-
PDF
- Split View
-
Views
-
Cite
Cite
Ali Rezaei-Matehkolaei, Hossein Mirhendi, Koichi Makimura, G. Sybren de Hoog, Kazuo Satoh, Mohammad Javad Najafzadeh, Mohammad Reza Shidfar, Nucleotide sequence analysis of beta tubulin gene in a wide range of dermatophytes, Medical Mycology, Volume 52, Issue 7, October 2014, Pages 674–688, https://doi.org/10.1093/mmy/myu033
Close - Share Icon Share
Abstract
We investigated the resolving power of the beta tubulin protein-coding gene (BT2) for systematic study of dermatophyte fungi. Initially, 144 standard and clinical strains belonging to 26 species in the genera Trichophyton, Microsporum, and Epidermophyton were identified by internal transcribe spacer (ITS) sequencing. Subsequently, BT2 was partially amplified in all strains, and sequence analysis performed after construction of a BT2 database that showed length ranged from approximately 723 (T. ajelloi) to 808 nucleotides (M. persicolor) in different species. Intraspecific sequence variation was found in some species, but T. tonsurans, T. equinum, T. concentricum, T. verrucosum, T. rubrum, T. violaceum, T. eriotrephon, E. floccosum, M. canis, M. ferrugineum, and M. audouinii were invariant. The sequences were found to be relatively conserved among different strains of the same species. The species with the closest resemblance were Arthroderma benhamiae and T. concentricum and T. tonsurans and T. equinum with 100% and 99.8% identity, respectively; the most distant species were M. persicolor and M. amazonicum. The dendrogram obtained from BT2 topology was almost compatible with the species concept based on ITS sequencing, and similar clades and species were distinguished in the BT2 tree. Here, beta tubulin was characterized in a wide range of dermatophytes in order to assess intra- and interspecies variation and resolution and was found to be a taxonomically valuable gene.
Introduction
Dermatophytes are currently classified in the genera Trichophyton, Microsporum, and Epidermophyton and are ecologically categorized as geophilic, zoophilic, or anthropophilic. About 30 of the known 40 species of dermatophytes can be listed as potential pathogens of humans and other mammals [1]. They have the ability to degrade keratin of hair, nails, and feathers; can produce superficial infections (dermatophytosis) of the skin of healthy human hosts; and can penetrate into deeper tissues in immunocompromised hosts [2]. Dermatophytosis is the most common fungal infection and one of the most common infectious diseases. Scales from the infected skin and nails transmit Anthropophilic species. In recent years, the incidence of nail dermatophytosis (onychomycosis) has increased and now accounts for up to 90% of toenail and 50% of fingernail disorders [3]. In industrialized nations, about 20% of the population aged >60 years and 50% of the population aged >70 years carry a fungal infection of the foot [4,5]. Treatment of onychomycosis imposes $43 million annually to the US healthcare system [6].
Identification of a dermatophyte to the species level is epidemiologically, ecologically, and therapeutically significant because of routes of infection (eg, M. canis infection originates from cats and dogs, while M. audouinii is transmitted between humans) and different treatment regimens (eg, tinea capitis caused by T. tonsurans requires shorter treatment than that caused by M. canis because the latter escapes drug exposure by producing arthroconidia outside the hair shaft [7]. Furthermore, there is a variable profile of in vitro antifungal susceptibility of microconidia and arthroconidia of T. rubrum, T. tonsurans, and T. equinum isolates [8–10].
Prior to the development of reliable molecular diagnostics, an accurate taxonomy of individual species was compulsory. During the last decade, progress has been made in the modern systematics of dermatophytes, nevertheless some pending issues remain. There are problems with accurate definition and characterization of dermatophytes that are not only relevant to phylogenetic analysis and taxonomic identity but also to clinical microbiology and epidemiology; one of these is the definition of species belonging to the T. mentagrophytes complex [11]. Different concepts of species based on phenotypical (morphological), ecological, and biological data have been compared with phylogeny [12]. Classic characteristics may be not only unstable or/and imprecise but also the genetic baseline for some phenotypic concepts may be unclear [7,12,13].
Phylogenetic analyses and the use of phylogenetic species concepts based on rDNA internal transcribe spacer (ITS) regions have improved the taxonomy, but confirmation and refinement using other genes are overdue [7]. Therefore, characterization of new genetic markers for dermatophyte group is needed. Beta tubulin is a monomeric globular protein involved in the generation of microfilaments; it has been used successfully for species delineation in other groups of fungi such as Aspergillus, Penicillium, Scedosporium, and Phaeoacremonium [14–17]. The locus includes some introns, which are known to be good estimators for distinction of closely related species.
Since the beta tubulin gene has not systematically been used in dermatophyte taxonomy, our aim was to investigate the beta tubulin protein-coding gene (BT2) as a new genetic marker in order to evaluate its intra- and interspecies variation in dermatophytes. Several reference strains and clinical isolates, including a wide range of common and rare pathogenic species, were used for this purpose.
Materials and methods
Strains and isolates
A total of 144 strains consisting of 26 species of dermatophytes were used in sequence analysis of partial beta tubulin, comprising 109 reference and 35 clinical isolates (Table 1). The reference strains were obtained from the Centraalbureau voor Schimmelcultures (CBS), Utrecht, the Netherlands, and the Teikyo University Institute of Medical Mycology (TIMM), Tokyo, Japan. The clinical isolates were recovered from a variety of specimens, including skin, nail, and hair, with proven dermatophytosis submitted to two medical mycology laboratories in Tehran, Iran. Species names used in this study were in concordance with the molecular-based taxonomy introduced by Gräser et al. [7].
Reference and clinical strains of dermatophytes used in this study for sequence analysis of partial beta tubulin gene.
| Species . | Strain . | Internal transcribe spacer . | BT2 accession no. . | Size of BT2 partial . |
|---|---|---|---|---|
| . | . | accession no. . | . | sequence (bp) . |
| Arthroderma vanbreuseghemii | JCM 1891 | JN134094 | JF731120 | 789 |
| Trichophyton interdigitale | NBRC 5974 | JN134103 | JF731087 | 790 |
| NBRC 5809 | JN134101 | JF731085 | 792 | |
| NBRC 5812 | JN134102 | JF731086 | 790 | |
| NBRC 5466 | JN134100 | JF731084 | 790 | |
| CBS 102.68 | - | - | 790 | |
| CBS 116916 | - | - | 788 | |
| CBS 119445 | - | - | 789 | |
| CBS 130816 | JN134009 | JF731046 | 792 | |
| CBS 130824 | JN134010 | JF731085 | 792 | |
| CBS 130930 | JN134015 | JF731049 | 792 | |
| CBS 130923 | JN134017 | JF731051 | 792 | |
| CBS 130810 | JN134021 | JF731052 | 790 | |
| CBS 130940 | JN134022 | JF731053 | 790 | |
| CBS 130928 | JN134024 | JF731079 | 792 | |
| CS 248 | JN133983 | JF731075 | 790 | |
| CS 365 | JN133985 | JF731076 | 790 | |
| CS 407 | JN133989 | JF731077 | 790 | |
| CS 437 | JN133991 | JF731043 | 790 | |
| CS 490 | JN133999 | JF731044 | 790 | |
| CS 558 | JN134005 | JF731045 | 790 | |
| CS 559 | JN134006 | JF731078 | 790 | |
| CS 584 | JN134011 | JF731048 | 790 | |
| CS 655 | JN134016 | JF731050 | 790 | |
| CS 759 | JN134023- | JF731054 | 792 | |
| CS 781 | - | JF731055 | 790 | |
| T. tonsurans | NBRC 5928 | JN134086 | JF731073 | 790 |
| NBRC 5945 | JN134087 | JF731074 | 790 | |
| CBS 130924 | JN134080 | JF731068 | 790 | |
| CBS 130822 | JN134085 | JF731071 | 790 | |
| CBS 238.33 | - | - | 790 | |
| CS 768 | JN134084 | JF731070 | 790 | |
| CS 782 | - | JF731072 | 790 | |
| T. equinum | NBRC 31610 | JN134108 | JF731092 | 790 |
| CBS 270.66 | - | - | 790 | |
| CBS 634.82 | - | - | 790 | |
| CBS 100080 | EF043273 | - | 790 | |
| T. mentagrophytes | CBS 106.67 | Z98000 | - | 796 |
| CBS 764.84 | - | - | 797 | |
| CBS 318.56 | Z97995 | - | 796 | |
| T. schoenleinii | NBRC 8191 | JN134098 | JF731082 | 798 |
| NBRC 8192 | JN134099 | JF731083 | 798 | |
| CBS 130812 | JN134097 | JF731081 | 797 | |
| CBS 855.71 | Z98011 | - | 797 | |
| CS 20 | JN134096 | JF731080 | 797 | |
| T. simii | CBS 417.65 | NR-103706 | - | 797 |
| CBS 448.65 | - | - | 796 | |
| CBS 150.66 | - | - | 794 | |
| T. rubrum | NBRC 5467 | JN134067 | JF731066 | 794 |
| NBRC 5808 | JN134068 | JF731067 | 794 | |
| CBS 360.62 | - | - | 794 | |
| CBS 734.88 | - | - | 794 | |
| CBS 100237 | - | - | 794 | |
| CBS 130927 | JN134027 | JF731056 | 794 | |
| CBS 130825 | JN134042 | JF731058 | 794 | |
| CBS 130817 | JN134058 | JF731060 | 794 | |
| CBS 130808 | JN134060 | JF731061 | 794 | |
| CBS 130933 | JN134063 | JF731063 | 794 | |
| CBS 130925 | JN134064 | JF731064 | 794 | |
| CBS 130821 | JN134065 | JF731065 | 794 | |
| CS 329 | JN134037 | JF731057 | 794 | |
| CS 534 | JN134048 | JF731059 | 794 | |
| CS 669 | JN134061 | JF731062 | 794 | |
| T. violaceum | NBRC 31064 | JN134106 | JF731090 | 794 |
| CBS 130937 | JN134104 | JF731088 | 794 | |
| CS 352 | JN134105 | JF731089 | 794 | |
