65 Da and its chemical formula is C29H39N5O8.24
Structure of tigecycline. The arrow indicates the addition of a 9-tert-butyl-glycylamido side chain on the D ring at the 9th position.
Ribosomal protection and active drug efflux are the major mechanisms conferring resistance to the tetracyclines.28,29 Resistance to tetracyclines can be acquired by other bacteria due to the spread of resistance genes, which are found in plasmids, integrons and conjugative transposons.25 Tigecycline appears to overcome these mechanisms of resistance because of steric hindrance due to a large substituent at position 9.30 Experiments conducted with dimethylsulphate modification of tetracycline and tigecycline binding sites, mutational analysis of 16S rRNA and analysis of tigecycline interacting with the 30S ribosomal subunit appear to confirm the role of steric hindrance.23
The activity of tigecycline has been assessed in Escherichia coli KAM3 (acrB) derivatives with plasmids containing different tetracycline-specific efflux transporter genes [tet(B), tet(C) and tet(K)]. The results of these studies showed the potent activity of tigecycline against all of the tested tetracycline-resistant bacterial strains. This was confirmed by the finding that MICs for the mutant strains were unchanged when compared with the original strains. The Tet efflux transporter does not take up tigecycline when present in low concentrations, and this probably explains the maintenance of tigecycline's antibacterial activity.31
Additionally, the actions of multidrug transporter genes, acrAB, acrEF and bcr, have been evaluated. Tigecycline appears to be a substrate of AcrAB and AcrEF. AcrAB and AcrEF are believed to be resistance-nodulation-division-type multicomponent efflux transporters. In experiments with strains producing these multidrug efflux proteins, the MICs of tigecycline increased fourfold.31 Recently, it has been reported that the AcrAB efflux pump is associated with reduced susceptibility to tigecycline among strains of P. mirabilis, Klebsiella pneumoniae, Morganella morganii and Enterobacter spp.32
Earlier studies evaluating potential development of glycylcycline resistance used laboratory-generated bacteria with mutations in the portion of the tetA(B) gene encoding a transmembrane spanning region of the efflux pump. The authors suggested that changes in the interdomain region of the efflux pump may be responsible for future glycylcycline resistance as these agents enter clinical use.33 In other laboratory studies, it has been difficult to generate mutations in bacteria engendering tigecycline resistance. Only small changes in tigecycline susceptibility have been noted among these mutant bacteria. Because of this, it has been hypothesized that significant bacterial resistance to tigecycline in the clinic will not occur easily, and the development of resistance will require more than trivial mutations in currently understood resistance genes.30
In vitro activity
There are currently no established susceptibility breakpoints for tigecycline (see Note added in proof). Tigecycline MIC breakpoints proposed by Wyeth Pharmaceuticals are susceptible, ≤2 mg/L; intermediate, 4 mg/L; and resistant, ≥8 mg/L, for streptococci (including S. pneumoniae), H. influenzae, N. gonorrhoeae and Acinetobacter baumannii.18,21,26,29,34–47
Tigecycline's broad spectrum of activity is reported in Table 1 (aerobic Gram-positive bacteria),18,21,26,29,35–47Table 2 (aerobic Gram-negative bacteria),18,21,26,27,29,35–37,47–52Table 3 (anaerobes)19,29,37 and Table 4 (atypical organisms).53–55
| Organism . | MIC range (mg/L) . | MIC50 range (mg/L) . | MIC90 range (mg/L) . |
|---|---|---|---|
| Staphylococcus aureus | ≤0.02–2 | 0.06–0.5 | 0.125–1 |
| S. aureus (OXAS) | 0.06–1 | ≤0.13–0.5 | 0.25–0.5 |
| S. aureus (OXAR) | ≤0.06–2 | ≤0.13–0.5 | 0.25–1 |
| S. aureus (VANI) | 0.06–2 | 0.25 | 0.5 |
| Coagulase-negative staphylococcus | ≤0.03–2 | 0.06–1 | 0.25–1 |
| CNS (OXAS) | ≤0.03–1 | 0.25–0.5 | 0.25–1 |
| CNS (OXAR) | ≤0.03–2 | 0.5–1 | 0.25–1 |
| Enterococcus species | ≤0.02–2 | 0.03–0.25 | 0.06–0.5 |
| E. faecalis | ≤0.02–2 | 0.13–0.25 | 0.13–0.5 |
| E. faecalis (VANR) | ≤0.03–0.5 | 0.13 | 0.13–0.5 |
| E. faecium | ≤0.03–0.5 | 0.06–0.25 | 0.13–0.25 |
| E. faecium (VANR) | ≤0.03–0.5 | 0.06–0.13 | 0.13 |
| E. avium | 0.06–0.13 | 0.06 | 0.06 |
| E. casseliflavus | ≤0.03–0.5 | 0.13–0.25 | 0.13–0.25 |
| E. fallinarum | 0.06–2 | 0.13 | 0.13–0.25 |
| E. raffinosus | 0.06–0.5 | 0.06 | 0.13 |
| Streptococcus pneumoniae | ≤0.01–1 | ≤0.02–0.25 | ≤0.02–0.5 |
| S. pneumoniae (PENS) | ≤0.02–0.5 | 0.03–0.25 | 0.13–0.25 |
| S. pneumoniae (PENI) | ≤0.02–1 | 0.03–0.25 | 0.06–0.5 |
| S. pneumoniae (PENR) | ≤0.02–1 | 0.06–0.25 | 0.13–0.25 |
| S. pneumoniae (TETS) | 0.01–0.13 | 0.03 | 0.03 |
| S. pneumoniae (TETR) | 0.02–0.5 | 0.03 | 0.03 |
| Group A streptococci | ≤0.02–0.5 | 0.06–0.13 | 0.06–0.25 |
| Group B streptococci | 0.03–0.5 | 0.06–0.13 | 0.06–0.25 |
| Viridans streptococci | 0.01–2 | ≤0.02–0.06 | 0.03–0.5 |
| Viridans streptococci (PENS) | 0.03–0.25 | 0.06 | 0.25 |
| Viridans streptococci (PENR) | 0.02–0.13 | 0.03 | 0.06 |
| Viridans streptococci (TETS) | 0.02–0.06 | 0.03 | 0.06 |
| Viridans streptococci (TETR) | 0.1–0.5 | 0.06 | 0.13 |
| Organism . | MIC range (mg/L) . | MIC50 range (mg/L) . | MIC90 range (mg/L) . |
|---|---|---|---|
| Staphylococcus aureus | ≤0.02–2 | 0.06–0.5 | 0.125–1 |
| S. aureus (OXAS) | 0.06–1 | ≤0.13–0.5 | 0.25–0.5 |
| S. aureus (OXAR) | ≤0.06–2 | ≤0.13–0.5 | 0.25–1 |
| S. aureus (VANI) | 0.06–2 | 0.25 | 0.5 |
| Coagulase-negative staphylococcus | ≤0.03–2 | 0.06–1 | 0.25–1 |
| CNS (OXAS) | ≤0.03–1 | 0.25–0.5 | 0.25–1 |
| CNS (OXAR) | ≤0.03–2 | 0.5–1 | 0.25–1 |
| Enterococcus species | ≤0.02–2 | 0.03–0.25 | 0.06–0.5 |
| E. faecalis | ≤0.02–2 | 0.13–0.25 | 0.13–0.5 |
| E. faecalis (VANR) | ≤0.03–0.5 | 0.13 | 0.13–0.5 |
| E. faecium | ≤0.03–0.5 | 0.06–0.25 | 0.13–0.25 |
| E. faecium (VANR) | ≤0.03–0.5 | 0.06–0.13 | 0.13 |
| E. avium | 0.06–0.13 | 0.06 | 0.06 |
| E. casseliflavus | ≤0.03–0.5 | 0.13–0.25 | 0.13–0.25 |
| E. fallinarum | 0.06–2 | 0.13 | 0.13–0.25 |
| E. raffinosus | 0.06–0.5 | 0.06 | 0.13 |
| Streptococcus pneumoniae | ≤0.01–1 | ≤0.02–0.25 | ≤0.02–0.5 |
| S. pneumoniae (PENS) | ≤0.02–0.5 | 0.03–0.25 | 0.13–0.25 |
| S. pneumoniae (PENI) | ≤0.02–1 | 0.03–0.25 | 0.06–0.5 |
| S. pneumoniae (PENR) | ≤0.02–1 | 0.06–0.25 | 0.13–0.25 |
| S. pneumoniae (TETS) | 0.01–0.13 | 0.03 | 0.03 |
| S. pneumoniae (TETR) | 0.02–0.5 | 0.03 | 0.03 |
| Group A streptococci | ≤0.02–0.5 | 0.06–0.13 | 0.06–0.25 |
| Group B streptococci | 0.03–0.5 | 0.06–0.13 | 0.06–0.25 |
| Viridans streptococci | 0.01–2 | ≤0.02–0.06 | 0.03–0.5 |
| Viridans streptococci (PENS) | 0.03–0.25 | 0.06 | 0.25 |
| Viridans streptococci (PENR) | 0.02–0.13 | 0.03 | 0.06 |
| Viridans streptococci (TETS) | 0.02–0.06 | 0.03 | 0.06 |
| Viridans streptococci (TETR) | 0.1–0.5 | 0.06 | 0.13 |
CNS, coagulase-negative staphylococcus; I, intermediate; OXA, oxacillin; PEN, penicillin; R, resistant; S, susceptible; TET, tetracycline; VAN, vancomycin.
