-
PDF
- Split View
-
Views
-
Cite
Cite
Nassim M. Amiali, Michael R. Mulvey, Brigitte Berger-Bächi, Jacqueline Sedman, Andrew E. Simor, Ashraf A. Ismail, Evaluation of Fourier transform infrared spectroscopy for the rapid identification of glycopeptide-intermediate Staphylococcus aureus, Journal of Antimicrobial Chemotherapy, Volume 61, Issue 1, January 2008, Pages 95–102, https://doi.org/10.1093/jac/dkm400
Close - Share Icon Share
Abstract
To evaluate Fourier transform infrared (FTIR) spectroscopy as a rapid method for distinguishing glycopeptide-intermediate Staphylococcus aureus (GISA) from glycopeptide-susceptible methicillin-resistant S. aureus (MRSA) and to compare three data analysis methods.
First-derivative normalized spectra of dried films of bacterial growth on Que-Bact® Universal Medium No. 2 were examined by singular value decomposition to identify key spectral regions. Region selection was analysed by principal component analysis (PCA), self-organizing maps (SOMs) and the K-nearest neighbour (KNN) algorithm. The initial data set included 35 GISA (including GISA Mu50 and heterogeneous GISA Mu3) and 25 epidemic MRSA. The regions were then tested using enlarged data sets that included 22 sporadic and 85 additional epidemic MRSA.
Epidemic MRSA and GISA/hGISA were separated into two distinct clusters on the basis of spectral data from regions 1352–1315 and 1480–1460 cm−1, the former providing 100% correct classification by all three analyses and the latter providing 96.67% correct by PCA, 98.34% by SOM and 100% by KNN. The 1480–1460 cm−1 region was more effective for distinguishing GISA/hGISA from a set combining sporadic and epidemic MRSA, with two GISA/hGISA and four sporadic MRSA misclassified by PCA and SOM (92.69% correct), while the KNN method misclassified three of the four sporadic MRSA (93.90% correct). The addition of 85 other epidemic MRSA this set increased the fraction of correctly classified isolates to 96.41% and 97.01% by PCA, SOM and KNN, respectively.
As only 6 of 167 isolates were misclassified, FTIR spectroscopy may provide means of rapid and accurate identification of GISA and hGISA among isolates of MRSA.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most common agents of community-acquired and nosocomial infections worldwide, causing high morbidity and mortality. For the past three decades, glycopeptides have been the antibiotics of choice for the treatment of MRSA infections. In recent years, however, MRSA with varying degrees of reduced susceptibility to glycopeptides have emerged, the so-called ‘glycopeptide-intermediate’ strains or GISA. These are characterized by MICs of 8–16 mg/L for teicoplanin and ≥8 mg/L for vancomycin. This situation is complicated by heterogeneous forms called hGISA, with MICs of 16 mg/L for teicoplanin and 1–4 mg/L for vancomycin. These appear to give rise to subpopulations able to grow in the presence of over 4 mg/L of vancomycin at a frequency of 10−6 or higher1–4 and may underlie some failures of vancomycin therapy.5–7 However, the true clinical incidence and significance of hGISA remain unclear.
The mechanism of glycopeptide resistance has not been fully defined and clinical detection continues to rely on growth and susceptibility in selective media, often giving ambiguous results.1,5,8,9 Thus there is a need in clinics for a more reliable method of screening MRSA for the glycopeptide resistance trait. Such a method should be simple and rapid and offer high reproducibility both within and between laboratories.
The GISA/hGISA phenotype may be confirmed by population analysis1 and/or by electron microscopy (thickened cell wall), both too complex and costly for routine clinical practice. The gold standard method, used in a number of surveillance studies for GISA and hGISA, is the population analysis profile (PAP)-area under the curve (AUC),10–12 with criteria specifically designed to discriminate between glycopeptide-susceptible S. aureus, hGISA and GISA. Ideally, PAP-AUC would be used in clinics to confirm the GISA/hGISA phenotypes when identified by a reliable and preferably rapid screening method.
Several screening methods and agars for GISA/hGISA have been reported.1,13,14 In a recent international study involving 12 laboratories,15 the macro-method Etest (MET) and Mueller–Hinton agar (MHA) with 5 mg/L teicoplanin provided correct identification of 82.5% and 85.9% of 48 test strains, whereas brain–heart infusion agar (BHIA) broth with 6 mg/L vancomycin identified only 11.5% of the hGISA in the set and gave the highest inter-laboratory variability for GISA in general.