| T. concentricum | CBS 109404 | - | - | 797 |
| CBS 109405 | - | - | 797 | |
| CBS 109407 | - | - | 797 | |
| CBS 173.40 | - | 797 | ||
| T. verrucosum | CBS 130944 | - | - | |
| CBS 130945 | - | - | ||
| CBS 130947 | - | - | ||
| T. erinacei | CS 379 | JN134091 | JF731117 | 796 |
| CBS 511.73 | JX122302 | - | 797 | |
| CBS 344.79 | Z97996 | - | 797 | |
| A. benhamiae | JCM 1885 | JN134093 | JF731118 | 797 |
| (Trichophyton sp. of A. benhamiae) | CS 269 | JN134088 | JF731114- | 797 |
| CBS 809.72 | - | - | 798 | |
| CBS 623.66 | Z98015 | - | 798 | |
| CBS 280.83 | Z98016 | 798 | ||
| T. eriotrephon | CS 361 | JN134089 | - | 798 |
| CS 363 | JN134090 | - | 798 | |
| T. ajelloi | IFO 31978 | JN134092 | JF731119 | 723 |
| CBS 130926 | JN134107 | JF731091 | 723 | |
| Microsporum cookei | NBRC 7862 | JN134140 | JF731110 | 770 |
| SS 779 | JN134139 | JF731109 | 770 | |
| CBS 129.67 | HQ825142 | - | 769 | |
| CBS 202.66 | DQ860714 | - | 769 | |
| CBS 337.74 | HQ825141 | - | 774 | |
| M. racemosum | CBS 130935 | JN134142 | JF731108 | 771 |
| CBS 423.74 | HQ825139 | - | 770 | |
| CBS 102175 | HQ825138 | - | 770 | |
| M. canis | NBRC 9182 | JN134129 | JF731103 | 770 |
| CBS 566.80 | - | - | 770 | |
| CBS 132.88 | - | - | 770 | |
| CBS 277.62 | - | - | 770 | |
| CBS 130811 | JN134125 | JF731099 | 770 | |
| CBS 130931 | JN134126 | JF731100 | 770 | |
| CS 674 | JN134128 | JF731101 | 770 | |
| CS 730 | - | JF731102 | 770 | |
| M. ferrugineum | NBRC 5831 | JN134137 | JF731106 | 770 |
| NBRC 6081 | JN134138 | JF731107 | 770 | |
| CBS 457.80 | - | - | 770 | |
| CBS 131111 | JN134136 | JF731105 | 770 | |
| CBS 427.63 | AJ252336 | - | 770 | |
| CBS 457.80 | AJ252335 | - | 770 | |
| CBS 118546 | - | - | 770 | |
| CBS 118547 | - | - | 770 | |
| CS 650 | JN134135 | JF731104 | 770 | |
| M. audouinii | NBRC 6074 | JN134145 | JF731113 | 771 |
| CBS 280.63 | - | - | 771 | |
| CBS 332.68 | - | - | 771 | |
| CBS 119448 | - | - | 771 | |
| M. nanum | JCM 1907 | JN134095 | JF731121 | 797 |
| CBS 321.61 | DQ860795 | - | 797 | |
| CBS 322.61 | - | - | 797 | |
| CBS 569.80 | DQ860796 | - | 796 | |
| CBS 633.82 | - | - | 796 | |
| CBS 728.88 | - | - | 796 | |
| M. persicolor | NBRC 5975 | JN134144 | JF731112 | 808 |
| CBS 421.74 | - | - | 808 | |
| CBS 422.74 | - | - | 808 | |
| CBS 871.70 | - | - | 808 | |
| M. gypseum (A. gypseum) | IFO 5948 | JN134133 | JF731097 | 798 |
| IFO 8228 | JN134134 | JF731098 | 798 | |
| CBS 130820 | JN134130 | JF731093 | 798 | |
| CBS 130936 | JN134131 | JF731094 | 798 | |
| CBS 130939 | JN134132 | JF731095 | 798 | |
| CBS 424.66 | - | - | 797 | |
| CS 718 | - | JF731096 | 798 | |
| M. gypseum (A. incurvatum) | CBS 548.82 | - | - | 793 |
| M. fulvum | CBS 599.66 | - | - | |
| CBS 287.55 | EF078472 | - | ||
| SS 672 | JN134143 | - | ||
| M. gallinae | CS 778 | JN134141 | JF731111 | 775 |
| Epidermophyton floccosum | CBS 767.73 | - | - | 789 |
| NBRC 9045 | JN134157 | JF731127 | 789 | |
| CS 657 | JN134154 | JF731122 | 789 | |
| CS 725 | - | JF731123 | 789 | |
| CS 739 | JN134155 | JF731124 | 789 | |
| CS 750 | JN134156 | JF731125 | 789 | |
| CS 752 | - | JF731126 | 789 | |
| M. amazonicum | CBS 222.75 | - | - | 762 |
| Species . | Strain . | Internal transcribe spacer . | BT2 accession no. . | Size of BT2 partial . |
|---|---|---|---|---|
| . | . | accession no. . | . | sequence (bp) . |
| Arthroderma vanbreuseghemii | JCM 1891 | JN134094 | JF731120 | 789 |
| Trichophyton interdigitale | NBRC 5974 | JN134103 | JF731087 | 790 |
| NBRC 5809 | JN134101 | JF731085 | 792 | |
| NBRC 5812 | JN134102 | JF731086 | 790 | |
| NBRC 5466 | JN134100 | JF731084 | 790 | |
| CBS 102.68 | - | - | 790 | |
| CBS 116916 | - | - | 788 | |
| CBS 119445 | - | - | 789 | |
| CBS 130816 | JN134009 | JF731046 | 792 | |
| CBS 130824 | JN134010 | JF731085 | 792 | |
| CBS 130930 | JN134015 | JF731049 | 792 | |
| CBS 130923 | JN134017 | JF731051 | 792 | |
| CBS 130810 | JN134021 | JF731052 | 790 | |
| CBS 130940 | JN134022 | JF731053 | 790 | |
| CBS 130928 | JN134024 | JF731079 | 792 | |
| CS 248 | JN133983 | JF731075 | 790 | |
| CS 365 | JN133985 | JF731076 | 790 | |
| CS 407 | JN133989 | JF731077 | 790 | |
| CS 437 | JN133991 | JF731043 | 790 | |
| CS 490 | JN133999 | JF731044 | 790 | |
| CS 558 | JN134005 | JF731045 | 790 | |
| CS 559 | JN134006 | JF731078 | 790 | |
| CS 584 | JN134011 | JF731048 | 790 | |
| CS 655 | JN134016 | JF731050 | 790 | |
| CS 759 | JN134023- | JF731054 | 792 | |
| CS 781 | - | JF731055 | 790 | |
| T. tonsurans | NBRC 5928 | JN134086 | JF731073 | 790 |
| NBRC 5945 | JN134087 | JF731074 | 790 | |
| CBS 130924 | JN134080 | JF731068 | 790 | |
| CBS 130822 | JN134085 | JF731071 | 790 | |
| CBS 238.33 | - | - | 790 | |
| CS 768 | JN134084 | JF731070 | 790 | |
| CS 782 | - | JF731072 | 790 | |
| T. equinum | NBRC 31610 | JN134108 | JF731092 | 790 |
| CBS 270.66 | - | - | 790 | |
| CBS 634.82 | - | - | 790 | |
| CBS 100080 | EF043273 | - | 790 | |
| T. mentagrophytes | CBS 106.67 | Z98000 | - | 796 |
| CBS 764.84 | - | - | 797 | |
| CBS 318.56 | Z97995 | - | 796 | |
| T. schoenleinii | NBRC 8191 | JN134098 | JF731082 | 798 |
| NBRC 8192 | JN134099 | JF731083 | 798 | |
| CBS 130812 | JN134097 | JF731081 | 797 | |
| CBS 855.71 | Z98011 | - | 797 | |
| CS 20 | JN134096 | JF731080 | 797 | |
| T. simii | CBS 417.65 | NR-103706 | - | 797 |
| CBS 448.65 | - | - | 796 | |
| CBS 150.66 | - | - | 794 | |
| T. rubrum | NBRC 5467 | JN134067 | JF731066 | 794 |
| NBRC 5808 | JN134068 | JF731067 | 794 | |
| CBS 360.62 | - | - | 794 | |
| CBS 734.88 | - | - | 794 | |
| CBS 100237 | - | - | 794 | |
| CBS 130927 | JN134027 | JF731056 | 794 | |
| CBS 130825 | JN134042 | JF731058 | 794 | |
| CBS 130817 | JN134058 | JF731060 | 794 | |
| CBS 130808 | JN134060 | JF731061 | 794 | |
| CBS 130933 | JN134063 | JF731063 | 794 | |
| CBS 130925 | JN134064 | JF731064 | 794 | |
| CBS 130821 | JN134065 | JF731065 | 794 | |
| CS 329 | JN134037 | JF731057 | 794 | |
| CS 534 | JN134048 | JF731059 | 794 | |
| CS 669 | JN134061 | JF731062 | 794 | |
| T. violaceum | NBRC 31064 | JN134106 | JF731090 | 794 |
| CBS 130937 | JN134104 | JF731088 | 794 | |
| CS 352 | JN134105 | JF731089 | 794 | |
| T. concentricum | CBS 109404 | - | - | 797 |
| CBS 109405 | - | - | 797 | |
| CBS 109407 | - | - | 797 | |
| CBS 173.40 | - | 797 | ||
| T. verrucosum | CBS 130944 | - | - | |
| CBS 130945 | - | - | ||
| CBS 130947 | - | - | ||
| T. erinacei | CS 379 | JN134091 | JF731117 | 796 |
| CBS 511.73 | JX122302 | - | 797 | |
| CBS 344.79 | Z97996 | - | 797 | |
| A. benhamiae | JCM 1885 | JN134093 | JF731118 | 797 |
| (Trichophyton sp. of A. benhamiae) | CS 269 | JN134088 | JF731114- | 797 |
| CBS 809.72 | - | - | 798 | |
| CBS 623.66 | Z98015 | - | 798 | |
| CBS 280.83 | Z98016 | 798 | ||
| T. eriotrephon | CS 361 | JN134089 | - | 798 |
| CS 363 | JN134090 | - | 798 | |
| T. ajelloi | IFO 31978 | JN134092 | JF731119 | 723 |
| CBS 130926 | JN134107 | JF731091 | 723 | |
| Microsporum cookei | NBRC 7862 | JN134140 | JF731110 | 770 |
| SS 779 | JN134139 | JF731109 | 770 | |
| CBS 129.67 | HQ825142 | - | 769 | |
| CBS 202.66 | DQ860714 | - | 769 | |
| CBS 337.74 | HQ825141 | - | 774 | |
| M. racemosum | CBS 130935 | JN134142 | JF731108 | 771 |
| CBS 423.74 | HQ825139 | - | 770 | |
| CBS 102175 | HQ825138 | - | 770 | |
| M. canis | NBRC 9182 | JN134129 | JF731103 | 770 |
| CBS 566.80 | - | - | 770 | |
| CBS 132.88 | - | - | 770 | |
| CBS 277.62 | - | - | 770 | |
| CBS 130811 | JN134125 | JF731099 | 770 | |
| CBS 130931 | JN134126 | JF731100 | 770 | |
| CS 674 | JN134128 | JF731101 | 770 | |
| CS 730 | - | JF731102 | 770 | |
| M. ferrugineum | NBRC 5831 | JN134137 | JF731106 | 770 |
| NBRC 6081 | JN134138 | JF731107 | 770 | |
| CBS 457.80 | - | - | 770 | |
| CBS 131111 | JN134136 | JF731105 | 770 | |
| CBS 427.63 | AJ252336 | - | 770 | |
| CBS 457.80 | AJ252335 | - | 770 | |
| CBS 118546 | - | - | 770 | |
| CBS 118547 | - | - | 770 | |
| CS 650 | JN134135 | JF731104 | 770 | |
| M. audouinii | NBRC 6074 | JN134145 | JF731113 | 771 |
| CBS 280.63 | - | - | 771 | |
| CBS 332.68 | - | - | 771 | |
| CBS 119448 | - | - | 771 | |
| M. nanum | JCM 1907 | JN134095 | JF731121 | 797 |
| CBS 321.61 | DQ860795 | - | 797 | |
| CBS 322.61 | - | - | 797 | |
| CBS 569.80 | DQ860796 | - | 796 | |
| CBS 633.82 | - | - | 796 | |
| CBS 728.88 | - | - | 796 | |
| M. persicolor | NBRC 5975 | JN134144 | JF731112 | 808 |
| CBS 421.74 | - | - | 808 | |
| CBS 422.74 | - | - | 808 | |
| CBS 871.70 | - | - | 808 | |
| M. gypseum (A. gypseum) | IFO 5948 | JN134133 | JF731097 | 798 |
| IFO 8228 | JN134134 | JF731098 | 798 | |
| CBS 130820 | JN134130 | JF731093 | 798 | |
| CBS 130936 | JN134131 | JF731094 | 798 | |
| CBS 130939 | JN134132 | JF731095 | 798 | |
| CBS 424.66 | - | - | 797 | |
| CS 718 | - | JF731096 | 798 | |
| M. gypseum (A. incurvatum) | CBS 548.82 | - | - | 793 |
| M. fulvum | CBS 599.66 | - | - | |
| CBS 287.55 | EF078472 | - | ||
| SS 672 | JN134143 | - | ||
| M. gallinae | CS 778 | JN134141 | JF731111 | 775 |
| Epidermophyton floccosum | CBS 767.73 | - | - | 789 |
| NBRC 9045 | JN134157 | JF731127 | 789 | |
| CS 657 | JN134154 | JF731122 | 789 | |
| CS 725 | - | JF731123 | 789 | |
| CS 739 | JN134155 | JF731124 | 789 | |
| CS 750 | JN134156 | JF731125 | 789 | |
| CS 752 | - | JF731126 | 789 | |
| M. amazonicum | CBS 222.75 | - | - | 762 |
All GenBank accession numbers listed belong to sequences achieved in the study.