| Organism . | MIC range (mg/L) . | MIC50 range (mg/L) . | MIC90 range (mg/L) . |
|---|---|---|---|
| Staphylococcus aureus | ≤0.02–2 | 0.06–0.5 | 0.125–1 |
| S. aureus (OXAS) | 0.06–1 | ≤0.13–0.5 | 0.25–0.5 |
| S. aureus (OXAR) | ≤0.06–2 | ≤0.13–0.5 | 0.25–1 |
| S. aureus (VANI) | 0.06–2 | 0.25 | 0.5 |
| Coagulase-negative staphylococcus | ≤0.03–2 | 0.06–1 | 0.25–1 |
| CNS (OXAS) | ≤0.03–1 | 0.25–0.5 | 0.25–1 |
| CNS (OXAR) | ≤0.03–2 | 0.5–1 | 0.25–1 |
| Enterococcus species | ≤0.02–2 | 0.03–0.25 | 0.06–0.5 |
| E. faecalis | ≤0.02–2 | 0.13–0.25 | 0.13–0.5 |
| E. faecalis (VANR) | ≤0.03–0.5 | 0.13 | 0.13–0.5 |
| E. faecium | ≤0.03–0.5 | 0.06–0.25 | 0.13–0.25 |
| E. faecium (VANR) | ≤0.03–0.5 | 0.06–0.13 | 0.13 |
| E. avium | 0.06–0.13 | 0.06 | 0.06 |
| E. casseliflavus | ≤0.03–0.5 | 0.13–0.25 | 0.13–0.25 |
| E. fallinarum | 0.06–2 | 0.13 | 0.13–0.25 |
| E. raffinosus | 0.06–0.5 | 0.06 | 0.13 |
| Streptococcus pneumoniae | ≤0.01–1 | ≤0.02–0.25 | ≤0.02–0.5 |
| S. pneumoniae (PENS) | ≤0.02–0.5 | 0.03–0.25 | 0.13–0.25 |
| S. pneumoniae (PENI) | ≤0.02–1 | 0.03–0.25 | 0.06–0.5 |
| S. pneumoniae (PENR) | ≤0.02–1 | 0.06–0.25 | 0.13–0.25 |
| S. pneumoniae (TETS) | 0.01–0.13 | 0.03 | 0.03 |
| S. pneumoniae (TETR) | 0.02–0.5 | 0.03 | 0.03 |
| Group A streptococci | ≤0.02–0.5 | 0.06–0.13 | 0.06–0.25 |
| Group B streptococci | 0.03–0.5 | 0.06–0.13 | 0.06–0.25 |
| Viridans streptococci | 0.01–2 | ≤0.02–0.06 | 0.03–0.5 |
| Viridans streptococci (PENS) | 0.03–0.25 | 0.06 | 0.25 |
| Viridans streptococci (PENR) | 0.02–0.13 | 0.03 | 0.06 |
| Viridans streptococci (TETS) | 0.02–0.06 | 0.03 | 0.06 |
| Viridans streptococci (TETR) | 0.1–0.5 | 0.06 | 0.13 |
| Organism . | MIC range (mg/L) . | MIC50 range (mg/L) . | MIC90 range (mg/L) . |
|---|---|---|---|
| Staphylococcus aureus | ≤0.02–2 | 0.06–0.5 | 0.125–1 |
| S. aureus (OXAS) | 0.06–1 | ≤0.13–0.5 | 0.25–0.5 |
| S. aureus (OXAR) | ≤0.06–2 | ≤0.13–0.5 | 0.25–1 |
| S. aureus (VANI) | 0.06–2 | 0.25 | 0.5 |
| Coagulase-negative staphylococcus | ≤0.03–2 | 0.06–1 | 0.25–1 |
| CNS (OXAS) | ≤0.03–1 | 0.25–0.5 | 0.25–1 |
| CNS (OXAR) | ≤0.03–2 | 0.5–1 | 0.25–1 |
| Enterococcus species | ≤0.02–2 | 0.03–0.25 | 0.06–0.5 |
| E. faecalis | ≤0.02–2 | 0.13–0.25 | 0.13–0.5 |
| E. faecalis (VANR) | ≤0.03–0.5 | 0.13 | 0.13–0.5 |
| E. faecium | ≤0.03–0.5 | 0.06–0.25 | 0.13–0.25 |
| E. faecium (VANR) | ≤0.03–0.5 | 0.06–0.13 | 0.13 |
| E. avium | 0.06–0.13 | 0.06 | 0.06 |
| E. casseliflavus | ≤0.03–0.5 | 0.13–0.25 | 0.13–0.25 |
| E. fallinarum | 0.06–2 | 0.13 | 0.13–0.25 |
| E. raffinosus | 0.06–0.5 | 0.06 | 0.13 |
| Streptococcus pneumoniae | ≤0.01–1 | ≤0.02–0.25 | ≤0.02–0.5 |
| S. pneumoniae (PENS) | ≤0.02–0.5 | 0.03–0.25 | 0.13–0.25 |
| S. pneumoniae (PENI) | ≤0.02–1 | 0.03–0.25 | 0.06–0.5 |
| S. pneumoniae (PENR) | ≤0.02–1 | 0.06–0.25 | 0.13–0.25 |
| S. pneumoniae (TETS) | 0.01–0.13 | 0.03 | 0.03 |
| S. pneumoniae (TETR) | 0.02–0.5 | 0.03 | 0.03 |
| Group A streptococci | ≤0.02–0.5 | 0.06–0.13 | 0.06–0.25 |
| Group B streptococci | 0.03–0.5 | 0.06–0.13 | 0.06–0.25 |
| Viridans streptococci | 0.01–2 | ≤0.02–0.06 | 0.03–0.5 |
| Viridans streptococci (PENS) | 0.03–0.25 | 0.06 | 0.25 |
| Viridans streptococci (PENR) | 0.02–0.13 | 0.03 | 0.06 |
| Viridans streptococci (TETS) | 0.02–0.06 | 0.03 | 0.06 |
| Viridans streptococci (TETR) | 0.1–0.5 | 0.06 | 0.13 |
CNS, coagulase-negative staphylococcus; I, intermediate; OXA, oxacillin; PEN, penicillin; R, resistant; S, susceptible; TET, tetracycline; VAN, vancomycin.
| Organism . | MIC range (mg/L) . | MIC50 range (mg/L) . | MIC90 range (mg/L) . |
|---|---|---|---|
| Escherichia coli | 0.06–2 | 0.13–0.5 | 0.25–1 |
| E. coli (non-ESBL) | 0.06–2 | 0.13–0.5 | 0.25–1 |
| E. coli (ESBL) | 0.06–2 | 0.13–0.25 | 0.5–1 |
| E. coli (CIPS) | 0.5–2 | 1 | 1 |
| E. coli (CIPR) | 0.5–2 | 1 | 1 |
| Klebsiella pneumoniae | 0.06–8 | 0.25–1 | 1–2 |
| K. pneumoniae (non-ESBL) | 0.06–4 | 0.25–1 | 1–2 |
| K. pneumoniae (ESBL) | 0.06–8 | 0.25–1 | 1–2 |
| Klebsiella oxytoca | 0.5–2 | 0.5–1 | 1 |
| Morganella morganii | 1–8 | 2–4 | 4 |
| Proteus mirabilis | 1–8 | 4 | 8 |
| Proteus vulgaris | 0.13–0.16 | 4 | 4 |
| Providencia species | 4–8 | 4 | 8 |
| Shigella species | 0.13–0.5 | 0.25 | 0.5 |
| Salmonella species | 0.25–2 | 1 | 1 |
| Citrobacter species | 0.25–16 | 0.5–1 | 0.5–2 |
| C. freundii | 0.25–16 | 0.5–1 | 2 |
| Enterobacter species | 0.25–8 | 1 | 1–2 |
| E. cloacae | 0.25–4 | 1 | 2 |
| E. aerogenes | 0.25–8 | 1 | 1 |
| Serratia marcescens | 0.5–8 | 2–4 | 2–4 |
| Stenotrophomonas maltophilia | 0.25–8 | 0.5–2 | 2–4 |
| Pseudomonas aeruginosa | 0.5–32 | 8–>16 | 16–32 |
| Acinetobacter species | ≤0.03–16 | 0.25–2 | 0.5–8 |
| A. baumannii | 0.03–16 | 0.5–2 | 2–8 |
| Burkholderia cepacia | 0.5–64 | 2–4 | 4–32 |
| Haemophilus influenzae | ≤0.13–4 | 0.25–1 | 0.5–2 |
| Moraxella species | ≤0.03–0.25 | 0.06–0.13 | 0.13–0.25 |
| M. catarrhalis | ≤0.03–2 | 0.06–0.13 | 0.13–0.5 |
| Neisseria gonorrhoeae | ≤0.02–1 | 0.06–0.5 | 0.13–1 |
| N. gonorrhoeae (TETS) | ≤0.02–0.25 | 0.06 | 0.13 |
| N. gonorrhoeae (TETI) | 0.06–0.25 | 0.13 | 0.25 |
| N. gonorrhoeae (TETR) | 0.06–1 | 0.25 | 0.5 |
| Eikenella corrodens | ≤0.06–4 | 0.5 | 2 |
| Organism . | MIC range (mg/L) . | MIC50 range (mg/L) . | MIC90 range (mg/L) . |
|---|---|---|---|
| Escherichia coli | 0.06–2 | 0.13–0.5 | 0.25–1 |
| E. coli (non-ESBL) | 0.06–2 | 0.13–0.5 | 0.25–1 |
| E. coli (ESBL) | 0.06–2 | 0.13–0.25 | 0.5–1 |
| E. coli (CIPS) | 0.5–2 | 1 | 1 |
| E. coli (CIPR) | 0.5–2 | 1 | 1 |
| Klebsiella pneumoniae | 0.06–8 | 0.25–1 | 1–2 |
| K. pneumoniae (non-ESBL) | 0.06–4 | 0.25–1 | 1–2 |
| K. pneumoniae (ESBL) | 0.06–8 | 0.25–1 | 1–2 |
| Klebsiella oxytoca | 0.5–2 | 0.5–1 | 1 |
| Morganella morganii | 1–8 | 2–4 | 4 |
| Proteus mirabilis | 1–8 | 4 | 8 |
| Proteus vulgaris | 0.13–0.16 | 4 | 4 |
| Providencia species | 4–8 | 4 | 8 |
| Shigella species | 0.13–0.5 | 0.25 | 0.5 |
| Salmonella species | 0.25–2 | 1 | 1 |
| Citrobacter species | 0.25–16 | 0.5–1 | 0.5–2 |
| C. freundii | 0.25–16 | 0.5–1 | 2 |
| Enterobacter species | 0.25–8 | 1 | 1–2 |
| E. cloacae | 0.25–4 | 1 | 2 |
| E. aerogenes | 0.25–8 | 1 | 1 |
| Serratia marcescens | 0.5–8 | 2–4 | 2–4 |
| Stenotrophomonas maltophilia | 0.25–8 | 0.5–2 | 2–4 |
| Pseudomonas aeruginosa | 0.5–32 | 8–>16 | 16–32 |
| Acinetobacter species | ≤0.03–16 | 0.25–2 | 0.5–8 |
| A. baumannii | 0.03–16 | 0.5–2 | 2–8 |
| Burkholderia cepacia | 0.5–64 | 2–4 | 4–32 |
| Haemophilus influenzae | ≤0.13–4 | 0.25–1 | 0.5–2 |
| Moraxella species | ≤0.03–0.25 | 0.06–0.13 | 0.13–0.25 |
| M. catarrhalis | ≤0.03–2 | 0.06–0.13 | 0.13–0.5 |
| Neisseria gonorrhoeae | ≤0.02–1 | 0.06–0.5 | 0.13–1 |
| N. gonorrhoeae (TETS) | ≤0.02–0.25 | 0.06 | 0.13 |
| N. gonorrhoeae (TETI) | 0.06–0.25 | 0.13 | 0.25 |
| N. gonorrhoeae (TETR) | 0.06–1 | 0.25 | 0.5 |
| Eikenella corrodens | ≤0.06–4 | 0.5 | 2 |
CIP, ciprofloxacin; ESBL, extended-spectrum β-lactamase; I, intermediate; R, resistant; S, susceptible; TET, tetracycline.