The GISA phenotype is associated with alterations in the cell-wall peptidoglycan synthesis pathway and increased levels of penicillin-binding proteins (PBP2), resulting in thickened cell walls.16 Biochemical characteristics such as these are detectable by Fourier transform infrared (FTIR) spectroscopy, a non-destructive technique that can be used to probe the total composition of intact microbial cells without the use of reagents. Complex yet distinct and reproducible spectral signatures or ‘fingerprints’ of microorganisms may be obtained for identification purposes, even down to the subspecies17–22 and antibiotic resistance phenotypes.23–25 On the basis of these findings, the present study was undertaken to evaluate its potential as a superior alternative to conventional clinical screening methods for distinguishing the GISA/hGISA phenotype from glycopeptide-susceptible S. aureus.
Materials and methods
Clinical specimens and microbiological analysis
The 35 GISA/hGISA isolates were obtained from the Network on Antimicrobial Resistance in S. aureus (NARSA) collection. These included 31 methicillin-resistant and 4 methicillin-susceptible GISA/hGISA isolates. Heterogeneous GISA strain Mu3 and GISA strain Mu50 were included among the 31 methicillin-resistant GISA/hGISA. Eighty-five isolates of epidemic MRSA and 22 isolates of sporadic MRSA were provided by the Health Canada National Microbiology Laboratory (NML). Another 25 epidemic MRSA were provided by the Royal Victoria Hospital (Montreal, QC, Canada).
Testing for glycopeptide-intermediate resistance in all GISA/hGISA was performed by NARSA using broth microdilution (BMD) with frozen and dried panels; conventional MET with MHA, 0.5 McFarland inoculum and 24 h incubation at 35°C (MHA-CMET); the modified MET with BHIA, 2.0 McFarland inoculum and 48 h incubation at 35°C (BHIA-MMET); the BHIA screen assay (24, 48 and 72 h growth of 10 µL spot on agar containing 6 mg/L vancomycin or BHIA-6V); and the conventional disc diffusion method (DD). In fact, identification of an isolate as GISA or hGISA by NARSA is based solely on the BHIA-MMET method.
FTIR spectroscopic methods
Sample preparation
All isolates were grown from frozen stocks kept at −70°C in BHI broth containing 15% glycerol, by overnight subculture on tryptic soy agar with sheep blood (Quelab Laboratories Inc., Montreal, QC, Canada) at 37°C. A single colony was collected, streaked in a four-quadrant pattern onto four plates of Que-Bact® Universal Medium No. 2 agar (Quelab Laboratories Inc.), which were then incubated for 18 h at 37°C. Cells were then carefully collected from a single quadrant of each plate using a 10 mm diameter soft plastic loop (10 µL, COP-S10, Copan Diagnostics Inc., Corona, CA, USA). All surfaces of the loop head are ultra-smooth for easy streaking or harvesting of growth, without damaging or scraping the agar surface. Collected colonies were suspended in the same 200 µL aliquot of sterile physiological saline (0.9% NaCl), which was then diluted 10-fold in saline to a cell concentration of ∼5 × 1011 cfu/mL. A 25 µL droplet of diluted suspension was deposited on a clean zinc selenide (ZnSe) optical window (13 mm diameter, 2 mm thick, two windows per aluminium slide) and oven-dried at 48°C for 1 h to form a thin and transparent homogeneous dried film for FTIR measurement. Four films were made for each isolate.
Spectral acquisition
All FTIR spectra were acquired in the transmission mode using a Bomem MB-104 (ABB-Bomem, Quebec, QC, Canada) FTIR spectrometer equipped with a non-hygroscopic ZnSe beamsplitter and a deuterated triglycine sulphate detector and operating under Bomem-Grams/386 software (Galactic, Salem, NH, USA). The spectrometer was purged with dry CO2-free air from a Balston dryer (Balston, Lexington, MA, USA) to minimize interference from atmospheric water vapour and CO2. To enhance the signal-to-noise ratio, 64 scans were co-added at 4 cm−1 resolution over the wavenumber range of 4000–400 cm−1 and ratioed against an open-beam background to obtain an absorbance spectrum. A single spectrum was thus obtained for each of the four windows made for each strain.