BT2, beta tubulin protein-coding gene; CBS, Centraalbureau voor Schimmelcultures; CS, clinical strain; JCM, Japan Collection of Microorganisms; NBRC, NITE Biological Resource Center; SS, soil strain
Reference and clinical strains of dermatophytes used in this study for sequence analysis of partial beta tubulin gene.
| Species . | Strain . | Internal transcribe spacer . | BT2 accession no. . | Size of BT2 partial . |
|---|---|---|---|---|
| . | . | accession no. . | . | sequence (bp) . |
| Arthroderma vanbreuseghemii | JCM 1891 | JN134094 | JF731120 | 789 |
| Trichophyton interdigitale | NBRC 5974 | JN134103 | JF731087 | 790 |
| NBRC 5809 | JN134101 | JF731085 | 792 | |
| NBRC 5812 | JN134102 | JF731086 | 790 | |
| NBRC 5466 | JN134100 | JF731084 | 790 | |
| CBS 102.68 | - | - | 790 | |
| CBS 116916 | - | - | 788 | |
| CBS 119445 | - | - | 789 | |
| CBS 130816 | JN134009 | JF731046 | 792 | |
| CBS 130824 | JN134010 | JF731085 | 792 | |
| CBS 130930 | JN134015 | JF731049 | 792 | |
| CBS 130923 | JN134017 | JF731051 | 792 | |
| CBS 130810 | JN134021 | JF731052 | 790 | |
| CBS 130940 | JN134022 | JF731053 | 790 | |
| CBS 130928 | JN134024 | JF731079 | 792 | |
| CS 248 | JN133983 | JF731075 | 790 | |
| CS 365 | JN133985 | JF731076 | 790 | |
| CS 407 | JN133989 | JF731077 | 790 | |
| CS 437 | JN133991 | JF731043 | 790 | |
| CS 490 | JN133999 | JF731044 | 790 | |
| CS 558 | JN134005 | JF731045 | 790 | |
| CS 559 | JN134006 | JF731078 | 790 | |
| CS 584 | JN134011 | JF731048 | 790 | |
| CS 655 | JN134016 | JF731050 | 790 | |
| CS 759 | JN134023- | JF731054 | 792 | |
| CS 781 | - | JF731055 | 790 | |
| T. tonsurans | NBRC 5928 | JN134086 | JF731073 | 790 |
| NBRC 5945 | JN134087 | JF731074 | 790 | |
| CBS 130924 | JN134080 | JF731068 | 790 | |
| CBS 130822 | JN134085 | JF731071 | 790 | |
| CBS 238.33 | - | - | 790 | |
| CS 768 | JN134084 | JF731070 | 790 | |
| CS 782 | - | JF731072 | 790 | |
| T. equinum | NBRC 31610 | JN134108 | JF731092 | 790 |
| CBS 270.66 | - | - | 790 | |
| CBS 634.82 | - | - | 790 | |
| CBS 100080 | EF043273 | - | 790 | |
| T. mentagrophytes | CBS 106.67 | Z98000 | - | 796 |
| CBS 764.84 | - | - | 797 | |
| CBS 318.56 | Z97995 | - | 796 | |
| T. schoenleinii | NBRC 8191 | JN134098 | JF731082 | 798 |
| NBRC 8192 | JN134099 | JF731083 | 798 | |
| CBS 130812 | JN134097 | JF731081 | 797 | |
| CBS 855.71 | Z98011 | - | 797 | |
| CS 20 | JN134096 | JF731080 | 797 | |
| T. simii | CBS 417.65 | NR-103706 | - | 797 |
| CBS 448.65 | - | - | 796 | |
| CBS 150.66 | - | - | 794 | |
| T. rubrum | NBRC 5467 | JN134067 | JF731066 | 794 |
| NBRC 5808 | JN134068 | JF731067 | 794 | |
| CBS 360.62 | - | - | 794 | |
| CBS 734.88 | - | - | 794 | |
| CBS 100237 | - | - | 794 | |
| CBS 130927 | JN134027 | JF731056 | 794 | |
| CBS 130825 | JN134042 | JF731058 | 794 | |
| CBS 130817 | JN134058 | JF731060 | 794 | |
| CBS 130808 | JN134060 | JF731061 | 794 | |
| CBS 130933 | JN134063 | JF731063 | 794 | |
| CBS 130925 | JN134064 | JF731064 | 794 | |
| CBS 130821 | JN134065 | JF731065 | 794 | |
| CS 329 | JN134037 | JF731057 | 794 | |
| CS 534 | JN134048 | JF731059 | 794 | |
| CS 669 | JN134061 | JF731062 | 794 | |
| T. violaceum | NBRC 31064 | JN134106 | JF731090 | 794 |
| CBS 130937 | JN134104 | JF731088 | 794 | |
| CS 352 | JN134105 | JF731089 | 794 | |
| T. concentricum | CBS 109404 | - | - | 797 |
| CBS 109405 | - | - | 797 | |
| CBS 109407 | - | - | 797 | |
| CBS 173.40 | - | 797 | ||
| T. verrucosum | CBS 130944 | - | - | |
| CBS 130945 | - | - | ||
| CBS 130947 | - | - | ||
| T. erinacei | CS 379 | JN134091 | JF731117 | 796 |
| CBS 511.73 | JX122302 | - | 797 | |
| CBS 344.79 | Z97996 | - | 797 | |
| A. benhamiae | JCM 1885 | JN134093 | JF731118 | 797 |
| (Trichophyton sp. of A. benhamiae) | CS 269 | JN134088 | JF731114- | 797 |
| CBS 809.72 | - | - | 798 | |
| CBS 623.66 | Z98015 | - | 798 | |
| CBS 280.83 | Z98016 | 798 | ||
| T. eriotrephon | CS 361 | JN134089 | - | 798 |
| CS 363 | JN134090 | - | 798 | |
| T. ajelloi | IFO 31978 | JN134092 | JF731119 | 723 |
| CBS 130926 | JN134107 | JF731091 | 723 | |
| Microsporum cookei | NBRC 7862 | JN134140 | JF731110 | 770 |
| SS 779 | JN134139 | JF731109 | 770 | |
| CBS 129.67 | HQ825142 | - | 769 | |
| CBS 202.66 | DQ860714 | - | 769 | |
| CBS 337.74 | HQ825141 | - | 774 | |
| M. racemosum | CBS 130935 | JN134142 | JF731108 | 771 |
| CBS 423.74 | HQ825139 | - | 770 | |
| CBS 102175 | HQ825138 | - | 770 | |
| M. canis | NBRC 9182 | JN134129 | JF731103 | 770 |
| CBS 566.80 | - | - | 770 | |
| CBS 132.88 | - | - | 770 | |
| CBS 277.62 | - | - | 770 | |
| CBS 130811 | JN134125 | JF731099 | 770 | |
| CBS 130931 | JN134126 | JF731100 | 770 | |
| CS 674 | JN134128 | JF731101 | 770 | |
| CS 730 | - | JF731102 | 770 | |
| M. ferrugineum | NBRC 5831 | JN134137 | JF731106 | 770 |
| NBRC 6081 | JN134138 | JF731107 | 770 | |
| CBS 457.80 | - | - | 770 | |
| CBS 131111 | JN134136 | JF731105 | 770 | |
| CBS 427.63 | AJ252336 | - | 770 | |
| CBS 457.80 | AJ252335 | - | 770 | |
| CBS 118546 | - | - | 770 | |
| CBS 118547 | - | - | 770 | |
| CS 650 | JN134135 | JF731104 | 770 | |
| M. audouinii | NBRC 6074 | JN134145 | JF731113 | 771 |
| CBS 280.63 | - | - | 771 | |
| CBS 332.68 | - | - | 771 | |
| CBS 119448 | - | - | 771 | |
| M. nanum | JCM 1907 | JN134095 | JF731121 | 797 |
| CBS 321.61 | DQ860795 | - | 797 | |
| CBS 322.61 | - | - | 797 | |
| CBS 569.80 | DQ860796 | - | 796 | |
| CBS 633.82 | - | - | 796 | |
| CBS 728.88 | - | - | 796 | |
| M. persicolor | NBRC 5975 | JN134144 | JF731112 | 808 |
| CBS 421.74 | - | - | 808 | |
| CBS 422.74 | - | - | 808 | |
| CBS 871.70 | - | - | 808 | |
| M. gypseum (A. gypseum) | IFO 5948 | JN134133 | JF731097 | 798 |
| IFO 8228 | JN134134 | JF731098 | 798 | |
| CBS 130820 | JN134130 | JF731093 | 798 | |
| CBS 130936 | JN134131 | JF731094 | 798 | |
| CBS 130939 | JN134132 | JF731095 | 798 | |
| CBS 424.66 | - | - | 797 | |
| CS 718 | - | JF731096 | 798 | |
| M. gypseum (A. incurvatum) | CBS 548.82 | - | - | 793 |
| M. fulvum | CBS 599.66 | - | - | |
| CBS 287.55 | EF078472 | - | ||
| SS 672 | JN134143 | - | ||
| M. gallinae | CS 778 | JN134141 | JF731111 | 775 |
| Epidermophyton floccosum | CBS 767.73 | - | - | 789 |
| NBRC 9045 | JN134157 | JF731127 | 789 | |
| CS 657 | JN134154 | JF731122 | 789 | |
| CS 725 | - | JF731123 | 789 | |
| CS 739 | JN134155 | JF731124 | 789 | |
| CS 750 | JN134156 | JF731125 | 789 | |
| CS 752 | - | JF731126 | 789 | |
| M. amazonicum | CBS 222.75 | - | - | 762 |
| Species . | Strain . | Internal transcribe spacer . | BT2 accession no. . | Size of BT2 partial . |
|---|---|---|---|---|
| . | . | accession no. . | . | sequence (bp) . |
| Arthroderma vanbreuseghemii | JCM 1891 | JN134094 | JF731120 | 789 |
| Trichophyton interdigitale | NBRC 5974 | JN134103 | JF731087 | 790 |
| NBRC 5809 | JN134101 | JF731085 | 792 | |
| NBRC 5812 | JN134102 | JF731086 | 790 | |
| NBRC 5466 | JN134100 | JF731084 | 790 | |
| CBS 102.68 | - | - | 790 | |
| CBS 116916 | - | - | 788 | |
| CBS 119445 | - | - | 789 | |
| CBS 130816 | JN134009 | JF731046 | 792 | |
| CBS 130824 | JN134010 | JF731085 | 792 | |
| CBS 130930 | JN134015 | JF731049 | 792 | |
| CBS 130923 | JN134017 | JF731051 | 792 | |
| CBS 130810 | JN134021 | JF731052 | 790 | |
| CBS 130940 | JN134022 | JF731053 | 790 | |
| CBS 130928 | JN134024 | JF731079 | 792 | |
| CS 248 | JN133983 | JF731075 | 790 | |
| CS 365 | JN133985 | JF731076 | 790 | |
| CS 407 | JN133989 | JF731077 | 790 | |
| CS 437 | JN133991 | JF731043 | 790 | |
| CS 490 | JN133999 | JF731044 | 790 | |
| CS 558 | JN134005 | JF731045 | 790 | |
| CS 559 | JN134006 | JF731078 | 790 | |
| CS 584 | JN134011 | JF731048 | 790 | |
| CS 655 | JN134016 | JF731050 | 790 | |
| CS 759 | JN134023- | JF731054 | 792 | |
| CS 781 | - | JF731055 | 790 | |
| T. tonsurans | NBRC 5928 | JN134086 | JF731073 | 790 |
| NBRC 5945 | JN134087 | JF731074 | 790 | |
| CBS 130924 | JN134080 | JF731068 | 790 | |
| CBS 130822 | JN134085 | JF731071 | 790 | |
| CBS 238.33 | - | - | 790 | |
| CS 768 | JN134084 | JF731070 | 790 | |
| CS 782 | - | JF731072 | 790 | |
| T. equinum | NBRC 31610 | JN134108 | JF731092 | 790 |
| CBS 270.66 | - | - | 790 | |
| CBS 634.82 | - | - | 790 | |
| CBS 100080 | EF043273 | - | 790 | |
| T. mentagrophytes | CBS 106.67 | Z98000 | - | 796 |
| CBS 764.84 | - | - | 797 | |
| CBS 318.56 | Z97995 | - | 796 | |
| T. schoenleinii | NBRC 8191 | JN134098 | JF731082 | 798 |