| Organism . | MIC range (mg/L) . | MIC50 range (mg/L) . | MIC90 range (mg/L) . |
|---|---|---|---|
| Escherichia coli | 0.06–2 | 0.13–0.5 | 0.25–1 |
| E. coli (non-ESBL) | 0.06–2 | 0.13–0.5 | 0.25–1 |
| E. coli (ESBL) | 0.06–2 | 0.13–0.25 | 0.5–1 |
| E. coli (CIPS) | 0.5–2 | 1 | 1 |
| E. coli (CIPR) | 0.5–2 | 1 | 1 |
| Klebsiella pneumoniae | 0.06–8 | 0.25–1 | 1–2 |
| K. pneumoniae (non-ESBL) | 0.06–4 | 0.25–1 | 1–2 |
| K. pneumoniae (ESBL) | 0.06–8 | 0.25–1 | 1–2 |
| Klebsiella oxytoca | 0.5–2 | 0.5–1 | 1 |
| Morganella morganii | 1–8 | 2–4 | 4 |
| Proteus mirabilis | 1–8 | 4 | 8 |
| Proteus vulgaris | 0.13–0.16 | 4 | 4 |
| Providencia species | 4–8 | 4 | 8 |
| Shigella species | 0.13–0.5 | 0.25 | 0.5 |
| Salmonella species | 0.25–2 | 1 | 1 |
| Citrobacter species | 0.25–16 | 0.5–1 | 0.5–2 |
| C. freundii | 0.25–16 | 0.5–1 | 2 |
| Enterobacter species | 0.25–8 | 1 | 1–2 |
| E. cloacae | 0.25–4 | 1 | 2 |
| E. aerogenes | 0.25–8 | 1 | 1 |
| Serratia marcescens | 0.5–8 | 2–4 | 2–4 |
| Stenotrophomonas maltophilia | 0.25–8 | 0.5–2 | 2–4 |
| Pseudomonas aeruginosa | 0.5–32 | 8–>16 | 16–32 |
| Acinetobacter species | ≤0.03–16 | 0.25–2 | 0.5–8 |
| A. baumannii | 0.03–16 | 0.5–2 | 2–8 |
| Burkholderia cepacia | 0.5–64 | 2–4 | 4–32 |
| Haemophilus influenzae | ≤0.13–4 | 0.25–1 | 0.5–2 |
| Moraxella species | ≤0.03–0.25 | 0.06–0.13 | 0.13–0.25 |
| M. catarrhalis | ≤0.03–2 | 0.06–0.13 | 0.13–0.5 |
| Neisseria gonorrhoeae | ≤0.02–1 | 0.06–0.5 | 0.13–1 |
| N. gonorrhoeae (TETS) | ≤0.02–0.25 | 0.06 | 0.13 |
| N. gonorrhoeae (TETI) | 0.06–0.25 | 0.13 | 0.25 |
| N. gonorrhoeae (TETR) | 0.06–1 | 0.25 | 0.5 |
| Eikenella corrodens | ≤0.06–4 | 0.5 | 2 |
| Organism . | MIC range (mg/L) . | MIC50 range (mg/L) . | MIC90 range (mg/L) . |
|---|---|---|---|
| Escherichia coli | 0.06–2 | 0.13–0.5 | 0.25–1 |
| E. coli (non-ESBL) | 0.06–2 | 0.13–0.5 | 0.25–1 |
| E. coli (ESBL) | 0.06–2 | 0.13–0.25 | 0.5–1 |
| E. coli (CIPS) | 0.5–2 | 1 | 1 |
| E. coli (CIPR) | 0.5–2 | 1 | 1 |
| Klebsiella pneumoniae | 0.06–8 | 0.25–1 | 1–2 |
| K. pneumoniae (non-ESBL) | 0.06–4 | 0.25–1 | 1–2 |
| K. pneumoniae (ESBL) | 0.06–8 | 0.25–1 | 1–2 |
| Klebsiella oxytoca | 0.5–2 | 0.5–1 | 1 |
| Morganella morganii | 1–8 | 2–4 | 4 |
| Proteus mirabilis | 1–8 | 4 | 8 |
| Proteus vulgaris | 0.13–0.16 | 4 | 4 |
| Providencia species | 4–8 | 4 | 8 |
| Shigella species | 0.13–0.5 | 0.25 | 0.5 |
| Salmonella species | 0.25–2 | 1 | 1 |
| Citrobacter species | 0.25–16 | 0.5–1 | 0.5–2 |
| C. freundii | 0.25–16 | 0.5–1 | 2 |
| Enterobacter species | 0.25–8 | 1 | 1–2 |
| E. cloacae | 0.25–4 | 1 | 2 |
| E. aerogenes | 0.25–8 | 1 | 1 |
| Serratia marcescens | 0.5–8 | 2–4 | 2–4 |
| Stenotrophomonas maltophilia | 0.25–8 | 0.5–2 | 2–4 |
| Pseudomonas aeruginosa | 0.5–32 | 8–>16 | 16–32 |
| Acinetobacter species | ≤0.03–16 | 0.25–2 | 0.5–8 |
| A. baumannii | 0.03–16 | 0.5–2 | 2–8 |
| Burkholderia cepacia | 0.5–64 | 2–4 | 4–32 |
| Haemophilus influenzae | ≤0.13–4 | 0.25–1 | 0.5–2 |
| Moraxella species | ≤0.03–0.25 | 0.06–0.13 | 0.13–0.25 |
| M. catarrhalis | ≤0.03–2 | 0.06–0.13 | 0.13–0.5 |
| Neisseria gonorrhoeae | ≤0.02–1 | 0.06–0.5 | 0.13–1 |
| N. gonorrhoeae (TETS) | ≤0.02–0.25 | 0.06 | 0.13 |
| N. gonorrhoeae (TETI) | 0.06–0.25 | 0.13 | 0.25 |
| N. gonorrhoeae (TETR) | 0.06–1 | 0.25 | 0.5 |
| Eikenella corrodens | ≤0.06–4 | 0.5 | 2 |
CIP, ciprofloxacin; ESBL, extended-spectrum β-lactamase; I, intermediate; R, resistant; S, susceptible; TET, tetracycline.
| Organism . | MIC range (mg/L) . | MIC50 range (mg/L) . | MIC90 range (mg/L) . |
|---|---|---|---|
| Bacteroides fragilis | 0.5–8 | 2 | 2 |
| Bacteroides fragilis group | 0.02–2 | 0.13–0.5 | 0.13–2 |
| Clostridium perfringens | 0.03–4 | 0.03–0.5 | 0.25–1 |
| Clostridium difficile | ≤0.02–0.25 | 0.03–0.13 | 0.03–0.13 |
| Proprionibacterium acnes | 0.03–0.13 | 0.03 | 0.06 |
| Peptostreptococcus species | ≤0.02–0.5 | 0.03–0.06 | 0.03–0.25 |
| Fusobacteium species | ≤0.02–0.25 | 0.02–0.06 | 0.06 |
| Prevotella species | 0.02–1 | 0.03–0.5 | 0.06–1 |
| Porphyromonas species | ≤0.02–0.13 | 0.03–0.06 | 0.06 |
| Organism . | MIC range (mg/L) . | MIC50 range (mg/L) . | MIC90 range (mg/L) . |
|---|---|---|---|
| Bacteroides fragilis | 0.5–8 | 2 | 2 |
| Bacteroides fragilis group | 0.02–2 | 0.13–0.5 | 0.13–2 |
| Clostridium perfringens | 0.03–4 | 0.03–0.5 | 0.25–1 |
| Clostridium difficile | ≤0.02–0.25 | 0.03–0.13 | 0.03–0.13 |
| Proprionibacterium acnes | 0.03–0.13 | 0.03 | 0.06 |
| Peptostreptococcus species | ≤0.02–0.5 | 0.03–0.06 | 0.03–0.25 |
| Fusobacteium species | ≤0.02–0.25 | 0.02–0.06 | 0.06 |
| Prevotella species | 0.02–1 | 0.03–0.5 | 0.06–1 |
| Porphyromonas species | ≤0.02–0.13 | 0.03–0.06 | 0.06 |
| Organism . | MIC range (mg/L) . | MIC50 range (mg/L) . | MIC90 range (mg/L) . |
|---|---|---|---|
| Bacteroides fragilis | 0.5–8 | 2 | 2 |
| Bacteroides fragilis group | 0.02–2 | 0.13–0.5 | 0.13–2 |
| Clostridium perfringens | 0.03–4 | 0.03–0.5 | 0.25–1 |
| Clostridium difficile | ≤0.02–0.25 | 0.03–0.13 | 0.03–0.13 |
| Proprionibacterium acnes | 0.03–0.13 | 0.03 | 0.06 |
| Peptostreptococcus species | ≤0.02–0.5 | 0.03–0.06 | 0.03–0.25 |
| Fusobacteium species | ≤0.02–0.25 | 0.02–0.06 | 0.06 |
| Prevotella species | 0.02–1 | 0.03–0.5 | 0.06–1 |
| Porphyromonas species | ≤0.02–0.13 | 0.03–0.06 | 0.06 |
| Organism . | MIC range (mg/L) . | MIC50 range (mg/L) . | MIC90 range (mg/L) . |
|---|---|---|---|
| Bacteroides fragilis | 0.5–8 | 2 | 2 |
| Bacteroides fragilis group | 0.02–2 | 0.13–0.5 | 0.13–2 |
| Clostridium perfringens | 0.03–4 | 0.03–0.5 | 0.25–1 |
| Clostridium difficile | ≤0.02–0.25 | 0.03–0.13 | 0.03–0.13 |
| Proprionibacterium acnes | 0.03–0.13 | 0.03 | 0.06 |
| Peptostreptococcus species | ≤0.02–0.5 | 0.03–0.06 | 0.03–0.25 |
| Fusobacteium species | ≤0.02–0.25 | 0.02–0.06 | 0.06 |
| Prevotella species | 0.02–1 | 0.03–0.5 | 0.06–1 |
| Porphyromonas species | ≤0.02–0.13 | 0.03–0.06 | 0.06 |
| Organism . | MIC range (mg/L) . | MIC50 range (mg/L) . | MIC90 range (mg/L) . | |||
|---|---|---|---|---|---|---|
| Mycobacterium abscessus | ||||||
| TETS | ≤0.06–1 | ≤0.13 | 0.25 | |||
| TETR | ≤0.06–1 | ≤0.13 | 0.25 | |||
| Mycobacterium chelonae | ||||||
| TETS | ≤0.25 | ≤0.06 | ≤0.13 | |||
| TETR | ≤0.25 | ≤0.06 | ≤0.13 | |||
| Mycobacterium fortuitum group | ||||||
| TETS | ≤0.25 | ≤0.06 | ≤0.13 | |||
| TETR | ≤0.25 | ≤0.06 | ≤0.13 | |||
| Mycobacterium avium complex | ≥32 | >32 | >32 | |||
| Mycobacterium lentiflavum | ≥32 | >32 | >32 | |||
| Mycobacterium marinum | 0.19–24 | 2–16 | 3–16 | |||
| Mycobacterium kansasii | 8–32 | 16 | 32 | |||
| Chlamydophilia pneumoniae | 0.13–0.25 | 0.13 | 0.13 | |||
| Mycoplasma hominis | ||||||
| TETS | 0.13–0.5 | 0.25 | 0.5 | |||
| TETR | 0.13–0.5 | 0.25 | 0.5 | |||
| Mycoplasma pneumoniae | 0.06–0.25 | 0.13 | 0.25 | |||
| Ureaplasma urealyticum | 1–16 | 4 | 8 | |||
| Organism . | MIC range (mg/L) . | MIC50 range (mg/L) . | MIC90 range (mg/L) . | |||
|---|---|---|---|---|---|---|
| Mycobacterium abscessus | ||||||
| TETS | ≤0.06–1 | ≤0.13 | 0.25 | |||
| TETR | ≤0.06–1 | ≤0.13 | 0.25 | |||
| Mycobacterium chelonae | ||||||
| TETS | ≤0.25 | ≤0.06 | ≤0.13 | |||
| TETR | ≤0.25 | ≤0.06 | ≤0.13 | |||
| Mycobacterium fortuitum group | ||||||
| TETS | ≤0.25 | ≤0.06 | ≤0.13 | |||
| TETR | ≤0.25 | ≤0.06 | ≤0.13 | |||
| Mycobacterium avium complex | ≥32 | >32 | >32 | |||
| Mycobacterium lentiflavum | ≥32 | >32 | >32 | |||
| Mycobacterium marinum | 0.19–24 | 2–16 | 3–16 | |||
| Mycobacterium kansasii | 8–32 | 16 | 32 | |||
| Chlamydophilia pneumoniae | 0.13–0.25 | 0.13 | 0.13 | |||
| Mycoplasma hominis | ||||||
| TETS | 0.13–0.5 | 0.25 | 0.5 | |||
| TETR | 0.13–0.5 | 0.25 | 0.5 | |||
| Mycoplasma pneumoniae | 0.06–0.25 | 0.13 | 0.25 | |||
| Ureaplasma urealyticum | 1–16 | 4 | 8 | |||
R, resistant; S, susceptible; TET, tetracycline.