Tentative assignments of spectral features based on the comparison of resolution-enhanced microbial IR spectra with IR spectra of known building blocks inherent in intact cells are described as follows.26
The spectral region of 3000–2800 cm−1 is dominated by broad features resulting from vibrations of –CH3, >CH2 and CH functional groups of fatty acid aliphatic chains. The region between 1800 and 1500 cm−1 contains bands corresponding to protein and peptide components such as amides I and II. The 1500–1200 cm−1 region is mixed and contains vibrations corresponding to carboxylic groups of proteins, free amino acids and polysaccharides as well as vibrations of phosphate, RNA/DNA and phospholipids. The 1200–900 cm−1 region is dominated by absorption bands corresponding to cell-wall carbohydrates. The 900–700 cm−1 region is yet to be associated with specific components.
Mathematical pre-processing and processing
Spectra acquired in Grams SPC format were converted into comma-separated value files and then into MATLAB files using MATLAB version 5.1 (The MathWorks, Inc., Natick, MA, USA). Because band intensity varies as a function of film thickness, the spectral data over the whole spectral range (4000–400 cm−1) were normalized to unit height by vector transformation to compensate for differences in film thickness and then transformed to the first derivative using the Savitzky–Golay algorithm to maximize peak separation, enhance apparent resolution and minimize problems arising from baseline shifts.
Prior to data processing, spectral feature selection was performed by singular value decomposition (SVD)27 using pairs of spectra and confirmed by visual examination of the normalized spectra. Data were processed using two exploratory data analysis methods, namely, principal component analysis or PCA (employing the non-linear iterative partial-least-squares algorithm28) and the self-organizing map (SOM) algorithm.29 Cluster analysis was performed using the K-nearest neighbours (KNN) algorithm.30 Programs were written in MATLAB version 5.1 to implement the data pre-processing and processing algorithms.
SVD algorithm
Spectral regions having features with the potential to distinguish all the strains were pre-selected using the SVD algorithm.27 The SVD algorithm inspects IR spectral region vectors (columns of matrix U and absorbance at specific wavenumbers) corresponding to the most significant features and compares each vector to a threshold value. If the threshold is exceeded, the corresponding spectral region is assigned to the clustering set.
Principal component analysis
PCA is a multivariate procedure that rotates data to maximize the variability projected onto axes. A set of correlated variables is thus transformed to a set of uncorrelated variables ranked by variability in the descending order.28 The resulting uncorrelated variables are linear combinations of the original variables and the last of these can be removed with minimal loss of real data. PCA is used mainly to reduce the dimensionality of a data set while retaining as much information as possible by computing a compact and optimal description of the data set. The first principal component (PC1) is the combination of variables that accounts for the greatest amount of the total variation. The second principal component (PC2) defines the largest amount of the remaining variation and is orthogonal to the first principal component. The number of principal components possible is equal to the number of variables. In addition to reducing the number of variables and detecting structural relationship between variables, a PCA plot shows spectral data distribution and hence the existence of clusters.
Self-organizing maps
K-nearest neighbour
The KNN classifier is a non-parametric similarity classification-based method that attempts to categorize unknown samples based on multivariate proximity to other samples of pre-assigned categories. The distance matrix is sorted and the distance of the unknown sample is compared with its KNNs. The identity of an unknown sample is based on the class of the nearest known samples, that is, by assigning the class shared by the majority of nearest neighbours to the unknown. Each class represents a bacterial strain type in the data set. The optimal number K of neighbours used to predict an unknown was determined from the highest number of correctly classified points obtained with K set at 1 through 10.30
Results and discussion
The emergence of S. aureus in many parts of the world with decreased susceptibility to glycopeptide antimicrobial agents such as vancomycin and teicoplanin is a major concern.9 It is particularly worrisome that hetero-resistant strains (hGISA) appear susceptible based on the conventional testing, but give rise to more resistant forms at a frequency of about one cell per million. Both GISA and hGISA confound routine methods used in diagnostic laboratories. The use of BHI agar with 6 mg/L vancomycin (BHIA-6V) or of Mueller–Hinton agar with5 mg/L teicoplanin and the MET appears to be <86% accurate and with considerable inter-laboratory variability.15 The true clinical incidence of GISA and in particular hGISA will not be known until a reliable detection method is available at a cost affordable by diagnostic laboratories. Such a method must minimize false-positive results, as it would be highly desirable to do the expensive confirmatory tests (PAP-AUC) when screening results are positive. Diagnostic laboratories will therefore assess the performance of any GISA/hGISA screening method in terms of cost-effectiveness. Reproducibility and variability of performance between laboratories is of concern, especially for public health and research. FTIR spectroscopy has the potential to offer a method that is affordable and reproducible both within and between laboratories.