| NBRC 8192 | JN134099 | JF731083 | 798 | |
| CBS 130812 | JN134097 | JF731081 | 797 | |
| CBS 855.71 | Z98011 | - | 797 | |
| CS 20 | JN134096 | JF731080 | 797 | |
| T. simii | CBS 417.65 | NR-103706 | - | 797 |
| CBS 448.65 | - | - | 796 | |
| CBS 150.66 | - | - | 794 | |
| T. rubrum | NBRC 5467 | JN134067 | JF731066 | 794 |
| NBRC 5808 | JN134068 | JF731067 | 794 | |
| CBS 360.62 | - | - | 794 | |
| CBS 734.88 | - | - | 794 | |
| CBS 100237 | - | - | 794 | |
| CBS 130927 | JN134027 | JF731056 | 794 | |
| CBS 130825 | JN134042 | JF731058 | 794 | |
| CBS 130817 | JN134058 | JF731060 | 794 | |
| CBS 130808 | JN134060 | JF731061 | 794 | |
| CBS 130933 | JN134063 | JF731063 | 794 | |
| CBS 130925 | JN134064 | JF731064 | 794 | |
| CBS 130821 | JN134065 | JF731065 | 794 | |
| CS 329 | JN134037 | JF731057 | 794 | |
| CS 534 | JN134048 | JF731059 | 794 | |
| CS 669 | JN134061 | JF731062 | 794 | |
| T. violaceum | NBRC 31064 | JN134106 | JF731090 | 794 |
| CBS 130937 | JN134104 | JF731088 | 794 | |
| CS 352 | JN134105 | JF731089 | 794 | |
| T. concentricum | CBS 109404 | - | - | 797 |
| CBS 109405 | - | - | 797 | |
| CBS 109407 | - | - | 797 | |
| CBS 173.40 | - | 797 | ||
| T. verrucosum | CBS 130944 | - | - | |
| CBS 130945 | - | - | ||
| CBS 130947 | - | - | ||
| T. erinacei | CS 379 | JN134091 | JF731117 | 796 |
| CBS 511.73 | JX122302 | - | 797 | |
| CBS 344.79 | Z97996 | - | 797 | |
| A. benhamiae | JCM 1885 | JN134093 | JF731118 | 797 |
| (Trichophyton sp. of A. benhamiae) | CS 269 | JN134088 | JF731114- | 797 |
| CBS 809.72 | - | - | 798 | |
| CBS 623.66 | Z98015 | - | 798 | |
| CBS 280.83 | Z98016 | 798 | ||
| T. eriotrephon | CS 361 | JN134089 | - | 798 |
| CS 363 | JN134090 | - | 798 | |
| T. ajelloi | IFO 31978 | JN134092 | JF731119 | 723 |
| CBS 130926 | JN134107 | JF731091 | 723 | |
| Microsporum cookei | NBRC 7862 | JN134140 | JF731110 | 770 |
| SS 779 | JN134139 | JF731109 | 770 | |
| CBS 129.67 | HQ825142 | - | 769 | |
| CBS 202.66 | DQ860714 | - | 769 | |
| CBS 337.74 | HQ825141 | - | 774 | |
| M. racemosum | CBS 130935 | JN134142 | JF731108 | 771 |
| CBS 423.74 | HQ825139 | - | 770 | |
| CBS 102175 | HQ825138 | - | 770 | |
| M. canis | NBRC 9182 | JN134129 | JF731103 | 770 |
| CBS 566.80 | - | - | 770 | |
| CBS 132.88 | - | - | 770 | |
| CBS 277.62 | - | - | 770 | |
| CBS 130811 | JN134125 | JF731099 | 770 | |
| CBS 130931 | JN134126 | JF731100 | 770 | |
| CS 674 | JN134128 | JF731101 | 770 | |
| CS 730 | - | JF731102 | 770 | |
| M. ferrugineum | NBRC 5831 | JN134137 | JF731106 | 770 |
| NBRC 6081 | JN134138 | JF731107 | 770 | |
| CBS 457.80 | - | - | 770 | |
| CBS 131111 | JN134136 | JF731105 | 770 | |
| CBS 427.63 | AJ252336 | - | 770 | |
| CBS 457.80 | AJ252335 | - | 770 | |
| CBS 118546 | - | - | 770 | |
| CBS 118547 | - | - | 770 | |
| CS 650 | JN134135 | JF731104 | 770 | |
| M. audouinii | NBRC 6074 | JN134145 | JF731113 | 771 |
| CBS 280.63 | - | - | 771 | |
| CBS 332.68 | - | - | 771 | |
| CBS 119448 | - | - | 771 | |
| M. nanum | JCM 1907 | JN134095 | JF731121 | 797 |
| CBS 321.61 | DQ860795 | - | 797 | |
| CBS 322.61 | - | - | 797 | |
| CBS 569.80 | DQ860796 | - | 796 | |
| CBS 633.82 | - | - | 796 | |
| CBS 728.88 | - | - | 796 | |
| M. persicolor | NBRC 5975 | JN134144 | JF731112 | 808 |
| CBS 421.74 | - | - | 808 | |
| CBS 422.74 | - | - | 808 | |
| CBS 871.70 | - | - | 808 | |
| M. gypseum (A. gypseum) | IFO 5948 | JN134133 | JF731097 | 798 |
| IFO 8228 | JN134134 | JF731098 | 798 | |
| CBS 130820 | JN134130 | JF731093 | 798 | |
| CBS 130936 | JN134131 | JF731094 | 798 | |
| CBS 130939 | JN134132 | JF731095 | 798 | |
| CBS 424.66 | - | - | 797 | |
| CS 718 | - | JF731096 | 798 | |
| M. gypseum (A. incurvatum) | CBS 548.82 | - | - | 793 |
| M. fulvum | CBS 599.66 | - | - | |
| CBS 287.55 | EF078472 | - | ||
| SS 672 | JN134143 | - | ||
| M. gallinae | CS 778 | JN134141 | JF731111 | 775 |
| Epidermophyton floccosum | CBS 767.73 | - | - | 789 |
| NBRC 9045 | JN134157 | JF731127 | 789 | |
| CS 657 | JN134154 | JF731122 | 789 | |
| CS 725 | - | JF731123 | 789 | |
| CS 739 | JN134155 | JF731124 | 789 | |
| CS 750 | JN134156 | JF731125 | 789 | |
| CS 752 | - | JF731126 | 789 | |
| M. amazonicum | CBS 222.75 | - | - | 762 |
All GenBank accession numbers listed belong to sequences achieved in the study.
BT2, beta tubulin protein-coding gene; CBS, Centraalbureau voor Schimmelcultures; CS, clinical strain; JCM, Japan Collection of Microorganisms; NBRC, NITE Biological Resource Center; SS, soil strain
DNA extraction
DNA was extracted and purified from fungal colonies using a previously described method [18]. Briefly, 10–20 mm3 of the fresh colonies grown on Sabouraud glucose agar (Difco, Detroit, MI, USA) were added to 1.5-ml tubes that contained 300 μl of lysis buffer (200 M Tris-HCl, pH 7.5; 25 mM ethylenediaminetetraacetic acid (EDTA); 0.5% w/v sodium dodecyl sulfate; and 250 mM NaCl) and crushed with a conical grinder (Micro Multi Mixer; IEDA Co. Ltd., Tokyo, Japan) for 1 min. The samples were incubated in a boiling water bath for 10 min, mixed with 150 μl of 3.0 M sodium acetate, kept at −20°C for 10 min, and centrifuged at 12 000 rpm for 10 min. The supernatant was extracted once with phenol/chloroform/isoamyl alcohol (25:24:1) and once more with chloroform. The DNA in supernatant was precipitated with 250 μl isopropanol, washed with 300 ml of 70% ethanol, air dried, and rehydrated in 50 μl ultrapure water, and stored at −20°C until use.
Polymerase chain reaction
For ITS polymerase chain reaction (PCR), each mixture contained 2.5 μl of 10× reaction buffer, 0.5 μM of the forward ITS1 and the reverse ITS4 universal fungal primers [19], 400 μM of deoxynucleoside triphosphate, 1.25 U of Taq DNA polymerase (Takara, Japan), 1 μl of extracted DNA, and enough ultrapure water to reach a final reaction volume of 25 μl. The PCRs were programmed for preheating at 96°C for 6 min followed by 35 cycles at 94°C for 1 min, 56°C for 1 min, and 72°C for 45 s, and a final extension step at 72°C for 10 min. Five microliters of the PCR products were electrophoresed onto 1.5% agarose gel in Tris-Acetate-EDTA (TAE) buffer (Tris 40 mM, acetic acid 20 mM, EDTA 1 mM), stained with 0.5 μg/ml of ethidium bromide, and observed and photographed under ultraviolet irradiation. Similar PCR conditions were used for amplification of beta tubulin gene except the oligonucleotide primers, which were the universal fungal primers T1 (5′-AACATGCGTGAGATTGTAAGT-3′) [20] as the forward and Bt2b (5′-ACCCTCA-GTGTAGTGACCCTTGGC-3′) [21] as the reverse primer.
Sequencing and sequence analysis
ITS and BT2 PCR products were purified using a QIAquick purification kit (Qiagen, Valencia, CA, USA), subjected to ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA) and sequenced in both directions using the same primers (individually) as for the primary PCR by an automated DNA sequencer (ABI Prism 3730 Genetic Analyzer, Applied Biosystems) and edited with MEGA5 [22] and Geneious (http://www.geneious.com) software. Levels of sequence identity matrix and sequence difference count in each target were calculated pairwise using BioEdit software (http://www.mbio.ncsu.edu/bioedit/bioedit.html) or evaluated using the following formula: D = 1 − (M/L), where M is the number of shared positions and L is the total number of alignment positions compared [23]. The sequences were deposited in GenBank (Table 1). ITS-rDNA was considered to be the molecular model for dermatophytes species designation [7]. Data were compared in GenBank using the BLASTn algorithm, and strain and sequence information provided with the reference strains from CBS (http://www.cbs.knaw.nl/dermatophytes/BioloMICSID.aspx) were evaluated.
Phylogenetic analyses
Multiple alignments of the beta tubulin gene sequences were made using BioEdit or Geneious software. The phylogenetic tree was built using MEGA5 software, and the tree topology was evaluated visually for congruence of species-rank clades with the following conditions: statistical method was maximum likelihood, test of phylogeny was bootstrap method with 1000 replication, substitution model was Tamura–Nei, and there were uniform rates among sites [23]. The bioinformatics data were analyzed and used to assess the inter- and intraspecies nucleotide variation of BT2 in all reference strains and clinical isolates used in this study.