| Organism . | MIC range (mg/L) . | MIC50 range (mg/L) . | MIC90 range (mg/L) . | |||
|---|---|---|---|---|---|---|
| Mycobacterium abscessus | ||||||
| TETS | ≤0.06–1 | ≤0.13 | 0.25 | |||
| TETR | ≤0.06–1 | ≤0.13 | 0.25 | |||
| Mycobacterium chelonae | ||||||
| TETS | ≤0.25 | ≤0.06 | ≤0.13 | |||
| TETR | ≤0.25 | ≤0.06 | ≤0.13 | |||
| Mycobacterium fortuitum group | ||||||
| TETS | ≤0.25 | ≤0.06 | ≤0.13 | |||
| TETR | ≤0.25 | ≤0.06 | ≤0.13 | |||
| Mycobacterium avium complex | ≥32 | >32 | >32 | |||
| Mycobacterium lentiflavum | ≥32 | >32 | >32 | |||
| Mycobacterium marinum | 0.19–24 | 2–16 | 3–16 | |||
| Mycobacterium kansasii | 8–32 | 16 | 32 | |||
| Chlamydophilia pneumoniae | 0.13–0.25 | 0.13 | 0.13 | |||
| Mycoplasma hominis | ||||||
| TETS | 0.13–0.5 | 0.25 | 0.5 | |||
| TETR | 0.13–0.5 | 0.25 | 0.5 | |||
| Mycoplasma pneumoniae | 0.06–0.25 | 0.13 | 0.25 | |||
| Ureaplasma urealyticum | 1–16 | 4 | 8 | |||
| Organism . | MIC range (mg/L) . | MIC50 range (mg/L) . | MIC90 range (mg/L) . | |||
|---|---|---|---|---|---|---|
| Mycobacterium abscessus | ||||||
| TETS | ≤0.06–1 | ≤0.13 | 0.25 | |||
| TETR | ≤0.06–1 | ≤0.13 | 0.25 | |||
| Mycobacterium chelonae | ||||||
| TETS | ≤0.25 | ≤0.06 | ≤0.13 | |||
| TETR | ≤0.25 | ≤0.06 | ≤0.13 | |||
| Mycobacterium fortuitum group | ||||||
| TETS | ≤0.25 | ≤0.06 | ≤0.13 | |||
| TETR | ≤0.25 | ≤0.06 | ≤0.13 | |||
| Mycobacterium avium complex | ≥32 | >32 | >32 | |||
| Mycobacterium lentiflavum | ≥32 | >32 | >32 | |||
| Mycobacterium marinum | 0.19–24 | 2–16 | 3–16 | |||
| Mycobacterium kansasii | 8–32 | 16 | 32 | |||
| Chlamydophilia pneumoniae | 0.13–0.25 | 0.13 | 0.13 | |||
| Mycoplasma hominis | ||||||
| TETS | 0.13–0.5 | 0.25 | 0.5 | |||
| TETR | 0.13–0.5 | 0.25 | 0.5 | |||
| Mycoplasma pneumoniae | 0.06–0.25 | 0.13 | 0.25 | |||
| Ureaplasma urealyticum | 1–16 | 4 | 8 | |||
R, resistant; S, susceptible; TET, tetracycline.
In vitro activity against Gram-negative bacteria
In several studies that assessed the in vitro activity of tigecycline against enteric Gram-negative bacteria, tigecycline was found to be highly active against Enterobacteriaceae, regardless of the presence or absence of ESBLs.20,35,36 Specifically, tigecycline was very active with MIC90 values of ≤2 mg/L,20,35,36 and >99% of all of the tested strains were inhibited with ≤4 mg/L of tigecycline.20 Uniformly reduced activity was observed for tigecycline against P. mirabilis and indole-positive Proteus spp.20,22,35,36 In P. mirabilis, reduction of in vitro activity of tigecycline is associated with the AcrAB multidrug efflux pump.56 Additionally, P. aeruginosa is not reliably inhibited by tigecycline.20,22,29 However, experiments with mutant bacteria containing the MexAB-OprM and MexCD-OprJ efflux pumps suggest that glycylcyclines, while still subject to efflux from P. aeruginosa, are generally inferior substrates with these efflux pumps than are earlier tetracyclines. This is supported by the observation that the MICs of doxycycline and minocycline increased to a greater extent than did those of tigecycline when tested against P. aeruginosa mutant strains overexpressing MexAB-OprM and MexCD-OprJ.57
Tigecycline is very active against other non-fermentative Gram-negative bacilli, which includes Acinetobacter spp. (generally >90% susceptibility, with MICs <8 mg/L) and Stenotrophomonas maltophilia (>90% susceptibility).20,34,35,36,48,49 One study found that 92% of A. baumannii strains were susceptible to tigecycline, but with only 20% susceptible to imipenem.34 Against S. maltophilia, tigecycline MICs were three to four dilutions lower than those of tetracycline and two dilutions higher than those of minocycline.49 Tigecycline was more active against S. maltophilia than amikacin, ceftazidime and ticarcillin/clavulanate.49 Overall, the above findings support those of two other studies which tested over 5000 Gram-negative clinical isolates obtained from patients with bacteraemia.18,58
Tigecycline also has potent in vitro activity against two commonly isolated respiratory tract pathogens—H. influenzae and M. catarrhalis. Against more than 10 000 clinical isolates, tigecycline MIC90 values ranged from ≤0.06 to ≥8 mg/L.59–61 Median MIC90 values for β-lactamase-positive and -negative H. influenzae and M. catarrhalis were 1 mg/L and 0.25 mg/L, respectively. Tigecycline was active against all of the isolates, without regard to the presence or absence of β-lactamases.
In vitro activity against Gram-positive bacteria
The activity of tigecycline against more than 20 000 clinical strains of oxacillin-susceptible and oxacillin-resistant S. aureus, glycopeptide-resistant S. aureus, coagulase-negative staphylococci, S. pneumoniae, β-haemolytic streptococci, and vancomycin-susceptible and -resistant enterococci has been investigated in numerous in vitro studies.18,36,38,39,40–45,59,62–68 Tigecycline exhibited potent activity, especially against the collected Gram-positive cocci when compared with the activities seen with vancomycin, linezolid or quinupristin/dalfopristin.