Spectral reproducibility
The fundamental requirement for IR analysis of microorganisms is that the variance among spectra of a single taxon be less than that among spectra of different taxa.31 In this context, it should be emphasized that IR spectroscopy is sensitive to phenotypic rather than genotypic differences; that is, it measures the biochemical expression of genes under a specific set of conditions rather than the presence of specific genes. As variations in biochemical composition among different taxa may be slight, spectral reproducibility depends on stringent control of sampling and measurement conditions. The composition of the growth medium and phase of growth (stationary versus exponential) are critical, as relative peak intensities may be affected much more (because of changes in metabolite pools) than peak positions.32 FTIR analysis thus depends very much on fidelity in the reproduction of growth media, culture handling techniques and culture conditions.31,33,34 An ‘IR grade’ standard blood-free synthetic growth medium, namely Que-Bact® Universal Medium No. 2 developed by Quelab Laboratories Inc., and strict observance of 18 h incubations at 37°C to obtain early stationary-phase cells on a consistent basis were employed in the present study.
There is a theoretical possibility of variance due to carry-over (from the medium used to reactivate the frozen strains) of sheep blood from different sources. Three years of practice have indicated that this does have any appreciable impact on spectral consistency (data not shown). It is also important to collect colonies uncontaminated by agar. This is ensured using a large plastic loop that gathers a large amount of bacterial growth without scraping the agar surface. The exact manner in which different individuals collect the growth and produce the cell suspension could introduce variance, but no spectral variations were attributable to this factor.
For spectra acquired in the transmission mode, reproducibility is affected by sample uniformity, in particular homogeneity, particle size and thickness or light path length.35 Drying a 25 µL droplet of diluted bacterial suspension for 1 h at 48°C yielded a transparent and uniform film and avoided baseline variations as well as scattering, diffraction and refraction of the infrared beam. Variations in the sample thickness cause band intensity to vary, although relative peak intensities may be maintained. Comparison of the spectra between 1800 and 800 cm−1 of the four films prepared for each isolate (from a suspension of colonies from four separate plates) yielded an average correlation coefficient of r = 0.97 (data not shown). Variations were further minimized by normalization to unit peak height and transformation to the first derivative, which highlights spectral shapes and contours and removes the effects of baseline shift, assuming that the spectra possess a sufficient signal-to-noise ratio. Finally, the use of a highly hygroscopic FTIR spectrometer equipped with a ZnSe beamsplitter improves reproducibility for long-term intra- and inter-laboratory comparisons.
Spectral differences between MRSA and GISA/hGISA
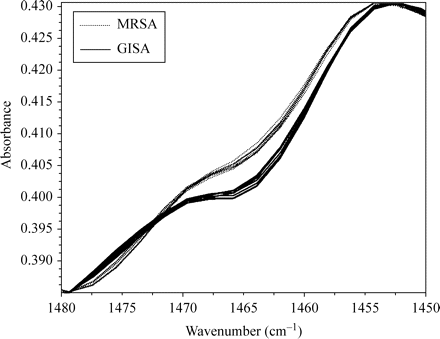
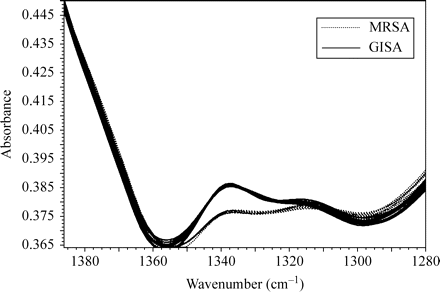
Infrared spectra of microorganisms have extensive overlap because of bio-molecules common to all cells, and the subtle differences among strains may be difficult to detect. The differentiation of GISA/hGISA and MRSA on the basis of their FTIR spectra may be categorized as a pattern recognition task. As in any such task, feature extraction using the SVD algorithm may assist in the selection of spectral regions for identification purposes. A random selection among the 240 spectra obtained for the 35 GISA/hGISA and 25 NML epidemic MRSA isolates allowed us to identify two narrow regions with clear differences using SVD: (i) 1480–1460 cm−1 (Figure 1), assignable to deformation vibrations of C-H bonds in methylene (CH2) groups of lipids and proteins; and (ii) 1352–1315 cm−1 (Figure 2), attributable to symmetric stretching vibrations of carboxyl groups of amino acid side chains or free fatty acids.