Results
Strain designations and corresponding GenBank accession numbers of 109 reference and 35 clinical isolates representing 26 dermatophyte species used in this study are listed in Table 1. PCR amplification of BT2 was successful for all strains, and a single band with overall length of approximately 750 base pairs (bp) was observed on electrophoresis. Sequencing results showed that length ranged from 723 to 808 nucleotides (nt; Table 1). The smallest size was found in T. ajelloi with 723 nt and the longest in M. persicolor with 808 nt. Some species had identical size, for example, 790 nt for T. tonsurans and T. equinum (Table 1). Despite significant variation in BT2 amplicon length, size polymorphism did not allow species differentiation by gel electrophoresis.
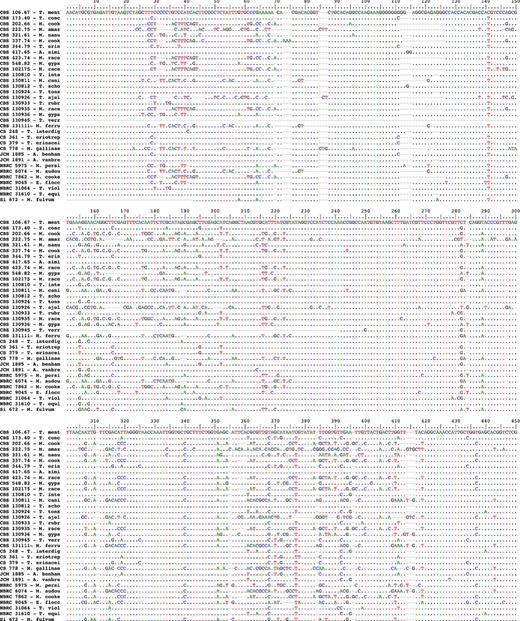
Intraspecific sequence variation was found in some species; however, T. tonsurans, T. equinum, T. concentricum, T. verrucosum, T. rubrum, T. violaceum, T. eriotrephon, E. floccosum, M. canis, M. ferrugineum, and M. audouinii were invariant. Figure 1 shows the multiple DNA sequence alignment of BT2 sequences in the most common pathogenic dermatophytes. The sequence alignment reveals significant divergences, including substitution, insertion/deletion, and gaps throughout the sequences. Nevertheless, confident alignment was possible over the entire dataset using several nearly invariant regions, which could be useful for reliable pan-dermatophyte primer and probe design.
Multiple sequence alignments of the partial beta tubulin gene sequence from various dermatophyte species and strains sequenced in this study. A dot indicates an identical nucleotide with respect to the top sequence; a dash indicates an insertion/deletion (indel) event.
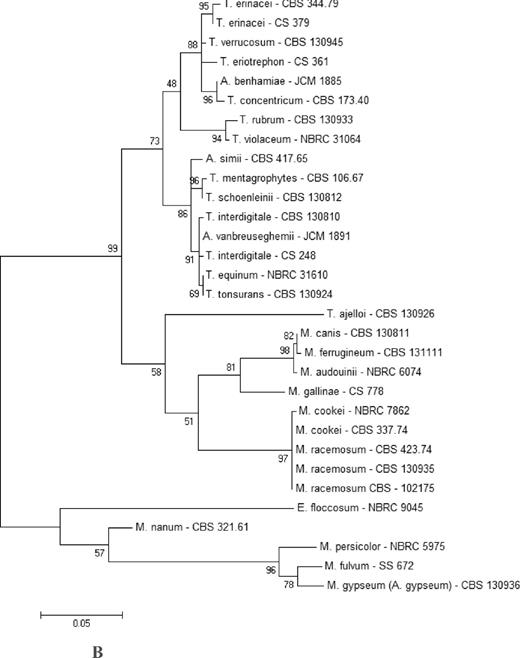
A sequence difference count matrix based on nucleotide pairwise comparison of the BT2 gene between representative dermatophyte species and strains is shown in Table 2. Sequence diversity among 26 dermatophyte species ranged from 0 to 229 nt. The species with the closest resemblance were Arthroderma benhamiae and T. concentricum and T. tonsurans and T. equinum with 100% and 99.8% identity, respectively, and the most distant species were M. persicolor and M. amazonicum. Strains of T. interdigitale (A. vanbreuseghemii) had the most intraspecies variability, 0–14 bp mismatching (98.2%–100% similarity); according to the results, six BT2 genotypes were found within the species. The distance range of BT2 sequence between T. interdigitale strains was 0.2%–1.8%. A low BT2 intrapopulation diversity of 0–2 bp was computed within the strains of M. gypseum with the genotype of A. gypseum. A significant BT2 interspecies diversity, ranging from 58 bp to 60 bp (7.3%–7.5% dissimilarity), was found among seven strains of A. gypseum and the only strain of M. gypseum with A. incurvatum genotype. The intraspecies pairwise distance variations as 1 nt (99.8% identity), 0–3 nt (99.6%–100% similarity), 0–4 nt (99.4%–100% similarity), 0–3 nt (99.6%–100% similarity), and 7–17 nt (99.7%–99%) were computed within the strains of T. ajelloi, M. cookei, M. nanum, M. persicolor, and M. racemosum, respectively. Trees constructed with BT2 and ITS sequences for representative strains of each species are shown in Figure 2. Some closely related species formed well-supported clades in the BT2 tree, such as T. interdigitale, T. tonsurans, and T. equinum (bootstrap value, 94%); A. simii, T. mentagrophytes, and T. schoenleinii (bootstrap value of 81%); T. rubrum and T. violaceum (bootstrap value, 100%); M. cookei and M. racemosum (bootstrap value, 100%); M. audouinii, M. canis, and M. ferrugineum (bootstrap value, 99%); and members of the A. benhamiae complex (bootstrap value, of 93%; Fig. 2A). Others, such as M. persicolor, E. floccosum, A. obtusum, and A. uncinatum, took relatively remote positions. Dermatophyte tree topologies were almost identical in BT2 and ITS (Fig. 1A, 1B).
(A) Phylogenetic tree of 54 representative dermatophyte species and strains (different species and the same species but with at least one nucleotide difference) based on analysis of beta tubulin gene sequences. The evolutionary history was inferred by using the maximum likelihood method based on the Tamura–Nei model. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree for the heuristic search was obtained automatically by applying neighbor–join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood approach and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA5 [22]. (B) A smaller phylogenetic tree of 31 representative dermatophytes species based on analysis of internal transcribe spacer sequences. Note that the position of the complexes and species are almost same in both trees.
Sequences difference count matrix based on pairwise sequence comparison of beta tubulin gene between representative dermatophytes investigated in this study.
| . | Species . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | 13 . | 14 . | 15 . | 16 . | 17 . | 18 . | 19 . | 20 . | 21 . | 22 . | 23 . | 24 . | 25 . | 26 . | 27 . | 28 . | 29 . | 30 . | 31 . | 32 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Trichophyton ment | ID | |||||||||||||||||||||||||||||||
| 2 | T. conce | 36 | ID | ||||||||||||||||||||||||||||||
| 3 | Microsporum cook | 131 | 140 | ID | |||||||||||||||||||||||||||||
| 4 | M. amaz | 217 | 215 | 210 | ID | ||||||||||||||||||||||||||||
| 5 | M. nanu | 90 | 94 | 134 | 218 | ID | |||||||||||||||||||||||||||
| 6 | M. cook | 133 | 142 | 18 | 203 | 133 | ID | ||||||||||||||||||||||||||
| 7 | T. erinac | 40 | 11 | 138 | 212 | 99 | 139 | ID | |||||||||||||||||||||||||
| 8 | Arthroderma simii | 3 | 39 | 134 | 221 | 93 | 136 | 43 | ID | ||||||||||||||||||||||||
| 9 | M. racem | 126 | 135 | 9 | 204 | 130 | 13 | 132 | 129 | ID | |||||||||||||||||||||||
| 10 | A. incur | 82 | 87 | 132 | 214 | 73 | 132 | 87 | 85 | 124 | ID | ||||||||||||||||||||||
| 11 | M. racem | 129 | 138 | 8 | 208 | 134 | 18 | 136 | 132 | 7 | 128 | ID | |||||||||||||||||||||
| 12 | T. interd | 22 | 35 | 132 | 211 | 93 | 134 | 39 | 25 | 127 | 83 | 130 | ID | ||||||||||||||||||||
| 13 | M. canis | 140 | 146 | 151 | 181 | 160 | 150 | 145 | 143 | 151 | 157 | 152 | 147 | ID | |||||||||||||||||||
| 14 | T. schoe | 1 | 37 | 132 | 219 | 91 | 134 | 41 | 2 | 127 | 83 | 130 | 23 | 142 | ID | ||||||||||||||||||
| 15 | T. tonsu | 23 | 36 | 129 | 210 | 96 | 131 | 40 | 26 | 124 | 86 | 127 | 3 | 147 | 24 | ID | |||||||||||||||||
| 16 | T. ajelloi | 126 | 196 | 167 | 145 | 192 | 165 | 196 | 195 | 161 | 182 | 164 | 187 | 167 | 192 | 187 | ID | ||||||||||||||||
| 17 | T. rubru | 31 | 39 | 134 | 209 | 98 | 138 | 42 | 34 | 131 | 87 | 132 | 36 | 149 | 32 | 37 | 195 | ID | |||||||||||||||
| 18 | M. racem | 132 | 137 | 13 | 207 | 137 | 17 | 133 | 135 | 14 | 131 | 17 | 130 | 151 | 133 | 127 | 163 | 136 | ID | ||||||||||||||