The potency of tigecycline was evident against both oxacillin-susceptible and oxacillin-resistant S. aureus with the majority of investigations reporting an MIC90 of 0.5 mg/L.18,36,39,41,59,62,66 In a study of glycopeptide-intermediate S. aureus strains (n = 47), the MIC90 for tigecycline was 0.5 mg/L (range, 0.0625–2 mg/L) and the MBC range was 0.25–2 mg/L.67 The investigators stated that these data suggest tigecycline may be a therapeutic option for multidrug-resistant S. aureus, including glycopeptide-intermediate strains, and that clinical investigations are warranted.67 Tigecycline also bacteriostatically inhibited the one strain of vancomycin-resistant S. aureus that was isolated at Hershey Medical Center, Hershey, PA, USA.45,66
Tigecycline is highly active against coagulase-negative staphylococci, including methicillin-resistant isolates, with MIC90 values ranging from 0.5 to 1.0 mg/L.45,59,66 An in vitro adherent- cell biofilm model was used to evaluate tigecycline activity against Staphylococcus epidermidis. In a biofilm of adherent cells, tigecycline MBCs were recorded as 1–8 mg/L. The MBCs were 0.12 to >32 mg/L for the freely growing cells of S. epidermidis. In this model, tigecycline's killing activity was more than four times better than that of vancomycin and daptomycin.68
Tigecycline demonstrates excellent activity against most streptococci, including S. pneumoniae with reduced susceptibility or resistance to macrolides or β-lactam antibiotics.18,36,40–42,44,59,62,63 In three large surveillance studies, tigecycline had an MIC90 ≤ 0.25 mg/L against >8500 isolates including both penicillin-susceptible and penicillin-resistant S. pneumoniae59,62,63 and 405 isolates of β-haemolytic streptococci.59 In another study, tetracycline, minocycline and doxycycline had less activity than tigecycline against 201 pneumococcal isolates and, against 11 of 12 tested strains, tigecycline was bactericidal.42 Tigecycline is also potent against β-haemolytic streptococci and viridans group streptococci (MIC90 ≤ 0.12–0.5 mg/L).59,62 In one study of 107 erythromycin-resistant isolates of Streptococcus pyogenes and 98 Streptococcus agalactiae, the most prevalent resistance genes identified were mef(A) (a gene encoding a commonly present macrolide efflux pump) in the S. pyogenes isolates (91.6%) and erm(B) in the S.agalactiae strains (65.3%).43 Fourteen (13.1%) S.pyogenes isolates and 88 (89.8%) S.agalactiae isolates exhibited resistance to tetracycline, of which most (76.5%) was due to tet(M), a gene that confers resistance to both tetracycline and minocycline.43 Tigecycline activity against all the isolates, including those resistant to tetracycline, was very good with an MIC90 of 0.06 mg/L.43
Several studies have reported that tigecycline was the most potent antibiotic tested against vancomycin-susceptible and vancomycin-resistant enterococci with MIC90s typically ranging from 0.25 to 0.5 mg/L.36,38,44,59,62,64 In addition, tigecycline was found to be equally active against E. faecium and E. faecalis.36 Activity (all MIC values ≤1 mg/L) was demonstrated against 37 clinical strains of vancomycin-resistant enterococci (including bacteria having the vanA, vanB, vanC-1 and vanC-2/3 genes).44 Time–kill experiments using vancomycin-resistant enterococci did not reveal any synergy or antagonism when tigecycline and quinupristin/dalfopristin were combined.44 A second study investigated tigecycline's ability to inhibit vancomycin-resistant enterococci, including E.faecalis (n = 25), E. faecium (n = 41), Enterococcus casseliflavus (n = 21) and Enterococcus gallinarum (n = 10).38 Twenty-eight of the strains were of the VanA phenotype, 38 expressed the VanB phenotype, and 31 were the VanC phenotype. Regardless of the presence or absence of tetracycline resistance, all enterococcal strains were inhibited by tigecycline at concentrations between ≤0.03 and 1 mg/L with an MIC90 of 0.12 mg/L. All 55 enterococcal isolates that were tetracycline-resistant (MIC > 8 mg/L) were inhibited by 0.5 mg/L of tigecycline. Against glycopeptide-resistant enterococci, tigecycline exhibited potent in vitro activity. However, tigecycline did not exhibit bactericidal activity (MBC > 32 mg/L) against three enterococcal isolates that were evaluated. The authors reported that tigecycline is mainly a bacteriostatic antibiotic with good activity against vancomycin-resistant enterococci. Tigecycline exhibited potent activity against the collected clinical bacteria regardless of resistance or susceptibility to tetracycline.38
The activity of tigecycline was studied in combination with other antimicrobial agents and by itself against highly resistant strains of E. faecium and S. aureus.65 The strains were two isolates of vancomycin-resistant E. faecium, three glycopeptide-intermediate S. aureus and one MRSA strain. Time–kill studies, MICs and MBCs were determined for tigecycline, vancomycin, gentamicin, rifampicin and doxycycline. Additional time–kill studies were done with tigecycline in combination with vancomycin, gentamicin, rifampicin and doxycycline. Tigecycline inhibited all of the tested isolates, and no enhanced killing of vancomycin-resistant E. faecium was elicited when tigecycline was tested with the other antimicrobial agents. However, the combination of gentamicin with tigecycline appeared to have improved effects when tested against the three strains of S. aureus.65
In vitro activity against anaerobic bacteria
The anti-anaerobe activity of tigecycline has been assessed in several studies.19,36,69 When using a breakpoint of 8 mg/L, tigecycline was reported to have more activity against the B. fragilis group than clindamycin, minocycline, trovafloxacin and cefoxitin, although imipenem and piperacillin–tazobactam were the most active agents tested.69 Specifically, most Bacteroides fragilis group isolates (B. thetaiotaomicron, B. vulgatus, B. ovatus, B. uniformis, B. distasonis and B. fragilis) were highly susceptible to tigecycline (MIC90 = 0.05 mg/L).19 In comparison, tigecycline exhibited some diminished activity against B. fragilis as evidenced by an MIC range of 0.064–16 mg/L (MIC90 = 0.5 mg/L).19
Tigecycline has excellent activity against most peptostreptococci (P. anaerobius, P. asaccharolyticus, P. micros, P. magnus, P. prevotii and P. indolicus) with MIC values ≤4 mg/L.19 Tigecycline also was uniformly active against all tested strains of Clostridium perfringens, Clostridium difficile, Prevotella spp., Propionibacterium acnes and Fusobacterium nucleatum.19 In one study, tigecycline inhibited 92.7% of C. difficile isolates at a concentration of 0.125 mg/L, which made tigecycline the most active of the antibiotics tested, which also included metronidazole.36
Tigecycline's excellent anaerobic activity has also been confirmed in a study of bacteria isolated from infected human and animal bite wounds inflicted upon humans (e.g. Pasteurella, Fusobacterium, Prevotella, Bacteroides).37 The MIC90s for tigecycline were ≤0.25 mg/L for all anaerobic species, including erythromycin-resistant and moxifloxacin-resistant fusobacteria (MIC90s for both = 0.06 mg/L). Tigecycline was not as active against Eikenella corrodens (MIC90 ≤ 4 mg/L). Another study reported that E. corrodens were inhibited by tigecycline at concentrations between ≤0.06 and 4 mg/L (MIC90 = 2 mg/L).50 For the other tested antibiotics, the MIC90s against E. corrodens were 1 mg/L for ampicillin, ≤0.5/0.25 mg/L for amoxicillin/clavulanate, 0.5 mg/L for cefotaxime, ≤0.12 mg/L for imipenem, ≤2 mg/L for chloramphenicol and 0.5 mg/L for ciprofloxacin.50
In vitro activity against atypical organisms
Several studies evaluated the in vitro activity of tigecycline against atypical bacterial pathogens. Tigecycline had more potent activity against Mycoplasma pneumoniae compared with minocycline or tetracycline (tigecycline MIC90 = 0.25 mg/L). Mycoplasma hominis strains were also inhibited by tigecycline, with an MIC90 of 0.5 mg/L. Tigecycline was less active against 25 Ureaplasma urealyticum isolates (MIC90 = 8 mg/L) when compared with minocycline (MIC90 = 0.25 mg/L) or tetracycline (MIC90 = 1 mg/L).70
In a study investigating the activity of tigecycline against Chlamydophilia species, 10 isolates of Chlamydophilia pneumoniae and five strains of Chlamydophilia trachomatis were evaluated. Based on the in vitro activity of tigecycline, the authors stated that tigecycline may have a future role in the therapy of chlamydial infections.53
Tigecycline has in vitro activity against rapidly, but not slowly growing mycobacteria. Against 76 rapidly growing mycobacterial strains (M. fortuitum group, M. abscessus, M. chelonae, M. immunogenum and the M. smegmatis group), tigecycline was very active, with MIC90 values of 0.25 mg/L for M. abscessus and <0.12 mg/L for M. chelonae and the M. fortuitum group. MICs for tigecycline were much better (4 to 11 times less) than those for the tetracyclines. None of the slowly growing non-tuberculous mycobacteria (M. avium complex, M. lentiflavum, M. kansasii, M. marinum, M. xenopi and M. simiae) were susceptible to tigecycline. Tigecycline was less active than minocycline when assessed against M. marinum and M. kansasii. The authors commented that tigecycline potentially may prove to be a useful agent for the rapidly growing mycobacteria, especially for M. chelonae and M. abscessus.54
In another survey of 37 strains of M. marinum, the most active antibiotic was trimethoprim/sulfamethoxazole, with activity against 91.9% of the tested strains. Minocycline was more active than tigecycline. Tigecycline had an MIC90 of 3 mg/L, and, in order to achieve inhibition of growth, one isolate required 24 mg/L of tigecycline.55
Post-antibiotic effect of tigecycline
For the most part, tetracyclines have been considered to be bacteriostatic agents, and area under the concentration–time curve has been considered a desirable therapeutic parameter to measure to help predict bacteriological cure. The post-antibiotic effect of tigecycline has been reported to be greater than that of minocycline in various strains of S. aureus and E. coli. When isolates were exposed to tigecycline or minocycline for 2 h at eightfold the MIC, the post-antibiotic effect of tigecycline exceeded that of minocycline by 0.6–2 h, depending upon the strain studied. The post-antibiotic effects of tigecycline were 4.1 h against a tetracycline-susceptible S. aureus strain, greater than 3.5 h against a tet(K)-expressing S. aureus strain and greater than 3 h against a tet(M)-expressing S. aureus strain. For E. coli strains that were tetracycline-susceptible, tet(B) expressing or tet(M) expressing, the post-antibiotic effects of tigecycline were 2.9, 2.6 and 1.8 h, respectively. Time–kill kinetic assays also looked for potential bactericidal activity when using four times the MIC of tigecycline. In these assays, tigecycline was bactericidal (defined as a greater than 3 log10 reduction in cfu) against the tested isolates of S. pneumoniae, H. influenzae and N. gonorrhoeae isolates, as well as some strains of E. coli. The long post-antibiotic effect of tigecycline supports the likelihood of less frequent dosing of this antimicrobial agent.30,71
The activity of tigecycline has also been evaluated in an in vitro pharmacodynamic model in which tigecycline's bacteriostatic activity remained unchanged with increasing concentrations of antibiotic to more than 1 mg/L. A post-antibiotic effect of 1–4.5 h at concentrations of 1 to 20 times the MIC was measured.72
Animal models assessing the in vivo activity of tigecycline
The therapeutic in vivo activity of tigecycline was compared with that of vancomycin in a rat endocarditis model caused by vancomycin-susceptible and vancomycin-resistant E. faecalis, and MRSA. Endocarditis was induced by a catheter placed across the aortic valve, followed 48 h later by an intravenous injection of bacteria. An additional 24–36 h later, treatment with tigecycline or vancomycin was started. Bacterial vegetation titres were reduced by greater than 2 log10 cfu with tigecycline administration when compared with untreated controls for both vancomycin-susceptible and vancomycin-resistant (VanA and VanB) E. faecalis strains. With the MRSA, a reduction of more than 4 log10 cfu was noted after the administration of tigecycline, which was also more effective, at a lower dose, than vancomycin. According to the authors, this in vivo animal study demonstrated the therapeutic potential of tigecycline.73
The activity of tigecycline was evaluated in an experimental rabbit endocarditis model. Endocarditis was induced with various strains of bacteria, including the susceptible E. faecalis JH2-2 strain, its VanA type transconjugant BM4316 or the tetracycline-resistant, VanA type E. faecium HB217. In vitro, tigecycline was highly active with MICs of 0.06 mg/L. In the rabbits, tigecycline homogeneously diffused into the vegetations. Additionally, the clearance of tigecycline from aortic vegetations was slower than from the serum. The elimination half-life from the serum ranged from 3.3 to 3.6 h. Tigecycline performed well in this rabbit endocarditis model. The positive attributes of tigecycline included homogeneous penetration into the bacterial vegetations, and a prolonged half-life and post-antibiotic effect.72
Tigecycline was administered intravenously in an intraperitoneal mouse model that used acute lethal injections with strains of E. coli, S. aureus and S. pneumoniae. Tigecycline was protective against S. aureus, including MRSA strains, and protection was also evident against strains containing tet(K) or tet(M) resistance determinants. In addition, tigecycline was active against those strains of E. coli that were susceptible to tetracycline, as well as those strains containing either tet(M) or the efflux determinant tet(A), tet(B) or tet(C).29