FTIR spectra of GISA/hGISA (solid lines) and MRSA (dotted lines) in the region 1480–1450 cm−1. A colour version of this figure is available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
FTIR spectra of GISA/hGISA (solid lines) and MRSA (dotted lines) in the region 1380–1280 cm−1. A colour version of this figure is available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Discrimination between MRSA and GISA/hGISA based on the 1480–1460 cm−1 region
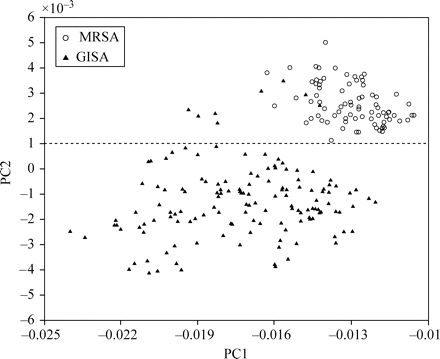
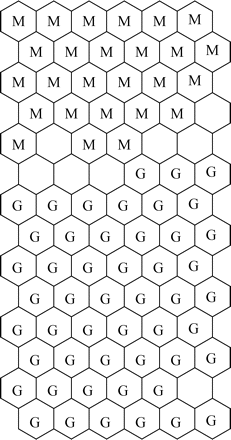
The first three principal components (PC1, PC2 and PC3) accounted for over 99.94% of the total variance, with PC1 and PC2 alone accounting for 97.92% and 1.84%, respectively. The PCA scores plot (PC1 versus PC2) gave two distinct clusters, corresponding to MRSA and GISA/hGISA (Figure 3). Two GISA/hGISA isolates, NRS4 and NRS68, were misclassified as MRSA, yielding an overall correct classification of 96.67%. Visual inspection of an SOM of size [14 × 6] generated by non-linear projection of the PC1 and PC2 scores and trained using a rough training phase of 4 iterations and a fine-tuning phase of 14 epochs also indicated two distinct clusters corresponding to MRSA and GISA/hGISA (Figure 4). The learning rate decreased linearly to zero during the fine-tuning phase with a final quantization error of 0.162 and a final topographic error of 0.048, yielding an overall correct classification of 98.34% with a single misclassified GISA/hGISA isolate, NRS68. This misclassified isolate cannot be visualized, as only the BMUs are shown. None of the four NRS68 replicates were classified as a BMU and therefore could be visualized only when plotting all data neighbours and BMUs at the same time, which would make the map difficult to visualize because of the large number of data. The supervised cluster analysis using the KNN algorithm with 120 spectra as the training set and the remaining 120 spectra as the test set gave 100% correct classification with K = 1 and K = 2.
Scores plot for the first two principal components obtained from FTIR spectra of GISA/hGISA (filled triangles) and MRSA (open circles) in the region 1480–1460 cm−1. A colour version of this figure is available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
SOM obtained using FTIR spectral profiles of GISA/hGISA (G) and MRSA (M) in the region 1480–1460 cm−1. A colour version of this figure is available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Discrimination between MRSA and GISA/hGISA based on the 1352–1315 cm−1 region
Using the region 1352–1315 cm−1, clustering of epidemic MRSA and GISA/hGISA was 100% correct by PCA, SOM and KNN. Two distinct clusters of GISA/hGISA and epidemic MRSA were clearly visible on the PCA plot and on an SOM of size [16 × 5] generated by non-linear projection of the PC1 and PC2 scores for this region using 4 iterations for rough training and 13 epochs for the fine-tuning phase (data not shown). The final quantization error was 0.128 with a final topological error of 0.008. Using half of the data set (n = 120) as the training set and the other half as the prediction set, the KNN algorithm was most effective for K = 1 and K = 2.