| 19 | M. gyps | 88 | 97 | 131 | 224 | 70 | 131 | 98 | 91 | 127 | 60 | 131 | 90 | 156 | 89 | 91 | 196 | 95 | 132 | ID | |||||||||||||
| 20 | M. ferru | 138 | 146 | 148 | 181 | 156 | 147 | 145 | 141 | 146 | 156 | 147 | 144 | 12 | 140 | 143 | 165 | 144 | 145 | 150 | ID | ||||||||||||
| 21 | T. interd | 22 | 41 | 134 | 213 | 100 | 135 | 45 | 27 | 129 | 91 | 132 | 8 | 150 | 23 | 5 | 190 | 42 | 130 | 96 | 148 | ID | |||||||||||
| 22 | T. eriotr | 38 | 11 | 136 | 214 | 91 | 140 | 14 | 41 | 131 | 86 | 132 | 41 | 151 | 39 | 42 | 197 | 42 | 136 | 98 | 151 | 47 | ID | ||||||||||
| 23 | T. erinac | 38 | 9 | 139 | 211 | 98 | 140 | 2 | 41 | 131 | 85 | 135 | 38 | 147 | 39 | 39 | 195 | 39 | 135 | 97 | 147 | 44 | 12 | ID | |||||||||
| 24 | M. gallin | 145 | 148 | 134 | 182 | 150 | 137 | 146 | 148 | 130 | 143 | 134 | 145 | 105 | 146 | 146 | 164 | 144 | 134 | 147 | 101 | 151 | 146 | 146 | ID | ||||||||
| 25 | A. benh | 36 | 0 | 140 | 215 | 95 | 142 | 11 | 40 | 133 | 87 | 136 | 35 | 146 | 38 | 36 | 196 | 40 | 137 | 97 | 146 | 41 | 11 | 9 | 149 | ID | |||||||
| 26 | A. vanbr | 26 | 38 | 130 | 212 | 95 | 135 | 41 | 29 | 128 | 85 | 128 | 8 | 150 | 27 | 9 | 186 | 38 | 130 | 92 | 147 | 12 | 42 | 39 | 142 | 38 | ID | ||||||
| 27 | M. persi | 77 | 84 | 129 | 229 | 69 | 129 | 88 | 80 | 125 | 70 | 129 | 90 | 170 | 78 | 91 | 197 | 88 | 131 | 79 | 162 | 90 | 83 | 86 | 156 | 84 | 94 | ID | |||||
| 28 | M. audo | 138 | 148 | 150 | 184 | 157 | 149 | 144 | 141 | 148 | 154 | 149 | 143 | 14 | 139 | 142 | 169 | 148 | 147 | 149 | 6 | 145 | 151 | 146 | 106 | 148 | 145 | 163 | ID | ||||
| 29 | M. cook | 129 | 139 | 3 | 208 | 132 | 17 | 136 | 132 | 8 | 130 | 7 | 130 | 152 | 130 | 127 | 167 | 133 | 12 | 129 | 147 | 132 | 135 | 139 | 132 | 139 | 128 | 127 | 151 | ID | |||
| 30 | Epidermophyton flocco | 79 | 89 | 126 | 216 | 80 | 123 | 90 | 82 | 119 | 79 | 124 | 94 | 158 | 80 | 95 | 188 | 86 | 126 | 77 | 153 | 100 | 90 | 87 | 139 | 89 | 96 | 78 | 152 | 124 | ID | ||
| 31 | T. violac | 33 | 39 | 136 | 210 | 100 | 140 | 44 | 36 | 133 | 89 | 134 | 38 | 149 | 34 | 39 | 193 | 2 | 136 | 97 | 144 | 44 | 44 | 41 | 146 | 39 | 40 | 90 | 147 | 135 | 88 | ID | |
| 32 | T. equin | 24 | 37 | 130 | 212 | 97 | 132 | 41 | 27 | 125 | 87 | 128 | 4 | 147 | 25 | 1 | 187 | 38 | 127 | 92 | 144 | 4 | 43 | 40 | 145 | 37 | 10 | 92 | 142 | 128 | 96 | 40 | ID |
| . | Species . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | 13 . | 14 . | 15 . | 16 . | 17 . | 18 . | 19 . | 20 . | 21 . | 22 . | 23 . | 24 . | 25 . | 26 . | 27 . | 28 . | 29 . | 30 . | 31 . | 32 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Trichophyton ment | ID | |||||||||||||||||||||||||||||||
| 2 | T. conce | 36 | ID | ||||||||||||||||||||||||||||||
| 3 | Microsporum cook | 131 | 140 | ID | |||||||||||||||||||||||||||||
| 4 | M. amaz | 217 | 215 | 210 | ID | ||||||||||||||||||||||||||||
| 5 | M. nanu | 90 | 94 | 134 | 218 | ID | |||||||||||||||||||||||||||
| 6 | M. cook | 133 | 142 | 18 | 203 | 133 | ID | ||||||||||||||||||||||||||
| 7 | T. erinac | 40 | 11 | 138 | 212 | 99 | 139 | ID | |||||||||||||||||||||||||
| 8 | Arthroderma simii | 3 | 39 | 134 | 221 | 93 | 136 | 43 | ID | ||||||||||||||||||||||||
| 9 | M. racem | 126 | 135 | 9 | 204 | 130 | 13 | 132 | 129 | ID | |||||||||||||||||||||||
| 10 | A. incur | 82 | 87 | 132 | 214 | 73 | 132 | 87 | 85 | 124 | ID | ||||||||||||||||||||||
| 11 | M. racem | 129 | 138 | 8 | 208 | 134 | 18 | 136 | 132 | 7 | 128 | ID | |||||||||||||||||||||
| 12 | T. interd | 22 | 35 | 132 | 211 | 93 | 134 | 39 | 25 | 127 | 83 | 130 | ID | ||||||||||||||||||||
| 13 | M. canis | 140 | 146 | 151 | 181 | 160 | 150 | 145 | 143 | 151 | 157 | 152 | 147 | ID | |||||||||||||||||||
| 14 | T. schoe | 1 | 37 | 132 | 219 | 91 | 134 | 41 | 2 | 127 | 83 | 130 | 23 | 142 | ID | ||||||||||||||||||
| 15 | T. tonsu | 23 | 36 | 129 | 210 | 96 | 131 | 40 | 26 | 124 | 86 | 127 | 3 | 147 | 24 | ID | |||||||||||||||||
| 16 | T. ajelloi | 126 | 196 | 167 | 145 | 192 | 165 | 196 | 195 | 161 | 182 | 164 | 187 | 167 | 192 | 187 | ID | ||||||||||||||||
| 17 | T. rubru | 31 | 39 | 134 | 209 | 98 | 138 | 42 | 34 | 131 | 87 | 132 | 36 | 149 | 32 | 37 | 195 | ID | |||||||||||||||
| 18 | M. racem | 132 | 137 | 13 | 207 | 137 | 17 | 133 | 135 | 14 | 131 | 17 | 130 | 151 | 133 | 127 | 163 | 136 | ID | ||||||||||||||
| 19 | M. gyps | 88 | 97 | 131 | 224 | 70 | 131 | 98 | 91 | 127 | 60 | 131 | 90 | 156 | 89 | 91 | 196 | 95 | 132 | ID | |||||||||||||
| 20 | M. ferru | 138 | 146 | 148 | 181 | 156 | 147 | 145 | 141 | 146 | 156 | 147 | 144 | 12 | 140 | 143 | 165 | 144 | 145 | 150 | ID | ||||||||||||
| 21 | T. interd | 22 | 41 | 134 | 213 | 100 | 135 | 45 | 27 | 129 | 91 | 132 | 8 | 150 | 23 | 5 | 190 | 42 | 130 | 96 | 148 | ID | |||||||||||
| 22 | T. eriotr | 38 | 11 | 136 | 214 | 91 | 140 | 14 | 41 | 131 | 86 | 132 | 41 | 151 | 39 | 42 | 197 | 42 | 136 | 98 | 151 | 47 | ID | ||||||||||
| 23 | T. erinac | 38 | 9 | 139 | 211 | 98 | 140 | 2 | 41 | 131 | 85 | 135 | 38 | 147 | 39 | 39 | 195 | 39 | 135 | 97 | 147 | 44 | 12 | ID | |||||||||
| 24 | M. gallin | 145 | 148 | 134 | 182 | 150 | 137 | 146 | 148 | 130 | 143 | 134 | 145 | 105 | 146 | 146 | 164 | 144 | 134 | 147 | 101 | 151 | 146 | 146 | ID | ||||||||
| 25 | A. benh | 36 | 0 | 140 | 215 | 95 | 142 | 11 | 40 | 133 | 87 | 136 | 35 | 146 | 38 | 36 | 196 | 40 | 137 | 97 | 146 | 41 | 11 | 9 | 149 | ID | |||||||
| 26 | A. vanbr | 26 | 38 | 130 | 212 | 95 | 135 | 41 | 29 | 128 | 85 | 128 | 8 | 150 | 27 | 9 | 186 | 38 | 130 | 92 | 147 | 12 | 42 | 39 | 142 | 38 | ID | ||||||
| 27 | M. persi | 77 | 84 | 129 | 229 | 69 | 129 | 88 | 80 | 125 | 70 | 129 | 90 | 170 | 78 | 91 | 197 | 88 | 131 | 79 | 162 | 90 | 83 | 86 | 156 | 84 | 94 | ID | |||||
| 28 | M. audo | 138 | 148 | 150 | 184 | 157 | 149 | 144 | 141 | 148 | 154 | 149 | 143 | 14 | 139 | 142 | 169 | 148 | 147 | 149 | 6 | 145 | 151 | 146 | 106 | 148 | 145 | 163 | ID | ||||
| 29 | M. cook | 129 | 139 | 3 | 208 | 132 | 17 | 136 | 132 | 8 | 130 | 7 | 130 | 152 | 130 | 127 | 167 | 133 | 12 | 129 | 147 | 132 | 135 | 139 | 132 | 139 | 128 | 127 | 151 | ID | |||
| 30 | Epidermophyton flocco | 79 | 89 | 126 | 216 | 80 | 123 | 90 | 82 | 119 | 79 | 124 | 94 | 158 | 80 | 95 | 188 | 86 | 126 | 77 | 153 | 100 | 90 | 87 | 139 | 89 | 96 | 78 | 152 | 124 | ID | ||
| 31 | T. violac | 33 | 39 | 136 | 210 | 100 | 140 | 44 | 36 | 133 | 89 | 134 | 38 | 149 | 34 | 39 | 193 | 2 | 136 | 97 | 144 | 44 | 44 | 41 | 146 | 39 | 40 | 90 | 147 | 135 | 88 | ID | |
| 32 | T. equin | 24 | 37 | 130 | 212 | 97 | 132 | 41 | 27 | 125 | 87 | 128 | 4 | 147 | 25 | 1 | 187 | 38 | 127 | 92 | 144 | 4 | 43 | 40 | 145 | 37 | 10 | 92 | 142 | 128 | 96 | 40 | ID |
Sequences difference count matrix based on pairwise sequence comparison of beta tubulin gene between representative dermatophytes investigated in this study.