Tigecycline and daptomycin were evaluated in another study also using a murine intraperitoneal infection model. Both agents demonstrated therapeutic activity against glycopeptide-intermediate S. aureus, MRSA, and methicillin-susceptible S. aureus strains. Tigecycline showed more activity than daptomycin when the glycopeptide-intermediate S. aureus strain was used.64 In another investigation, three E. faecalis and four E. faecium strains were used in a mouse peritonitis model. Tigecycline was noted to be very active, and even with only one subcutaneous dose, tigecycline showed protection against all of the tested strains, including those that contained the VanA, VanB or tet(M) resistance determinants.74
Tigecycline was almost as effective as erythromycin against intracellular Legionella pneumophila in a Legionnaires' disease guinea pig animal model. The authors predicted that tigecycline should be effective for therapy of mild Legionnaires' disease; however, a longer course of administration (14–21 days) would be required for a cure, assuming that guinea pig pharmacokinetics are similar to human pharmacokinetics. However, tigecycline may not be the best drug for severe Legionnaires' disease, especially for immunocompromised patients and individuals who require hospitalization.75
The activities of tigecycline and vancomycin with and without rifampicin were evaluated against MRSA in a rabbit osteomyelitis model. One group of rabbits received 28 days of antibiotics and an untreated group served as controls. Antibiotic therapy was initiated 2 weeks after the intramedullary injection of MRSA. A 100% bacterial clearance rate occurred in the 14 animals that had received tigecycline and oral rifampicin. The bacterial clearance rate was 90% in the 10 rabbits that had received tigecycline alone, 81.8% in the rabbits receiving vancomycin (n = 11), and 26% in the untreated controls (n = 15). The authors concluded that tigecycline may be an effective alternative to vancomycin in the therapy of MRSA osteomyelitis.76
In a murine model of P. aeruginosa pneumonia, the activities of tigecycline, gentamicin, and piperacillin, alone and in combination were evaluated. When tigecycline was combined with gentamicin, a greater reduction in P. aeruginosa colony counts occurred than with the use of either agent alone. The combination of piperacillin and tigecycline or gentamicin did not increase the therapeutic antimicrobial activity. None of the study combinations created antagonism. The authors stated that further studies were warranted as the tigecycline–gentamicin combination had potential value in improving therapy.77
Neutropenic mice have been used in an experimental murine thigh infection model in which tigecycline and another experimental glycylcycline were further evaluated. Infection was induced with different bacteria including S. pneumoniae, S. aureus, E. coli, or K. pneumoniae. Most therapy was administered twice daily. A maximum-effect dose–response model was created in order to determine a dose that produced a net bacteriostatic effect over a 24 h period. Tigecycline elimination half-lives varied from 1.05 to 2.34 h, and serum protein binding was 59% in this animal model. The time above the MIC was a better predictor of in vivo efficacy than Cmax or area under the curve (AUC) in most of the experiments. The authors suggested that the concentration of unbound serum tigecycline should be maintained above the MIC for at least 50% of the time to reach maximum efficacy of 80%.46
Another murine model has been developed to assess antibiotic efficacy with thigh and lung infections. In this model, tigecycline activity was tested against methicillin-resistant and -susceptible S. aureus, penicillin-resistant and penicillin-susceptible S. pneumoniae, E. faecium, vancomycin-resistant E. faecalis, E. coli and K. pneumoniae. Tigecycline exhibited excellent activity against the broad spectrum of organisms. In the penicillin-resistant S. pneumoniae pneumonia model, tigecycline was three times as active as vancomycin.77
Additionally, tigecycline's activity was tested in a rabbit meningitis model due to a penicillin-resistant S. pneumoniae strain previously isolated from the CSF of a patient diagnosed with meningitis. Tigecycline had a long half-life in this model, and tigecycline exhibited bactericidal activity as manifested by a 2 to 3 log10 reduction in cfu per mL of CSF. When vancomycin and tigecycline were administered together, greater clearance of the bacteria occurred. These results support the further investigation of tigecycline as a possible therapeutic option for penicillin-resistant S. pneumoniae meningitis.78
Pharmacokinetics and pharmacodynamics of tigecycline
For human therapy, tigecycline is administered twice daily over a 30 to 60 min infusion. Tigecycline is only available as an injectable antibiotic as its oral bioavailability is very limited. Most tigecycline pharmacokinetic studies have been performed in normal healthy volunteers as single dose administrations. With a 100 mg intravenous dose administered over 1 h, the Cmax was 0.85–1 mg/L, the AUC0–∞ was 4.2–5.8 mg · h/L, and the half-life was measured as 16–24 h.77 In addition, a linear dose–response was noted for the Cmax and AUC0–∞. In some of the reported studies, no significant differences were seen in tigecycline pharmacokinetics based on food ingestion or gender.25,79,80
A randomized, double-blind, placebo-controlled, Phase I, ascending single dose study in healthy male subjects evaluated the safety, tolerability and pharmacokinetics of tigecycline. The study also measured any food effects and dose proportionality. The Cmax and AUC0–12 were linear and food ingestion improved the ability to tolerate tigecycline.81 Tigecycline has a large and variable volume of distribution that has been reported to be dose proportional and significantly greater than that of other tetracyclines.25,79–81
Tigecycline human protein binding has been reported to be 68%. The half-life of tigecycline is 36 h, and less than 30% of tigecycline is excreted unchanged in the faeces and urine. Tigecycline is metabolized primarily by liver glucuronidation. The AUC and Cmax of tigecycline have been higher in patients with renal dysfunction. There are no data suggesting how to dose tigecycline in patients with hepatic disease.25,82 In another pharmacokinetic study of tigecycline performed with healthy adults and in individuals with renal impairment, slight trends for different tigecycline pharmacokinetics were noted, possibly due to age, gender or race. The pharmacokinetics of tigecycline were not changed by severe renal disease or haemodialysis. The authors stated, however, that further pharmacokinetic studies should be performed.83
In accordance with previously performed animal studies, the pharmacodynamic parameter that appeared best at predicting bacteriological cure was the total time above the MIC.25 Additionally, the combination of tigecycline and linezolid performed best in an in vitro pharmacodynamic infection model assessing the activity of tigecycline, linezolid, quinupristin–dalfopristin, arbekacin, and daptomycin alone and in combination against vancomycin-resistant S. aureus.84
Limited human data are available concerning adverse events occurring with the administration of tigecycline. More patients are being studied in clinical trials, and current information from a few of these trials suggests that tigecycline is generally safe and well tolerated. Nausea, vomiting and headache are the most frequently reported adverse events.25–27
Clinical trials
Human Phase I, II and III trials evaluating tigecycline have been performed in adults. Patients have been administered intravenous tigecycline 25 or 50 mg every 12 h for 7 to 14 days in a Phase II trial of complicated skin and skin-structure infections. In this randomized, open-label trial from 1999 to 2001 in 14 United States medical centres, the two doses of tigecycline were assessed for tolerability, differences in pharmacokinetics, and clinical and microbiological efficacy. The observed clinical cure rate for patients completing the test-of-cure visit was the primary end point. Secondary end points were the clinical cure rate at the end of therapy and the bacteriological response. At least one tigecycline dose was administered to 160 patients. Of these patients, 109 were clinically evaluable and 91 patients were microbiologically evaluable. For the primary end point, the clinical cure rates were 67% (95% CI, 53.3–79.3%) in the 25 mg group and 74% (95% CI, 60.3–85.0%) in the 50 mg group. Bacteriologic responses occurred in 56% (95% CI, 40.0–70.4%) of the patients in the 25 mg group and in 69% (95% CI, 54.2–82.3%) of the patients in the 50 mg group. Based in part on these data, Phase III studies were initiated. Nausea was the most commonly reported adverse event occurring in 22% of the intent-to-treat patients in the 25 mg group and in 35% of the 50 mg group. Vomiting was the second most common adverse event in this clinical trial, occurring in 13% of the intent-to-treat patients in the 25 mg group and in 19% of the 50 mg group.26 Tigecycline exhibited a good pharmacokinetic profile in patients requiring hospitalization due to complicated skin and skin-structure infections.26
In addition, tigecycline activity was evaluated in a Phase II, multicentre, open-label trial of therapy for complicated intra-abdominal infections that required surgery. All patients received 100 mg of tigecycline as a loading dose, followed by the administration of 50 mg of tigecycline every 12 h for 5 to 14 days. Patients were diagnosed with perforated and gangrenous appendicitis, complicated cholecystitis, or perforated diverticulitis and peritonitis. Of the 111 patients entered into this trial, 66 met the inclusion criteria and were evaluated. Cure rates were 67% (44 patients; 95% CI, 54.0–77.8%) at the test-of-cure visit and 76% (50 patients; 95% CI, 63.6–85.5%) at the end of treatment visit. In the intent-to-treat analyses, at the test-of-cure visit the cure rate was 55% (61/111 patients; 95% CI, 45.2–64.45%) and the end of treatment cure rate was 72% (80/111 patients; 95% CI, 62.8–80.2%). Nausea and vomiting were the most common adverse events. In patients with complicated intra-abdominal infections, tigecycline was safe and effacious.27
Two Phase III, multinational, randomized, double-blind studies have been performed in hospitalized patients with complicated skin and skin-structure infections comparing the safety and efficacy of tigecycline with combined therapy with vancomycin and aztreonam. Infections included deep soft tissue infections, infected ulcers with evidence of an acute infection, major abscesses (complicated or extensive), infected burns, infected human or animal bites and superficial infections or abscesses with a high risk of infection due to anaerobic or Gram-negative pathogens. The most common diagnosis was deep soft tissue infection with cellulitis. Nausea (25%) and vomiting (12%) were the most frequent adverse events associated with tigecycline administration. The individual studies and the pooled analysis demonstrated that the clinical efficacy and safety of tigecycline were comparable to combined vancomycin and aztreonam therapy for the treatment of complicated skin and skin-structure infections in hospitalized patients.85,86
Early results from a double-blind, Phase III, multinational trial revealed that tigecycline was comparable to imipenem in activity for patients with complicated intra-abdominal infections, which primarily consisted of complicated appendicitis (41%), cholecystitis (22%) and intra-abdominal abscess (11%). Tigecycline was administered intravenously as a 100 mg initial dose, followed by 50 mg every 12 h, and 500 mg of imipenem was administered intravenously every 6 h. The mean duration of treatment was ∼8 days.87
Clinical responses at test-of-cure for microbiological evaluable and microbiological modified intent-to-treat populations were the co-primary efficacy end points. Eight hundred and seventeen patients received at least one dose of the study drug, 641 (78%) formed the microbiological modified intent-to-treat cohort and 523 (64%) were included in the microbiologically evaluable cohort. In the microbiologically evaluable group, clinical cure rates at the test-of-cure determination were 91.3% (242/265) in the tigecycline group and 89.9% (232/258) in the imipenem group (95% CI = −4.6 to 6.8; P < 0.001). Clinical cure rates in the microbiological modified intent-to-treat cohorts were 86.6% (279/322) in the tigecycline group and 84.6% (270/319) in the imipenem group (95% CI = −3.7 to 7.7; P < 0.001).87
The most common adverse events were nausea (17.6% in the tigecycline group and 13.3% in the imipenem group) and vomiting (12.6% and 9.2%, tigecycline and imipenem groups, respectively). The authors concluded that tigecycline was safe and effective for therapy of patients with complicated intra-abdominal infections.87
The promising results using tigecycline in Phase II studies have led to Phase III trials, which are necessary to further assess the safety and therapeutic efficacy of tigecycline. Early results from multicentre Phase III clinical trials are also promising. The results of these trials will help guide the future usage of tigecycline.