Discrimination between epidemic MRSA/sporadic MRSA and GISA/hGISA
Twenty-two sporadic MRSA isolates were added to investigate the ability of FTIR to distinguish GISA/hGISA from both sporadic and epidemic MRSA. The expanded data set contained four replicate spectra for each isolate for a total of 328 spectra. The region 1352–1315 cm−1, although suitable for discriminating between epidemic MRSA and GISA/hGISA, proved unsuitable for discriminating between sporadic MRSA and GISA/hGISA, yielding an overall correct classification of only 84.15% by PCA, with 13 sporadic MRSA misclassified (data not shown). Some improvement was obtained using PCA with the region 1480–1460 cm−1. The first three PCs contained over 99.9% of the overall total variance, with totals of 98.18%, 99.75% and 99.93% for PC1, PC2 and PC3, respectively, the scores plot of PC1 versus PC2 revealing two distinct clusters corresponding to GISA/hGISA and MRSA (data not shown). However, three sporadic MRSA isolates (715, 864 and 693) were misclassified as GISA/hGISA strains and a single sporadic MRSA (105) fell between the GISA/hGISA and MRSA cluster. The same two GISA/hGISA strains already mentioned (NRS4 and NRS68) fell within the MRSA cluster, yielding 92.69% overall correct classification. The SOM algorithm sorted the scores of PCs 1 and 2 into clear clusters of GISA/hGISA and MRSA (data not shown) with sporadic MRSA isolates 182, 715, 864 and 693 as well as GISA/hGISA isolates NRS4 and NRS68 misclassified, yielding 92.69% overall correct classification. The map of size [13 × 7] was trained using a short rough training phase of three iterations that corresponded to the first stage in which the initial order is formed. The initial weight vector, a large neighbourhood radius and a large learning rate were then applied and the remaining iteration stages constituted a fine-tuning phase of 11 epochs in which both learning rate and neighbourhood radius began at small values and were gradually reduced at each iteration. The learning rate decreased linearly to zero during the fine-tuning phase with a final quantization error of 0.173 and a final topographic error of 0.009. Supervised classification performed with the KNN algorithm using half of the data set (n = 170) as the training set and the other half as the prediction set gave a slightly higher correct classification of 93.9% using K = 7. The same three sporadic MRSA (715, 864 and 693) and the same two GISA/hGISA (NRS4 and NRS68) misclassified by the PCA scores plot were also misclassified by KNN. The single sporadic MRSA (105) that fell between the GISA/hGISA and MRSA clusters was correctly classified on the basis of the class of the majority of its nearest neighbouring known samples.
Validation of the FTIR method
The robustness of the differentiation using the 1480–1460 cm−1 region was assessed by challenging it with another collection of epidemic MRSA isolates. Sixty of these were collected by NML from various Canadian sources, whereas the other 25 were provided by a single hospital (Montreal Royal Victoria). Classification of a set containing these plus the original 35 GISA/hGISA strains was 98.34% correct by PCA with the same GISA/hGISA isolates as mentioned above misclassified (NRS4 and NRS68) and 99.37% correct by SOM and KNN for K = 3 with a single strain of GISA (NRS 68) misclassified. When these 85 epidemic MRSA were combined with the initial expanded set of 82 isolates (i.e. including the sporadic MRSA), the same four sporadic MRSA (715, 693, 864 and 105) and two GISA/hGISA (NRS4 and NRS68) strains remained misclassified by PCA but the larger set increased the percent correct classification from 92.69% to 96.41%. For SOM, sporadic MRSA isolates 715, 693, 864 and 182 were again misclassified as well as GISA/hGISA isolates NRS4 and NRS68, thus increasing the correct classification from 92.69% to 96.41% as for PCA. Correct classification by the KNN analysis also increased, from 93.90% to 97.01% for K = 7, with the same three sporadic MRSA (715, 693 and 864) and the same GISA/hGISA (NRS4 and NRS68) misclassified.