| . | Species . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | 13 . | 14 . | 15 . | 16 . | 17 . | 18 . | 19 . | 20 . | 21 . | 22 . | 23 . | 24 . | 25 . | 26 . | 27 . | 28 . | 29 . | 30 . | 31 . | 32 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Trichophyton ment | ID | |||||||||||||||||||||||||||||||
| 2 | T. conce | 36 | ID | ||||||||||||||||||||||||||||||
| 3 | Microsporum cook | 131 | 140 | ID | |||||||||||||||||||||||||||||
| 4 | M. amaz | 217 | 215 | 210 | ID | ||||||||||||||||||||||||||||
| 5 | M. nanu | 90 | 94 | 134 | 218 | ID | |||||||||||||||||||||||||||
| 6 | M. cook | 133 | 142 | 18 | 203 | 133 | ID | ||||||||||||||||||||||||||
| 7 | T. erinac | 40 | 11 | 138 | 212 | 99 | 139 | ID | |||||||||||||||||||||||||
| 8 | Arthroderma simii | 3 | 39 | 134 | 221 | 93 | 136 | 43 | ID | ||||||||||||||||||||||||
| 9 | M. racem | 126 | 135 | 9 | 204 | 130 | 13 | 132 | 129 | ID | |||||||||||||||||||||||
| 10 | A. incur | 82 | 87 | 132 | 214 | 73 | 132 | 87 | 85 | 124 | ID | ||||||||||||||||||||||
| 11 | M. racem | 129 | 138 | 8 | 208 | 134 | 18 | 136 | 132 | 7 | 128 | ID | |||||||||||||||||||||
| 12 | T. interd | 22 | 35 | 132 | 211 | 93 | 134 | 39 | 25 | 127 | 83 | 130 | ID | ||||||||||||||||||||
| 13 | M. canis | 140 | 146 | 151 | 181 | 160 | 150 | 145 | 143 | 151 | 157 | 152 | 147 | ID | |||||||||||||||||||
| 14 | T. schoe | 1 | 37 | 132 | 219 | 91 | 134 | 41 | 2 | 127 | 83 | 130 | 23 | 142 | ID | ||||||||||||||||||
| 15 | T. tonsu | 23 | 36 | 129 | 210 | 96 | 131 | 40 | 26 | 124 | 86 | 127 | 3 | 147 | 24 | ID | |||||||||||||||||
| 16 | T. ajelloi | 126 | 196 | 167 | 145 | 192 | 165 | 196 | 195 | 161 | 182 | 164 | 187 | 167 | 192 | 187 | ID | ||||||||||||||||
| 17 | T. rubru | 31 | 39 | 134 | 209 | 98 | 138 | 42 | 34 | 131 | 87 | 132 | 36 | 149 | 32 | 37 | 195 | ID | |||||||||||||||
| 18 | M. racem | 132 | 137 | 13 | 207 | 137 | 17 | 133 | 135 | 14 | 131 | 17 | 130 | 151 | 133 | 127 | 163 | 136 | ID | ||||||||||||||
| 19 | M. gyps | 88 | 97 | 131 | 224 | 70 | 131 | 98 | 91 | 127 | 60 | 131 | 90 | 156 | 89 | 91 | 196 | 95 | 132 | ID | |||||||||||||
| 20 | M. ferru | 138 | 146 | 148 | 181 | 156 | 147 | 145 | 141 | 146 | 156 | 147 | 144 | 12 | 140 | 143 | 165 | 144 | 145 | 150 | ID | ||||||||||||
| 21 | T. interd | 22 | 41 | 134 | 213 | 100 | 135 | 45 | 27 | 129 | 91 | 132 | 8 | 150 | 23 | 5 | 190 | 42 | 130 | 96 | 148 | ID | |||||||||||
| 22 | T. eriotr | 38 | 11 | 136 | 214 | 91 | 140 | 14 | 41 | 131 | 86 | 132 | 41 | 151 | 39 | 42 | 197 | 42 | 136 | 98 | 151 | 47 | ID | ||||||||||
| 23 | T. erinac | 38 | 9 | 139 | 211 | 98 | 140 | 2 | 41 | 131 | 85 | 135 | 38 | 147 | 39 | 39 | 195 | 39 | 135 | 97 | 147 | 44 | 12 | ID | |||||||||
| 24 | M. gallin | 145 | 148 | 134 | 182 | 150 | 137 | 146 | 148 | 130 | 143 | 134 | 145 | 105 | 146 | 146 | 164 | 144 | 134 | 147 | 101 | 151 | 146 | 146 | ID | ||||||||
| 25 | A. benh | 36 | 0 | 140 | 215 | 95 | 142 | 11 | 40 | 133 | 87 | 136 | 35 | 146 | 38 | 36 | 196 | 40 | 137 | 97 | 146 | 41 | 11 | 9 | 149 | ID | |||||||
| 26 | A. vanbr | 26 | 38 | 130 | 212 | 95 | 135 | 41 | 29 | 128 | 85 | 128 | 8 | 150 | 27 | 9 | 186 | 38 | 130 | 92 | 147 | 12 | 42 | 39 | 142 | 38 | ID | ||||||
| 27 | M. persi | 77 | 84 | 129 | 229 | 69 | 129 | 88 | 80 | 125 | 70 | 129 | 90 | 170 | 78 | 91 | 197 | 88 | 131 | 79 | 162 | 90 | 83 | 86 | 156 | 84 | 94 | ID | |||||
| 28 | M. audo | 138 | 148 | 150 | 184 | 157 | 149 | 144 | 141 | 148 | 154 | 149 | 143 | 14 | 139 | 142 | 169 | 148 | 147 | 149 | 6 | 145 | 151 | 146 | 106 | 148 | 145 | 163 | ID | ||||
| 29 | M. cook | 129 | 139 | 3 | 208 | 132 | 17 | 136 | 132 | 8 | 130 | 7 | 130 | 152 | 130 | 127 | 167 | 133 | 12 | 129 | 147 | 132 | 135 | 139 | 132 | 139 | 128 | 127 | 151 | ID | |||
| 30 | Epidermophyton flocco | 79 | 89 | 126 | 216 | 80 | 123 | 90 | 82 | 119 | 79 | 124 | 94 | 158 | 80 | 95 | 188 | 86 | 126 | 77 | 153 | 100 | 90 | 87 | 139 | 89 | 96 | 78 | 152 | 124 | ID | ||
| 31 | T. violac | 33 | 39 | 136 | 210 | 100 | 140 | 44 | 36 | 133 | 89 | 134 | 38 | 149 | 34 | 39 | 193 | 2 | 136 | 97 | 144 | 44 | 44 | 41 | 146 | 39 | 40 | 90 | 147 | 135 | 88 | ID | |
| 32 | T. equin | 24 | 37 | 130 | 212 | 97 | 132 | 41 | 27 | 125 | 87 | 128 | 4 | 147 | 25 | 1 | 187 | 38 | 127 | 92 | 144 | 4 | 43 | 40 | 145 | 37 | 10 | 92 | 142 | 128 | 96 | 40 | ID |
| . | Species . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | 13 . | 14 . | 15 . | 16 . | 17 . | 18 . | 19 . | 20 . | 21 . | 22 . | 23 . | 24 . | 25 . | 26 . | 27 . | 28 . | 29 . | 30 . | 31 . | 32 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Trichophyton ment | ID | |||||||||||||||||||||||||||||||
| 2 | T. conce | 36 | ID | ||||||||||||||||||||||||||||||
| 3 | Microsporum cook | 131 | 140 | ID | |||||||||||||||||||||||||||||
| 4 | M. amaz | 217 | 215 | 210 | ID | ||||||||||||||||||||||||||||
| 5 | M. nanu | 90 | 94 | 134 | 218 | ID | |||||||||||||||||||||||||||
| 6 | M. cook | 133 | 142 | 18 | 203 | 133 | ID | ||||||||||||||||||||||||||
| 7 | T. erinac | 40 | 11 | 138 | 212 | 99 | 139 | ID | |||||||||||||||||||||||||
| 8 | Arthroderma simii | 3 | 39 | 134 | 221 | 93 | 136 | 43 | ID | ||||||||||||||||||||||||
| 9 | M. racem | 126 | 135 | 9 | 204 | 130 | 13 | 132 | 129 | ID | |||||||||||||||||||||||
| 10 | A. incur | 82 | 87 | 132 | 214 | 73 | 132 | 87 | 85 | 124 | ID | ||||||||||||||||||||||
| 11 | M. racem | 129 | 138 | 8 | 208 | 134 | 18 | 136 | 132 | 7 | 128 | ID | |||||||||||||||||||||
| 12 | T. interd | 22 | 35 | 132 | 211 | 93 | 134 | 39 | 25 | 127 | 83 | 130 | ID | ||||||||||||||||||||
| 13 | M. canis | 140 | 146 | 151 | 181 | 160 | 150 | 145 | 143 | 151 | 157 | 152 | 147 | ID | |||||||||||||||||||
| 14 | T. schoe | 1 | 37 | 132 | 219 | 91 | 134 | 41 | 2 | 127 | 83 | 130 | 23 | 142 | ID | ||||||||||||||||||
| 15 | T. tonsu | 23 | 36 | 129 | 210 | 96 | 131 | 40 | 26 | 124 | 86 | 127 | 3 | 147 | 24 | ID | |||||||||||||||||
| 16 | T. ajelloi | 126 | 196 | 167 | 145 | 192 | 165 | 196 | 195 | 161 | 182 | 164 | 187 | 167 | 192 | 187 | ID | ||||||||||||||||
| 17 | T. rubru | 31 | 39 | 134 | 209 | 98 | 138 | 42 | 34 | 131 | 87 | 132 | 36 | 149 | 32 | 37 | 195 | ID | |||||||||||||||
| 18 | M. racem | 132 | 137 | 13 | 207 | 137 | 17 | 133 | 135 | 14 | 131 | 17 | 130 | 151 | 133 | 127 | 163 | 136 | ID | ||||||||||||||
| 19 | M. gyps | 88 | 97 | 131 | 224 | 70 | 131 | 98 | 91 | 127 | 60 | 131 | 90 | 156 | 89 | 91 | 196 | 95 | 132 | ID | |||||||||||||
| 20 | M. ferru | 138 | 146 | 148 | 181 | 156 | 147 | 145 | 141 | 146 | 156 | 147 | 144 | 12 | 140 | 143 | 165 | 144 | 145 | 150 | ID | ||||||||||||
| 21 | T. interd | 22 | 41 | 134 | 213 | 100 | 135 | 45 | 27 | 129 | 91 | 132 | 8 | 150 | 23 | 5 | 190 | 42 | 130 | 96 | 148 | ID | |||||||||||
| 22 | T. eriotr | 38 | 11 | 136 | 214 | 91 | 140 | 14 | 41 | 131 | 86 | 132 | 41 | 151 | 39 | 42 | 197 | 42 | 136 | 98 | 151 | 47 | ID | ||||||||||
| 23 | T. erinac | 38 | 9 | 139 | 211 | 98 | 140 | 2 | 41 | 131 | 85 | 135 | 38 | 147 | 39 | 39 | 195 | 39 | 135 | 97 | 147 | 44 | 12 | ID | |||||||||
| 24 | M. gallin | 145 | 148 | 134 | 182 | 150 | 137 | 146 | 148 | 130 | 143 | 134 | 145 | 105 | 146 | 146 | 164 | 144 | 134 | 147 | 101 | 151 | 146 | 146 | ID | ||||||||
| 25 | A. benh | 36 | 0 | 140 | 215 | 95 | 142 | 11 | 40 | 133 | 87 | 136 | 35 | 146 | 38 | 36 | 196 | 40 | 137 | 97 | 146 | 41 | 11 | 9 | 149 | ID | |||||||
| 26 | A. vanbr | 26 | 38 | 130 | 212 | 95 | 135 | 41 | 29 | 128 | 85 | 128 | 8 | 150 | 27 | 9 | 186 | 38 | 130 | 92 | 147 | 12 | 42 | 39 | 142 | 38 | ID | ||||||
| 27 | M. persi | 77 | 84 | 129 | 229 | 69 | 129 | 88 | 80 | 125 | 70 | 129 | 90 | 170 | 78 | 91 | 197 | 88 | 131 | 79 | 162 | 90 | 83 | 86 | 156 | 84 | 94 | ID | |||||
| 28 | M. audo | 138 | 148 | 150 | 184 | 157 | 149 | 144 | 141 | 148 | 154 | 149 | 143 | 14 | 139 | 142 | 169 | 148 | 147 | 149 | 6 | 145 | 151 | 146 | 106 | 148 | 145 | 163 | ID | ||||
| 29 | M. cook | 129 | 139 | 3 | 208 | 132 | 17 | 136 | 132 | 8 | 130 | 7 | 130 | 152 | 130 | 127 | 167 | 133 | 12 | 129 | 147 | 132 | 135 | 139 | 132 | 139 | 128 | 127 | 151 | ID | |||
| 30 | Epidermophyton flocco | 79 | 89 | 126 | 216 | 80 | 123 | 90 | 82 | 119 | 79 | 124 | 94 | 158 | 80 | 95 | 188 | 86 | 126 | 77 | 153 | 100 | 90 | 87 | 139 | 89 | 96 | 78 | 152 | 124 | ID | ||
| 31 | T. violac | 33 | 39 | 136 | 210 | 100 | 140 | 44 | 36 | 133 | 89 | 134 | 38 | 149 | 34 | 39 | 193 | 2 | 136 | 97 | 144 | 44 | 44 | 41 | 146 | 39 | 40 | 90 | 147 | 135 | 88 | ID | |
| 32 | T. equin | 24 | 37 | 130 | 212 | 97 | 132 | 41 | 27 | 125 | 87 | 128 | 4 | 147 | 25 | 1 | 187 | 38 | 127 | 92 | 144 | 4 | 43 | 40 | 145 | 37 | 10 | 92 | 142 | 128 | 96 | 40 | ID |
Discussion
Since a high degree of phylogenetic resolution is the primary benchmark used to determine the suitability of a locus for molecular systematic analysis [24], the suitability of partial beta tubulin gene sequencing for specifying the species boundaries among dermatophytes was targeted in this study. The taxonomy of dermatophytes has been changed fundamentally recently [7,18,25–28]; however, there are still some areas of conflict [29–31]. The recently proposed taxonomy principally relied on molecular species recognition inferred from ITS sequences [7]. In main traits, congruence with phenotypic species concepts was found, but sequence data have resulted in some controversies [30–32]. Previous studies indicated that protein-coding loci provide high resolution and support for fungal systematics [33,34], especially the BT2 gene [14–17]. A recent BT2 sequence–based study led to discovery of a new morphologically cryptic phylogenetic species within the M. cookei clade, M. mirabile [35]. Moreover, our recent partial sequencing of BT2 provided evidence that this locus is more useful for species discrimination of Arthroderma otae members [36] and it has the same value as ITS for differentiation of T. tonsurans and T. equinum [37]. Here we extended the BT2 partial sequencing to a spectrum of 26 dermatophyte species in order to perform species delineation.
It is known that most Trichophyton spp., especially the human-adapted species, are similar to each other in ITS. Sequence mismatching may be as low as a single nucleotide (SNP) exchange, as observed in ITS1 of the closely related species of the A. vanbreuseghemii complex, T. tonsurans and T. equinum [7,37,38]. However, the pairwise sequence comparison of BT2 in the ecologically differentiated species T. tonsurans and T. equinum also showed a single SNP difference. ITS sequence homology between A. vanbreuseghemii (anamorph T. interdigitale) and T. tonsurans/T. equinum strains was noted to be >98.6% [38]. Our BT2 pairwise comparison in this study revealed a similar range of 98.4% to 99.4% between T. interdigitale and T. equinum and 98.6% to 99.6% between T. interdigitale and T. tonsurans (Table 2). We attribute these findings to the significant variation among T. interdigitale strains of 0.2%–1.8% intraspecies dissimilarity in BT2. Among dermatophytes, T. interdigitale is the only dermatophyte taxon that unifies strains of both human- and animal-associated lineages [7] and, additionally, shows a high frequency of intragenomic polymorphism in ITS [39]. Recently, the taxonomic status of T. interdigitale was revisited by Beguin et al. [31]. Based on the phylogram inferred from a combined sequences dataset of three loci, including ITS, partial BT2, and Actin genes, they concluded that T. interdigitale cannot be considered to belong to A. vanbreuseghemii. In their analysis, the two taxa differed by one to six nucleotides across the ITS-rDNA. However, such minor genetic polymorphisms may be insufficient for separation of the mentioned taxa. In addition, there was no relationship between genotypes of ITS and mating behavior of T. interdigitale, including strains ranging from sexually active to inactive [29].
Our BT2-based phylogenic analysis showed that the genealogical relationships of species in the A. benhamiae complex were compatible with that inferred from the ITS cladogram. The intraspecies divergences were found to be 0, 0–1, 0, and 1–3 bp in ITS vs. 0, 0–8, 0, and 3 bp in BT2 for strains of T. concentricum, A. benhamiae, T. eriotrephon, and T. erinacei, respectively. The ITS sequence diversity between the strains of T. erinacei and A. benhamiae was reported to be up to 21 substitutions [25]. In the present study, sequence variation between two species ranged from 10 to 15 bp in ITS and 11 to 13 bp in BT2. In general, both ITS and BT2 regions could discriminate between the strains of all species in the complex at almost the same level of diversity (data not shown). BT2 sequencing could not distinguish between A. benhamiae (T. anamorph) and T. concentricum (99.9%–100% identity), two species that also were the nearest relatives to each other in our ITS genealogy (98.5%–99.7% similarity). These results corroborate the findings of Heidemann et al., who observed T. concentricum as the closest relative of A. benhamiae [26], and of Kawasaki et al., who found that the two mentioned species were not indistinguishable from each other by actin and topoisomerase II gene sequencing [32].
The status of our two strains of a very rare species, T. eriotrephon, on BT2 and ITS sequence analysis was interesting. Until recently, there was only a single clinical strain of this species (CBS 220.25), isolated in 1925 in Amsterdam from a patient with tinea corporis. In 2012 we encountered two new cases of this species in Iran [40]. Our strains were identical in both ITS and BT2 sequencing and showed complete ITS sequence homology to T. eriotrephon strain CBS 220.25. Together with those of T. erinacei and T. verrucosum, the sequences constituted a separate internode on the BT2 phylogram. Additionally, they had close similarity to T. erinacei, strain CS 379, in both ITS and BT2 regions and were remote from T. concentricum. This finding supports the possibility that the species has a zoophilic origin [26]. In the BT2 pairwise comparison of T. eriotrephon and its relatives (T. concentricum, A. benhamiae, T. verrucosum, and T. erinacei), five signature nucleotides unique to the taxon were identified.
Unfortunately, we did not obtain the full length of the BT2 sequence in the cattle-associated species, T. verrucosum; after alignment, a shorter 509-bp fragment was obtained in all three strains of the species. While the strains had intraspecies variation in ITS marker, no intraspecific variation was found in BT2; moreover, two species-specific barcode nucleotides were determined for the species. The location of T. verrucosum in both ITS and BT2 trees at a node with T. erinacei was almost the same and in consonance with those inferred from earlier studies [25,26]. Although the natural distribution of Arthroderma simii (anamorph T. simii), a monkey-associated species, is probably not as restricted as has previously been proposed [41], it is considered to be prevalent only in the Indian subcontinent where it was isolated from soil as well as from human and animal infections. The species was poorly studied, and its level of genetic biodiversity is unclear [7,42]. Sequence analysis of the ITS region and other genetic markers indicates that it is closely related to T. mentagrophytes sensu stricto and T. schoenleinii [18,43]. Based on new species concepts, the former zoophilic taxa, including T. mentagrophytes var. quinckeanum (associated with rodents), T. langeronii, and T. sarkisovii (both associated with camels), are now conspecific with T. mentagrophytes sensu stricto [7,25]. In our BT2 and ITS analyses, strains of these species were found in the same node and had intraspecies variation levels of 0–1 nt, except for A. simii strains, which had 2–6 nt variation in BT2. While the interspecies dissimilarities in ITS sequences varied with 5 bp between T. mentagrophytes and T. schoenleinii and with 15 bp between A. simii and T. schoenleinii (data not shown), a high degree of similarity was found in BT2 of three species, especially between T. mentagrophytes and T. schoenleinii (99.8% identity). These findings indicate that ITS-rDNA is more heterogeneous than BT2 in the strains of the A. simii complex, on the one hand, and supports the theory that T. schoenleinii possibly originated from camels, rather than rodents, on the other hand [26,43].
The phylogeny of the closely related species within the Arthroderma otae complex was discussed in our recent multilocus-based study [36]. Microsporum canis and M. ferrugineum were distinguished by 2 bp in ITS2, while M. audouinii differentiated from these two species by polymorphisms in both ITS1 and ITS2 [36,44]. The average pairwise sequence divergence between the three tested species was 5.33 nt for the ITS region, in comparison with 10.67 nt for BT2, suggesting that BT2 is more useful than ITS for species delineation in the A. otae complex.
The taxa around T. rubrum consisted of significant numbers of phenotypic species and varieties that were recently reclassified and found to be conspecific with T. rubrum or T. violaceum [28]. Trichophyton rubrum strains have relatively identical ITS sequences, and characteristic sites were found in both ITS1 and ITS2 [44]. In contrast, despite the presence of species-specific sequences in ITS1, T. violaceum strains have intraspecies diversity, especially in the number of TA motifs at the end of ITS2 region [28,44,45]. Our ITS sequencing from T. violaceum strains provided complementary support for this fact. The length of the consensus sequence for BT2 of T. rubrum and T. violaceum was the same (794 nt), but the two species differed in two signature nucleotides, that is, transitions in positions 173 and 376. Based on the fact that no intraspecies variation was found among strains of each species, the mentioned SNPs can act as molecular autapomorphies for species assignment of T. rubrum and T. violaceum isolates.
Epidermophyton floccosum, the type species of the genus Epidermophyton, has received limited attention in genetic analyses. Kawasaki et al., using restriction fragment length polymorphism of mitochondrial DNA, concluded that Epidermophyton could not be separated from the genera Trichophyton and Microsporum [46]. In contrast, Kano et al. found <1% intraspecies variation in the CHS1 gene among strains of E. floccosum and suggested that the species is genetically distinct from Trichophyton and Microsporum [47]. It was stated that E. floccosum is close to the anthropophilic Trichophyton species [25], while in our study, the taxon was paraphyletic to Microsporum in both ITS and BT2 trees. No intraspecific variation was found in the BT2 locus of our E. floccosum strains.
Intraspecies diversities with three BT2 genotypes were found in strains of each zoophilic species studied, M. nanum (teleomorph A. obtusum) and M. persicolor. Nonetheless, the species were found to have enough variation locating them at relevant clades on BT2 tree. Similarly, M. gallinae, which is located at a separate position, showed some BT2 heterogeneity. The so-called M. gypseum complex comprised two morphologically similar species, M. fulvum and M. gypseum. Microsporum fulvum (teleomorph A. fulvum) rarely causes infection [48]. Microsporum gypseum, the most common geophilic species implicated in dermatophytosis, has been linked to two teleomorphs, A. gypseum and A. incurvatum, which morphologically cannot be differentiated either from each other or from M. fulvum; however, genetically the three taxa are distinct [7,49]. Our BT2 analysis by maximum likelihood showed significant interspecies distances. We also investigated ITS with CHS1 partial gene sequences retrieved from GenBank (data not shown). Similar to BT2, large phylogenetic distances were found in ITS/CHS1 analyses, and the branching patterns were concordant in all trees, meaning that phylogenetically, three different species are involved. Members of the geophilic dermatophytes, which uncommonly cause infection in humans and animals, are known to show more genetic intraspecies variation and to have the highest mutual ITS distances [7,35]. In concordance with earlier studies, the soilborne species M. amazonicum and T. ajelloi were most distant in all trees. Investigated strains of M. cookei and M. racemosum showed three BT2 genotypes. Delimitation of the two species remains ambiguous because both ITS and BT2 datasets showed low resolution between the taxa, confirming results of Choi et al. [35]. In concordance with their findings, strain CBS 102175 was found once in M. cookei (BT2) and once in M. racemosum (ITS). Furthermore, CBS 337.74 was close to M. racemosum in BT2 but clustered with M. cookei in data from Choi et al. [35]. This discrepancy may be explained by the fact that the part of BT2 used for analysis by Choi et al. [35] was shorter (almost 450 bp) than the fragment analyzed in our study (nearly 770 bp).
The phylogeny derived from BT2 in this study aimed to provide a novel genetic marker as an alternative to ITS-rDNA and to evaluate its utility as a DNA barcoding tool. In general, phylogenetic species concepts of dermatophytes based on BT2 and ITS proved to be similar, with only very small inconsistencies found between the two phylogenies. The resolution of the two markers was in the same range. Closely related anthropophilic taxa are differentiated by limited numbers of mutations in both markers, but their congruence has reinforced existing species concepts. Study of other loci as well as more extensive taxon sampling of zoologic and geophilic species is necessary. Until a consensus classification system that is accepted by all mycologists is reached, it is recommended that the current taxonomy of dermatophytes based on ITS sequence data be retained.
This work was financially supported by Tehran University of Medical Sciences (grant 90–04–27–15673), Tehran, Iran and Teikyo University TIMM, Tokyo, Japan. We thank all personnel in the Molecular Biology Laboratory at TIMM and CBS.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and the writing of the paper.




![(A) Phylogenetic tree of 54 representative dermatophyte species and strains (different species and the same species but with at least one nucleotide difference) based on analysis of beta tubulin gene sequences. The evolutionary history was inferred by using the maximum likelihood method based on the Tamura–Nei model. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree for the heuristic search was obtained automatically by applying neighbor–join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood approach and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA5 [22]. (B) A smaller phylogenetic tree of 31 representative dermatophytes species based on analysis of internal transcribe spacer sequences. Note that the position of the complexes and species are almost same in both trees.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/mmy/52/7/10.1093_mmy_myu033/2/m_myu033fig2a.jpeg?Expires=1716360404&Signature=wA1HifyrinumpD-1AZr80DLRsLxhHqmRHFt6iEHQsf252-aa43uyNXEYXlTfvMwiS3g3hpAEBfqFU-1A1skaM~nR5UorO9Yd9n5w9G7VnVWm~difZEYyXXCCz8~qgLl14P5tSgSWSjx9jLOlKUhPG-d9Mw0cm8WuG~QTMWSKn2i-ZVOPtBGl8B6WMNnZm-4ctZBdlXfPexZTimSXQgnWWCaSt4IdMqto0VupABO~NlOgVv63mTtgqgNTax0ajiHUfaG3HGZFPEj1MgtdS25zsCIC4GLgqFGzpS8hSlZoIoN1etstVWguZYUFFG10o61iu7BMfjar74jgiemIuS3HWA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)