Note added in proof
Tigecycline for injection was approved by the US FDA on 16 June 2005 for adults with `complicated skin and skin structure infections caused by Escherichia coli, Enterococcus faecalis (vancomycin-susceptible isolates only), Staphylococcus aureus (methicillin-susceptible and -resistant isolates), Streptococcus agalactiae, Streptococcus anginosus grp. (includes S. anginosus, S. intermedius and S. constellatus), Streptococcus pyogenes and Bacteroides fragilis' and `complicated intra-abdominal infections caused by Citrobacter freundii, Enterobacter cloacae, Escherichia coli, Klebsiella pneumoniae, Enterococcus faecalis (vancomycin-susceptible isolates only), Staphylococcus aureus (methicillin-susceptible isolates only), Streptococcus anginosus grp. (includes S. anginosus, S. intermedius and S. constellatus), Bacteroides fragilis, Bacteroides thetaiotaomicron, Bacteroides uniformis, Bacteroides vulgatus, Clostridium perfringens, and Peptostreptococcus micros.'
The FDA has established MIC susceptibility breakpoints for tigecycline for Staphylococcus aureus (including methicillin-resistant isolates) ≤0.5 mg/L; Streptococcus spp. other than S. pneumoniae ≤0.25 mg/L; and Enterococcus faecalis (vancomycin-susceptible isolates only) ≤0.25 mg/L. The established breakpoints for Enterobacteriaceae are susceptibility ≤2 mg/L, intermediate 4 mg/L, and resistant ≥8 mg/L. The established breakpoints for anaerobes are susceptibility ≤4 mg/L, intermediate 8 mg/L, and resistant ≥16 mg/L. [Source: Tigecycline package insert. Wyeth Pharmaceuticals, Philadelphia, PA, USA.]
Transparency declaration
I performed a tigecycline in vitro surveillance study for Wyeth Laboratories.
This manuscript was edited by Marion Stafford at Ochsner Clinic Foundation in New Orleans, LA, USA. Unconditional grant support was given from Upside Endeavors, LLC, Berwyn, PA, USA.
References
Shlaes DM, Projan SJ, Edwards JE. Antibiotic discovery: state of the state.
Spellberg B, Powers JH, Brass EP et al. Trends in antimicrobial drug development: implications for the future.
Infectious Diseases Society of America. Bad Bugs, No Drugs as Antibiotic Discovery Stagnates … A Public Health Crisis Brews, July
Ehrhardt AF, Russo R. Clinical resistance encountered in the respiratory surveillance program (RESP) study: a review of the implications for the treatment of community-acquired respiratory tract infections.
Buckingham SC, McDougal LK, Cathey LD et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus at a Memphis, Tennessee Children's Hospital.
Charlebois ED, Perdreau-Remington F, Kreiswirth B et al. Origins of community strains of methicillin-resistant Staphylococcus aureus.
Cazzola M, Blasi F, Centanni S et al. Advances in the research and development of chemotherapeutic agents for respiratory tract bacterial infections.
Jones RN. Resistance patterns among nosocomial pathogens: trends over the past few years.
Johnson AP, Livermore DM, Tillotson GS. Antimicrobial susceptibility of Gram-positive bacteria: what's current, what's anticipated?
Kang C-I, Kim S-H, Park WB et al. Bloodstream infections caused by Enterobacter species: predictors of 30-day mortality rate and impact of broad-spectrum cephalosporin resistance on outcome.
Melzer M, Eykyn SJ, Gransden WR et al. Is methicillin-resistant Staphylococcus aureus more virulent than methicillin-susceptible S. aureus? A comparative cohort study of British patients with nosocomial infection and bacteremia.
Bouchillon SK, Johnson BM, Hobana DJ et al. Determining incidence of extended spectrum β-lactamase producing Enterobacteriaceae, vancomycin-resistant Enterococcus faecium and methicillin-resistant Staphylococcus aureus in 38 centres from 17 countries: the PEARLS study 2001–2002.
Hiramatsu K, Okuma K, Ma XX et al. New trends in Staphylococcus aureus infections: glycopeptide resistance in hospital and methicillin resistance in the community.
Hiramatsu K. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance.
Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance.
Sum PE, Sum FW, Projan SJ. Recent developments in tetracycline antibiotics.
Biedenbach DJ, Beach ML, Jones RN. In vitro antimicrobial activity of GAR-936 tested against antibiotic-resistant Gram-positive blood stream infection isolates and strains producing extended-spectrum β-lactamases.
Edlund C, Nord CE. In-vitro susceptibility of anaerobic bacteria to GAR-936, a new glycylcycline.
Jones R, Fritsche T, Sader H et al. Antimicrobial activity of tigecycline (GAR-936) tested against Enterobacteriaceae, and selected non-fermentative Gram-negative bacilli, a worldwide sample. Abstract P939.
Gales AC, Jones RN. Antimicrobial activity and spectrum of the new glycylcycline, GAR-936 tested against 1,203 recent clinical bacterial isolates.
Jones RN. Disk diffusion susceptibility test development for the new glycylcycline, GAR-936.
Bauer G, Berens C, Projan SJ et al. Comparison of tetracycline and tigecycline binding to ribosomes mapped by dimethylsulphate and drug-directed Fe2+ cleavage of 16S rRNA.
Chopra I. Glycylcyclines: third-generation tetracycline antibiotics.
Zhanel GG, Homenuik K, Nichol K et al. The glycylcyclines: a comparative review with the tetracyclines.
Postier RG, Green SL, Klein SR et al. Tigecycline 200 Study Group. Results of a multicenter, randomized, open-label efficacy and safety study of two doses of tigecycline for complicated skin and skin-structure infections in hospitalized patients.
Murray J, Wilson S, Klein S et al. The clinical response to tigecycline in the treatment of complicated intra-abdominal infections in hospitalized patients, a phase 2 clinical trial. In: Programs and Abstracts of the Forty-third Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL,
Connell SR, Tracz DM, Nierhaus KH et al. Ribosomal protection proteins and their mechanism of tetracycline resistance.
Petersen PJ, Jacobus NV, Weiss WJ et al. In vitro and in vivo antibacterial activities of a novel glycylcycline, the 9-t-butylglycylamido derivative of minocycline (GAR-936).
Projan SJ. Preclinical pharmacology of GAR-936, a novel glycylcycline antibacterial agent.
Hirata T, Saito A, Nishino K et al. Effects of efflux transporter genes on susceptibility of Escherichia coli to tigecycline (GAR-936).
Ruzin A, Keeney D, Visalli MA et al. AcrAB efflux-mediated decreased susceptibility to tigecycline in Enterobacter spp. In: Programs and Abstracts of the Forty-fourth Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC, 2004. Abstract 1061, p. 81. American Society for Microbiology, Washington, DC, USA.
Tuckman M, Petersen PJ, Projan SJ. Mutations in the interdomain loop region of the tetA(A) tetracycline resistance gene increase efflux of minocycline and glycylcyclines.
Pachon-Ibanez ME, Jimenez-Mejias ME, Pichardo C et al. Activity of tigecycline (GAR-936) against Acinetobacter baumannii strains, including those resistant to imipenem.
Milatovic D, Schmitz FJ, Verhoef J et al. Activities of the glycylcycline tigecycline (GAR-936) against 1,924 recent European clinical bacterial isolates.
Betriu C, Rodriguez-Avial I, Sanchez BA et al; Spanish Group of Tigecycline. In vitro activities of tigecycline (GAR-936) against recently isolated clinical bacteria in Spain.
Goldstein EJ, Citron DM, Merriam CV et al. Comparative in vitro activities of GAR-936 against aerobic and anaerobic animal and human bite wound pathogens.
Cercenado E, Cercenado S, Gomez JA et al. In vitro activity of tigecycline (GAR-936), a novel glycylcycline, against vancomycin-resistant enterococci and staphylococci with diminished susceptibility to glycopeptides (letter).
Low DE, Kreiswirth BN, Weiss K et al. Activity of GAR-936 and other antimicrobial agents against North American isolates of Staphylococcus aureus.
Boucher HW, Wennersten CB, Eliopoulos GM. In vitro activities of the glycylcycline GAR-936 against Gram-positive bacteria.
Kitzis MD, Ly A, Goldstein FW. In vitro activities of tigecycline (GAR-936) against multidrug-resistant Staphylococcus aureus and Streptococcus pneumoniae (letter).