Other work has shown that Enterococcus casseliflavus and Enterococcus gallinarum with intrinsic low-level vancomycin resistance were correctly identified by FTIR spectroscopy18 using a combination of equally weighted broad spectral ranges (1500–1200, 1200–900, 900–700 cm−1) of first-derivative spectra. However, these species were not distinguished from vancomycin-susceptible strains of identical species. Our present study is the first demonstration of reliable identification of glycopeptide-intermediate S. aureus by FTIR spectroscopy.
The decrease in the discrimination between epidemic MRSA and GISA/hGISA from 100% to 84.15% with 13 sporadic MRSA misclassified indicates poor specificity in the 1352–1315 cm−1 region. This may indicate heterogeneity of this phenotype, resulting from the acquisition of SCCmec by strains of different genetic background. In comparison, discrimination using the spectral region of 1480–1460 cm−1, containing primarily absorption bands that may be assigned to CH2 asymmetric bending vibrations of lipids and proteins,36,37 was not affected by the inclusion of the 22 sporadic MRSA isolates. On the basis of the interpretations for MIC breakpoints and recommendations of the CDC38 and CLSI,39 the misclassified GISA/hGISA NRS4 has shown intermediate resistance to vancomycin and teicoplanin by BHIA-MMET and a susceptible-resistant pattern by all other screening methods (BMD, MHA-CMET, BHIA-6V and DD), except for intermediate teicoplanin resistance using MHA-CMET. The other misclassified GISA/hGISA (NRS68) has shown intermediate resistance to vancomycin and susceptibility to teicoplanin using BHIA-MMET. All the other screening methods used (BMD, MHA-CMET, BHIA-6V and DD) have indicated no resistance to either glycopeptide. The four sporadic MRSA strains (715, 864, 693 and 105) apparently misclassified by FTIR as glycopeptide intermediate using PCA, as well as the sporadic MRSA 182 misclassified by SOM, have never in fact been tested for glycopeptide susceptibility.
An explanation for the repeated misclassification of GISA/hGISA isolates NRS4 and NRS68 by PCA and KNN could not be advanced on the basis of their vancomycin and teicoplanin susceptibility patterns. However, it should be noted that the glycopeptide-intermediate resistance phenotype may be unstable and affected by subculture methodology, repeated subculture or by different growth media used prior to susceptibility testing. Uniform decreases in vancomycin and teicoplanin MICs occurred after 15 days of serial passage on non-selective medium,40–42 a situation that provides an opportunity for reversion to occur. Reversion of the glycopeptide-intermediate resistance phenotype can occur in all types of GISA/hGISA isolates,40,42,43 although it appears to depend on the genetic background of the bacterial strain to some extent.40 This reversion may account for some of the difficulties in identifying them in the clinical laboratory and may explain the misidentification of isolates NRS4 and NRS68. All isolates in the present study were activated in the blood agar and cultured on Que-Bact® Universal Medium No. 2, both without glycopeptides, prior to FTIR analysis.
The identification of GISA/hGISA isolates NRS4 (HIP5836) and NRS68 should be confirmed by quantitative susceptibility and confirmatory testing using PAPs, whereas the sporadic MRSA strains 715, 864, 693, 105 and 182 should be tested for glycopeptide resistance by conventional screening methods to determine whether or not our FTIR method did in fact misclassify them.
Conclusions
On the basis of the results of this study, FTIR spectroscopy combined with the use of growth medium Que-Bact® Universal Medium No. 2 and chemometric analysis of spectral data appear to offer a promising alternative to conventional susceptibility testing methods for routine identification of GISA/hGISA and accordingly warrant further validation studies to confirm the suitability of this technique for rapid screening of these strains. In addition, further investigation of the spectral differences between MRSA and GISA/hGISA isolates observed in this study may aid in the elucidation of the mechanisms of glycopeptide resistance in MRSA.
Funding
This work was funded by Quelab Laboratories Inc.
Acknowledgements
We thank the Canadian Nosocomial Infection Surveillance Program (CNISP) for participating in this research. GISA/hGISA isolates were obtained through the Network on Antimicrobial Resistance in S. aureus (NARSA) programme, supported by the US National Institute for Allergy and Infectious Diseases (US National Institutes of Health, contract N01-AI-95359).
Transparency declarations
None to declare.