Hoellman DB, Pankuch GA, Jacobs MR et al. Antipneumococcal activities of GAR-936 (a new glycylcycline) compared to those of nine other agents against penicillin-susceptible and -resistant pneumococci.
Betriu C, Culebras E, Rodriguez-Avial I et al. In vitro activities of tigecycline against erythromycin-resistant Streptococcus pyogenes and Streptococcus agalactiae: mechanisms of macrolide and tetracycline resistance.
Patel R, Rouse MS, Piper KE et al. In vitro activity of GAR-936 against vancomycin-resistant enterococci, methicillin-resistant Staphylococcus aureus and penicillin-resistant Streptococcus pneumoniae.
Bozdogan B, Esel D, Whitener C et al. Antibacterial susceptibility of a vancomycin-resistant Staphylococcus aureus strain isolated at the Hershey Medical Center.
van Ogtrop ML, Andes D, Stamstad TJ et al. In vivo pharmacodynamic activities of two glycylcyclines (GAR-936 and WAY 152,288) against various Gram-positive and Gram-negative bacteria.
Deshpande LM, Gales AC, Jones RN. GAR-936 (9-t-butylglycylamido- minocycline) susceptibility test development for streptococci, Haemophilus influenzae and Neisseria gonorrhoeae: preliminary guidelines and interpretive criteria.
Henwood CJ, Gatward T, Warner M et al. Antibiotic resistance among clinical isolates of Acinetobacter in the UK, and in vitro evaluation of tigecycline (GAR-936).
Betriu C, Rodriguez-Avial I, Sanchez BA et al. Comparative in vitro activities of tigecycline (GAR-936) and other antimicrobial agents against Stenotrophomonas maltophilia (letter).
Cercenado E, Cercenado S, Bouza E. In vitro activities of tigecycline (GAR-936) and 12 other antimicrobial agents against 90 Eikenella corrodens clinical isolates.
Johnson B, Stevens T, Bouchillon S et al. In vitro study of tigecycline against 776 clinical isolates of non-Enterobacteriaceae from hospitals across Europe. In: Programs and Abstracts of the Forty-third Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL,
Johnson B, Stevens T, Bouchillon S et al. In vitro antibacterial activity of tigecycline a novel glycylcycline against clinical isolates of Enterobacteriaceae. In: Programs and Abstracts of the Forty-third Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL,
Roblin PM, Hammerschlag MR. In vitro activity of GAR-936 against Chlamydia pneumoniae and Chlamydia trachomatis.
Wallace RJ Jr, Brown-Elliott BA, Crist CJ et al. Comparison of the in vitro activity of the glycylcycline tigecycline (formerly GAR-936) with those of tetracycline, minocycline, and doxycycline against isolates of nontuberculous mycobacteria.
Rhomberg PR, Jones RN. In vitro activity of 11 antimicrobial agents, including gatifloxacin and GAR936, tested against clinical isolates of Mycobacterium marinum.
Visalli MA, Murphy E, Projan SJ et al. AcrAB multidrug efflux pump is associated with reduced levels of susceptibility to tigecycline (GAR-936) in Proteus mirabilis.
Dean CR, Visalli MA, Projan SJ et al. Efflux-mediated resistance to tigecycline (GAR-936) in Pseudomonas aeruginosa PAO1.
Reynolds R, Potz N, Colman M et al. Antimicrobial susceptibility of the pathogens of bacteraemia in the UK and Ireland 2001–2002: the BSAC Bacteraemia Resistance Surveillance Programme.
Fritsche TR, Kirby JT, Jones RN. In vitro activity of tigecycline (GAR-936) tested against 11,859 recent clinical isolates associated with community-acquired respiratory tract and Gram-positive cutaneous infections.
Zhanel GG, Palatnick L, Nichol KA et al. Cross Study Group. Antimicrobial resistance in Haemophilus influenzae and Moraxella catarrhalis respiratory tract isolates: results of the Canadian Respiratory Organism Susceptibility Study, 1997 to 2002.
Sader HS, Fritsche TR, Kirby JT et al. Activity of tigecycline tested against clinical isolates of Haemophilus influenzae, Moraxella catarrhalis, and Neisseria meningitidis, a worldwide perspective. In: Programs and Abstracts of the European Congress of Clinical Microbiology and Infectious Diseases, Prague, Czech Republic, 2004. Abstract 940, p. 247.
Fritsche TR, Sader HS, Kirby JT et al. In vitro activity of the glycylcycline tigecycline tested against a worldwide collection of 10,127 contemporary staphylococci, streptococci and enterococci. Abstract P937.
Zhanel GG, Palatnick L, Nichol KA et al. Antimicrobial resistance in respiratory tract Streptococcus pneumoniae isolates: results of the Canadian Respiratory Organism Susceptibility Study, 1997 to 2002.
Petersen PJ, Bradford PA, Weiss WJ et al. In vitro and in vivo activities of tigecycline (GAR-936), daptomycin, and comparative antimicrobial agents against glycopeptide-intermediate Staphylococcus aureus and other resistant Gram-positive pathogens.
Mercier RC, Kennedy C, Meadows C. Antimicrobial activity of tigecycline (GAR-936) against Enterococcus faecium and Staphylococcus aureus used alone and in combination.
Appelbaum PC, Kelly LM, Ednie LM et al. Comparative activities of dalbavancin against staphylococci, including a vancomycin-resistant strain of Staphylococcus aureus. In: Programs and Abstracts of the Forty-first Annual Meeting of the Infectious Diseases Society of America, San Diego, CA,
Tedesco KL, Huang V, Rybak MJ. In vitro activities of daptomycin, arbekacin & tigecycline (GAR-936) against several clinical glycopeptide-intermediate Staphylococcus aureus (GISA) pathogens. In: Programs and Abstracts of the Forty-first Annual Meeting of the Infectious Diseases Society of America, San Diego, CA,
Labthavikul P, Petersen PJ, Bradford PA. In vitro activity of tigecycline against Staphylococcus epidermidis growing in an adherent-cell biofilm model.
Jacobus NV, McDermott LA, Ruthazer R et al. In vitro activities of tigecycline against the Bacteroides fragilis group.
Kenny GE, Cartwright FD. Susceptibilities of Mycoplasma hominis, M. pneumoniae, and Ureaplasma urealyticum to GAR-936, dalfopristin, dirithromycin, evernimicin, gatifloxacin, linezolid, moxifloxacin, quinupristin–dalfopristin, and telithromycin compared to their susceptibilities to reference macrolides, tetracyclines, and quinolones.
Petersen PJ, Weiss WJ, Labthavikul P et al. The post-antibiotic effect and time–kill kinetics of the glycylcyclines, GAR-936 (TBG-MINO) and (PAM-MINO). In: Programs and Abstracts of the Thirty-eighth Interscience Conference on Antimicrobial Agents and Chemotherapy, San Diego, CA,
Lefort A, Lafaurie M, Massias L et al. Activity and diffusion of tigecycline (GAR-936) in experimental enterococcal endocarditis.
Murphy TM, Deitz JM, Petersen PJ et al. Therapeutic efficacy of GAR-936, a novel glycylcycline, in a rat model of experimental endocarditis.
Nannini EC, Pai SR, Singh KV et al. Activity of tigecycline (GAR-936), a novel glycylcycline, against Enterococci in the mouse peritonitis model.
Edelstein PH, Weiss WJ, Edelstein MA. Activities of tigecycline (GAR-936) against Legionella pneumophila in vitro and in guinea pigs with L. pneumophila pneumonia.
Yin L, Lazzarini L, Bortolo S et al. Comparative evaluation of tigecycline and vancomycin in the treatment of methicillin-resistant Staphylococcus aureus experimental osteomyelitis. In: Programs and Abstracts of the Forty-first Annual Meeting of the Infectious Diseases Society of America, San Diego, CA,
Mikels SM, Brown AS, Breden L et al. In vivo activities of GAR-936 (GAR), gentamicin (GEN), piperacillin (PIP), alone and in combination in a murine model of Pseudomonas aeruginosa pneumonia. In: Programs and Abstracts of the Thirty-ninth Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA,
Fang GD, Weiss WJ, Scheld WM. Comparative efficacy of GAR-936 (GAR), a novel glycylcycline, alone and in combination with vancomycin against highly penicillin-resistant Streptococcus pneumoniae (PRSP) experimental meningitis in rabbits. In: Programs and Abstracts of the Fortieth Interscience Conference on Antimicrobial Agents and Chemotherapy, Toronto, Canada,
Sesoko S, Umemura K, Nakashima M. Pharmacokinetics (PK), safety, and tolerability of tigecycline (GAR-936) in healthy Japanese males. In: Programs and Abstracts of the Forty-second Interscience Conference on Antimicrobial Agents and Chemotherapy, San Diego, CA,
Muralidharan G, Mojaverian P, Micalizzi M et al. The effects of age and gender on the pharmacokinetics, safety and tolerability of GAR-936, a novel glycylcycline antibiotic, in healthy subjects. In: Programs and Abstracts of the Fortieth Interscience Conference on Antimicrobial Agents and Chemotherapy, Toronto, Canada,
Muralidharan G, Getsy J, Mayer P et al. Pharmacokinetics (PK), safety and tolerability of GAR-936, a novel glycycline antibiotic, in healthy subjects. In: Programs and Abstracts of the Thirty-ninth Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA,
Troy SM, Muralidharan G, Micalizzi M et al. The effects of renal disease on the pharmacokinetics of tigecycline (GAR-936). In: Programs and Abstracts of the Forty-third Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL,
Meagher AK, Cirincione BB, Liolios KA et al. Pharmacokinetics of tigecycline in healthy adult volunteers and in subjects with renal impairment. Abstract P1023.
Huang V, Brown WJ, Rybak MJ. Evaluation of tigecycline, linezolid, quinupristin–dalfopristin, arbekacin, and daptomycin alone and in combination against two clinical strains of vancomycin-resistant Staphylococcus aureus in an in vitro pharmacodynamic infection model. Abstract P1040.
Ellis-Grosse EJ, Loh E. Tigecycline is safe and effective in the treatment of skin and skin structure infections: results of two double-blind Phase 3 comparison studies with vancomycin/aztreonam. In: Programs and Abstracts of the 9th Western Pacific Congress on Chemotherapy and Infectious Diseases, Bangkok, Thailand,
Dartois N. Results of a Phase 3, double-blind, safety and efficacy study comparing tigecycline with vancomycin/aztreonam to treat complicated skin and skin structure infections. In: Programs and Abstracts of the Forty-fourth Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC,
Dartois N, Gioud-Paquet M, Ellis-Grosse EJ et al. Tigecycline vs imipenem/cilastatin for treatment of complicated intra-abdominal infections. In: Programs and Abstracts of the Forty-fourth Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC,