-
PDF
- Split View
-
Views
-
Cite
Cite
Laszlo Irinyi, Carolina Serena, Dea Garcia-Hermoso, Michael Arabatzis, Marie Desnos-Ollivier, Duong Vu, Gianluigi Cardinali, Ian Arthur, Anne-Cécile Normand, Alejandra Giraldo, Keith Cassia da Cunha, Marcelo Sandoval-Denis, Marijke Hendrickx, Angela Satie Nishikaku, Analy Salles de Azevedo Melo, Karina Bellinghausen Merseguel, Aziza Khan, Juliana Alves Parente Rocha, Paula Sampaio, Marcelo Ribeiro da Silva Briones, Renata Carmona e Ferreira, Mauro de Medeiros Muniz, Laura Rosio Castañón-Olivares, Daniel Estrada-Barcenas, Carole Cassagne, Charles Mary, Shu Yao Duan, Fanrong Kong, Annie Ying Sun, Xianyu Zeng, Zuotao Zhao, Nausicaa Gantois, Françoise Botterel, Barbara Robbertse, Conrad Schoch, Walter Gams, David Ellis, Catriona Halliday, Sharon Chen, Tania C. Sorrell, Renaud Piarroux, Arnaldo L. Colombo, Célia Pais, Sybren de Hoog, Rosely Maria Zancopé-Oliveira, Maria Lucia Taylor, Conchita Toriello, Célia Maria de Almeida Soares, Laurence Delhaes, Dirk Stubbe, Françoise Dromer, Stéphane Ranque, Josep Guarro, Jose F. Cano-Lira, Vincent Robert, Aristea Velegraki, Wieland Meyer, International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database—the quality controlled standard tool for routine identification of human and animal pathogenic fungi, Medical Mycology, Volume 53, Issue 4, May 2015, Pages 313–337, https://doi.org/10.1093/mmy/myv008
Close - Share Icon Share
Abstract
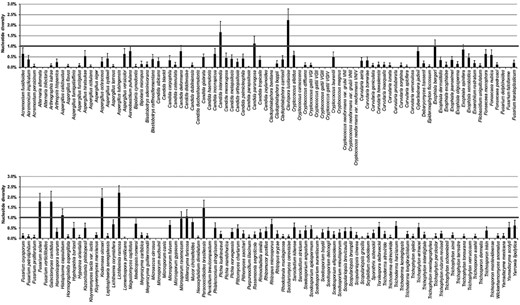
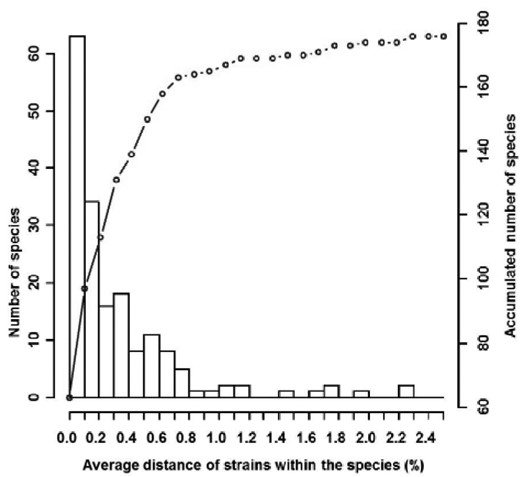
Human and animal fungal pathogens are a growing threat worldwide leading to emerging infections and creating new risks for established ones. There is a growing need for a rapid and accurate identification of pathogens to enable early diagnosis and targeted antifungal therapy. Morphological and biochemical identification methods are time-consuming and require trained experts. Alternatively, molecular methods, such as DNA barcoding, a powerful and easy tool for rapid monophasic identification, offer a practical approach for species identification and less demanding in terms of taxonomical expertise. However, its wide-spread use is still limited by a lack of quality-controlled reference databases and the evolving recognition and definition of new fungal species/complexes. An international consortium of medical mycology laboratories was formed aiming to establish a quality controlled ITS database under the umbrella of the ISHAM working group on “DNA barcoding of human and animal pathogenic fungi.” A new database, containing 2800 ITS sequences representing 421 fungal species, providing the medical community with a freely accessible tool at http://www.isham.org/ and http://its.mycologylab.org/ to rapidly and reliably identify most agents of mycoses, was established. The generated sequences included in the new database were used to evaluate the variation and overall utility of the ITS region for the identification of pathogenic fungi at intra-and interspecies level. The average intraspecies variation ranged from 0 to 2.25%. This highlighted selected pathogenic fungal species, such as the dermatophytes and emerging yeast, for which additional molecular methods/genetic markers are required for their reliable identification from clinical and veterinary specimens.
Introduction
The number of human and animal fungal infections, ranging from superficial infections of the nails and skin, through mucocutaneous candidiasis to invasive fungal infections, have significantly increased over the last three decades, causing serious public health burdens and increased risk of biodiversity loss among animal species [1,2]. In humans, superficial infections affect an estimated 25% ( = 1.7 billion) individuals world-wide. Oropharyngeal or genital mucosal infections are also common and can be disabling. For example, an estimated 75% of women of childbearing age suffering from vulvovaginitis, mainly caused by Candida species [3], which are the third most common opportunistic fungal disease agents after Aspergillus spp. worldwide [1]. Invasive fungal diseases are of great concern, due to their high mortality that can exceed 50%. More than 90% of fungal-related deaths are caused by four fungal genera: Aspergillus, Candida, Cryptococcus, and Pneumocystis [1,4,5]. Delays in diagnosis are not only associated with high mortality but also severe organ dysfunction, for example, respiratory failure (endemic fungal infections and chronic pulmonary aspergillosis), neurologic deficits (endemic fungal infections and cryptococcosis) [6], blindness and visual impairment (fungal keratitis) [7]. To better understand, control, and treat these diseases, more rapid and accurate identification of the causal agents is essential.
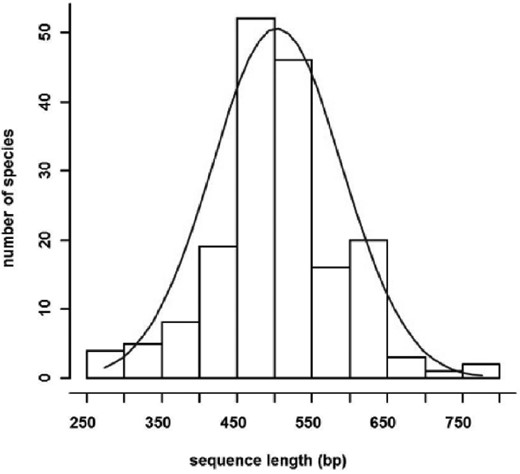
DNA barcoding, first proposed by Hebert et al. [8], utilizes DNA sequences to standardize the identification of organisms from all kingdoms to the species level by comparison to a reference collection of well-identified species. The principle behind barcoding is that species identification must be accurate, fast, cost-effective, culture independent, universally accessible, and feasible for nonexperts [9]. As a consequence, its popularity as a species identification tool has drastically increased. Barcodes are short diverse genetic sequences (500–800 bp) that are flanked by conserved regions allowing for the design of universal primers. From a pragmatic perspective, a universal sequence suitable for all kingdoms would be ideal, but the identification of a universal genetic region for a wide range of taxa remains elusive. The key concepts underlying barcoding are that the interspecies distances should exceed intraspecies distances, creating a barcoding gap [10], and that identification is straightforward when a sequence is unique to a single species and constant within each species [8,11,12]. The most important question in barcoding is: How accurate and reliable are the delineation and identification of a species using a single gene?
The correct identification of fungi is essential for many biological purposes, such as the assessment of biodiversity, taxonomy and species conservation [9,13]. It is mandatory for clinical diagnosis and early initiation of appropriate antifungal therapy. Traditional identification based on morphology and biochemistry of pathogenic fungi is time-consuming and requires a certain level of morphological and taxonomical expertise. To overcome these limitations, DNA barcoding was evaluated in fungi, targeting numerous genetic loci, including COX1 [14], protein-coding genes like RNA polymerase I and II [15–19], partial translation elongation factor 1-α [20–22], β-tubulin [23], and the internal transcribed spacer (ITS) regions [24,25]. The protein coding genes have proven to be a powerful tool for species delimitation, providing a high level of phylogenetic resolution and information [21,26,27]. However, the primers used to amplify these regions are usually restricted to specific taxa and amplification can often be problematic [16]. In contrast, the ITS regions are easily amplified with universal primers that are compatible among most fungal species. It has shown sufficient genetic variability for identification at interspecies level, and has been adopted as the official standard barcoding region for fungi [28]. However, use of the ITS region as a barcode has been criticized by Kiss [29] because of its inability to distinguish many closely related fungal species. In addition, for some fungi, the ITS regions alone do not provide accurate identification to species level [30]. In some groups of fungi (Aspergillus, Colletotrichum) the interspecies variation is insignificant [31,32] and in other groups (Glomeromycota, Chytridiomycota) the diversity within species is too high [33,34] Fungal genomes may contain more than 200 copies of the ribosomal region [35,36] dispersed over one or more chromosomal locations [37]. This results in polymorphism within a genome of one individual [38,39]. Intragenomic diversity is mainly explained by concerted evolutionary processes, for example, unequal crossing over between repeat units, gene conversion or gene amplification [39,40].
Despite these limitations the ITS region has been used in molecular identification and phylogenetic studies of human pathogenic fungi [41–48] long before its selection as the official fungal DNA barcode. The ITS sequences in publicly accessible databases are used routinely by the medical community to identify fungi at the species level on the basis of matching sequences. However, its widespread application has been compromised by the deposition of incorrectly identified or incomplete sequences in the commonly used public databases of the International Nucleotide Sequence Database Collaboration (INSDC) [49]. This includes GenBank [50], at the National Center for Biotechnology Information (NCBI), which is the major nucleotide sequence depository and is widely utilised by clinical microbiologists and the scientific community [51,52]. Because GenBank acts primarily as an archive, many sequences submitted have been annotated with incorrect or poorly defined species names. It has also been shown that more than 10% of the publicly available fungal ITS sequences were annotated incorrectly at species level [53]. As a consequence, a number of curated ITS databases have been created to ensure the correct identification of fungal species, for example, within the Barcode of Life Data System (BOLD) [54] and UNITE [55]. Partially in response to requests to allow third party annotation of GenBank records NCBI has also initiated a curated database RefSeq Targeted Loci (RTL) [56] that will provide a limited set of curated sequences obtained from type and verified material [57]. In a second, broader approach NCBI is currently annotating the type material associated with taxonomic names. This will allow type related searches to be conducted across multiple sequence markers or whole genomes [58]. Other reference databases are available for specific taxonomic groups, for example, Fusarium [59] and Aspergillus [60]. The deficiency of these reference databases with respect to human pathogenic fungi is the limited number of medically important fungal species contained within them. The demand for curated, reliable reference databases has increased significantly due to diminishing expertise in fungal morphology and its increasing replacement by the use of sequencing in fungal diagnostic laboratories.
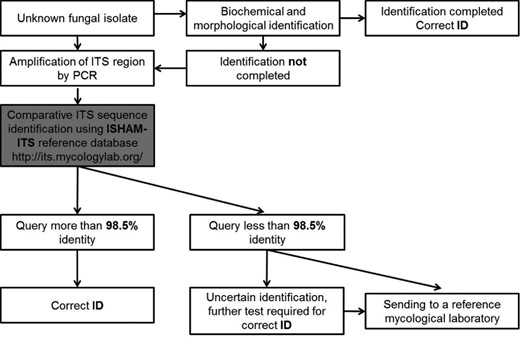
To address these issues, a working group of the International Society for Human and Animal Mycology (ISHAM) on “Barcoding of Medical Fungi” was established in 2011 [61]. The working group identified the need to: (a) generate a medical barcode database by incorporating existing fungal group-specific databases; (b) extend the number of quality-controlled ITS sequences to cover all medically important fungal species; (c) evaluate the value of ITS as a barcode at intra-and interspecies level; (d) eventually incorporate these sequences into the BOLD database; (e) UNITE; and (f) achieve a species status as “quality controlled reference sequences” for those sequences within RTL at NCBI.
The main objective of this study was to generate a publicly available, quality-controlled, ITS reference database for human and animal pathogenic fungal species and to evaluate the applicability of ITS sequences (the official barcode for fungi) as a genetic marker for species identification. The secondary aim was to highlight fungal taxa where additional genetic sequence information is recommended beyond the ITS for a more accurate identification.
Materials and methods
Generating the database
The ISHAM-ITS reference database is a result of an international collaboration between 14 medical mycology laboratories representing three continents (Table 1). The contributors provided a total of 2945 ITS sequences. Species were identified based on polyphasic identification including morphology, biochemical and physiological tests when appropriate and sequencing. After collecting all the data, the overall identity of sequences obtained from more than two strains per species was determined, including available type strains. In the case of species with less than two strains, trace files were checked for the quality and integrity of sequences. A total of 145 sequences that did not meet the inclusion criteria were discarded, as well as sequences that were misidentified or not identified to species level. Each taxon was provided with the taxonomic name, taking into account the “One Name = One Fungus” concept of the International Code of Nomenclature for algae, fungi and plants (ICN) [62]. The current taxonomical names were provided by using online nomenclature data resources such as MycoBank [63,64], Index Fungorum [65], the latest edition of The Yeasts [66], as well as the latest publications and consulting taxonomical experts of specific taxa. Where possible, former anamorph or teleomorph names and the most-used synonyms were also listed to facilitate reading for clinicians.
Institutions, number of quality controlled ITS sequences, and represented number of species contributed to the ISHAM-ITS reference database.
| . | Number of . | Number of . |
|---|---|---|
| Institutions . | strains . | species . |
| Molecular Mycology Research Laboratory, CIDM, Sydney Medical School-Westmead Hospital, The University of Sydney, WMI, Australia | 663 | 173 |
| Mycology Research Laboratory, Department of Microbiology, Medical School, the University of Athens Hellenic Collection of Pathogenic Fungi (UOA/HCPF), National and Kapodistrian University of Athens, Athens, Greece | 417 | 117 |
| Unitat de Microbiologia, Facultat de Medicina i Ciències de la Salut, IISPV, Universitat Rovira i Virgili, Reus, Spain | 360 | 52 |
| CBS-KNAW, Fungal Biodiversity Centre, Utrecht, The Netherlands | 352 | 33 |
| BCCM/IHEM, Biomedical fungi and yeasts collection, Scientific Institute of Public Health, Brussels, Belgium | 289 | 92 |
| Institut Pasteur, National Reference Center of Invasive Mycosis and Antifungals, Molecular Mycology Unit, CNRS URA 3012, Paris, France | 223 | 106 |
| Parasitology - Mycology, APHM, CHU Timone-Adultes, Marseille, France; Aix-Marseille University, UMR MD3 IP-TPT, Marseille, France | 146 | 55 |
| Mycology Laboratory, Department of Microbiology and Infectious Diseases, PathWest Laboratory Medicine WA, QEII Medical Centre, Nedlands, Western Australia, Australia | 99 | 31 |
| BDEEP-EA4547, CIIL, Institut Pasteur de Lille, CHU de Lille, Université de Lille2, Lille, France | 73 | 18 |
| Laboratório Especial de Micologia, Escola Paulista de Medicina, Universidade Federal de São Paulo, São Paulo, Brazil | 58 | 18 |
| Instituto de Pesquisa Clínica Evandro Chagas (IPEC) - Fundação Oswaldo Cruz (Fiocruz), Rio de Janeiro, Brazil | 50 | 1 |
| Facultad de Medicina, Departamento de Microbiología y Parasitología (Unidad de Micología), Universidad Nacional Autónoma de México, Ciudad de México, México | 39 | 3 |
| Centre of Molecular and Environmental Biology (CBMA), Biology Department, School of Sciences, University of Minho, Braga, Portugal | 22 | 10 |
| Universidade Federal de Goiás, Instituto de Ciências Biológicas, Laboratório de Biologia Molecular, Goiânia, Goiás, Brazil | 9 | 2 |
| . | Number of . | Number of . |
|---|---|---|
| Institutions . | strains . | species . |
| Molecular Mycology Research Laboratory, CIDM, Sydney Medical School-Westmead Hospital, The University of Sydney, WMI, Australia | 663 | 173 |
| Mycology Research Laboratory, Department of Microbiology, Medical School, the University of Athens Hellenic Collection of Pathogenic Fungi (UOA/HCPF), National and Kapodistrian University of Athens, Athens, Greece | 417 | 117 |
| Unitat de Microbiologia, Facultat de Medicina i Ciències de la Salut, IISPV, Universitat Rovira i Virgili, Reus, Spain | 360 | 52 |
| CBS-KNAW, Fungal Biodiversity Centre, Utrecht, The Netherlands | 352 | 33 |
| BCCM/IHEM, Biomedical fungi and yeasts collection, Scientific Institute of Public Health, Brussels, Belgium | 289 | 92 |
| Institut Pasteur, National Reference Center of Invasive Mycosis and Antifungals, Molecular Mycology Unit, CNRS URA 3012, Paris, France | 223 | 106 |
| Parasitology - Mycology, APHM, CHU Timone-Adultes, Marseille, France; Aix-Marseille University, UMR MD3 IP-TPT, Marseille, France | 146 | 55 |
| Mycology Laboratory, Department of Microbiology and Infectious Diseases, PathWest Laboratory Medicine WA, QEII Medical Centre, Nedlands, Western Australia, Australia | 99 | 31 |
| BDEEP-EA4547, CIIL, Institut Pasteur de Lille, CHU de Lille, Université de Lille2, Lille, France | 73 | 18 |
| Laboratório Especial de Micologia, Escola Paulista de Medicina, Universidade Federal de São Paulo, São Paulo, Brazil | 58 | 18 |
| Instituto de Pesquisa Clínica Evandro Chagas (IPEC) - Fundação Oswaldo Cruz (Fiocruz), Rio de Janeiro, Brazil | 50 | 1 |
| Facultad de Medicina, Departamento de Microbiología y Parasitología (Unidad de Micología), Universidad Nacional Autónoma de México, Ciudad de México, México | 39 | 3 |
| Centre of Molecular and Environmental Biology (CBMA), Biology Department, School of Sciences, University of Minho, Braga, Portugal | 22 | 10 |
| Universidade Federal de Goiás, Instituto de Ciências Biológicas, Laboratório de Biologia Molecular, Goiânia, Goiás, Brazil | 9 | 2 |
Institutions, number of quality controlled ITS sequences, and represented number of species contributed to the ISHAM-ITS reference database.
| . | Number of . | Number of . |
|---|---|---|
| Institutions . | strains . | species . |
| Molecular Mycology Research Laboratory, CIDM, Sydney Medical School-Westmead Hospital, The University of Sydney, WMI, Australia | 663 | 173 |
| Mycology Research Laboratory, Department of Microbiology, Medical School, the University of Athens Hellenic Collection of Pathogenic Fungi (UOA/HCPF), National and Kapodistrian University of Athens, Athens, Greece | 417 | 117 |
| Unitat de Microbiologia, Facultat de Medicina i Ciències de la Salut, IISPV, Universitat Rovira i Virgili, Reus, Spain | 360 | 52 |
| CBS-KNAW, Fungal Biodiversity Centre, Utrecht, The Netherlands | 352 | 33 |
| BCCM/IHEM, Biomedical fungi and yeasts collection, Scientific Institute of Public Health, Brussels, Belgium | 289 | 92 |
| Institut Pasteur, National Reference Center of Invasive Mycosis and Antifungals, Molecular Mycology Unit, CNRS URA 3012, Paris, France | 223 | 106 |
| Parasitology - Mycology, APHM, CHU Timone-Adultes, Marseille, France; Aix-Marseille University, UMR MD3 IP-TPT, Marseille, France | 146 | 55 |
| Mycology Laboratory, Department of Microbiology and Infectious Diseases, PathWest Laboratory Medicine WA, QEII Medical Centre, Nedlands, Western Australia, Australia | 99 | 31 |
| BDEEP-EA4547, CIIL, Institut Pasteur de Lille, CHU de Lille, Université de Lille2, Lille, France | 73 | 18 |
| Laboratório Especial de Micologia, Escola Paulista de Medicina, Universidade Federal de São Paulo, São Paulo, Brazil | 58 | 18 |
| Instituto de Pesquisa Clínica Evandro Chagas (IPEC) - Fundação Oswaldo Cruz (Fiocruz), Rio de Janeiro, Brazil | 50 | 1 |
| Facultad de Medicina, Departamento de Microbiología y Parasitología (Unidad de Micología), Universidad Nacional Autónoma de México, Ciudad de México, México | 39 | 3 |
| Centre of Molecular and Environmental Biology (CBMA), Biology Department, School of Sciences, University of Minho, Braga, Portugal | 22 | 10 |
| Universidade Federal de Goiás, Instituto de Ciências Biológicas, Laboratório de Biologia Molecular, Goiânia, Goiás, Brazil | 9 | 2 |
| . | Number of . | Number of . |
|---|---|---|
| Institutions . | strains . | species . |
| Molecular Mycology Research Laboratory, CIDM, Sydney Medical School-Westmead Hospital, The University of Sydney, WMI, Australia | 663 | 173 |
| Mycology Research Laboratory, Department of Microbiology, Medical School, the University of Athens Hellenic Collection of Pathogenic Fungi (UOA/HCPF), National and Kapodistrian University of Athens, Athens, Greece | 417 | 117 |
| Unitat de Microbiologia, Facultat de Medicina i Ciències de la Salut, IISPV, Universitat Rovira i Virgili, Reus, Spain | 360 | 52 |
| CBS-KNAW, Fungal Biodiversity Centre, Utrecht, The Netherlands | 352 | 33 |
| BCCM/IHEM, Biomedical fungi and yeasts collection, Scientific Institute of Public Health, Brussels, Belgium | 289 | 92 |
| Institut Pasteur, National Reference Center of Invasive Mycosis and Antifungals, Molecular Mycology Unit, CNRS URA 3012, Paris, France | 223 | 106 |
| Parasitology - Mycology, APHM, CHU Timone-Adultes, Marseille, France; Aix-Marseille University, UMR MD3 IP-TPT, Marseille, France | 146 | 55 |
| Mycology Laboratory, Department of Microbiology and Infectious Diseases, PathWest Laboratory Medicine WA, QEII Medical Centre, Nedlands, Western Australia, Australia | 99 | 31 |
| BDEEP-EA4547, CIIL, Institut Pasteur de Lille, CHU de Lille, Université de Lille2, Lille, France | 73 | 18 |
| Laboratório Especial de Micologia, Escola Paulista de Medicina, Universidade Federal de São Paulo, São Paulo, Brazil | 58 | 18 |
| Instituto de Pesquisa Clínica Evandro Chagas (IPEC) - Fundação Oswaldo Cruz (Fiocruz), Rio de Janeiro, Brazil | 50 | 1 |
| Facultad de Medicina, Departamento de Microbiología y Parasitología (Unidad de Micología), Universidad Nacional Autónoma de México, Ciudad de México, México | 39 | 3 |
| Centre of Molecular and Environmental Biology (CBMA), Biology Department, School of Sciences, University of Minho, Braga, Portugal | 22 | 10 |
| Universidade Federal de Goiás, Instituto de Ciências Biológicas, Laboratório de Biologia Molecular, Goiânia, Goiás, Brazil | 9 | 2 |
DNA isolation, amplification and sequencing
DNA was isolated and purified from cultures using the methods routinely used in the contributing laboratories. A number of fungal-specific universal primers (Table 2) were used to amplify the ITS region, polymerase chain reaction (PCR), and sequencing protocols varied from laboratory to laboratory according to the primers, chemical reagents, and thermocyclers used. Primers used differed depending on the fungal species investigated or starting material used. In general ITS1, ITS3, ITS4 and ITS5 [67] are universal ribosomal primers, which are recommended being used if the amplification is based on pure fungal cultures. The primers SR6R and LR1 [68], V9D, V9G and LS266 [69] and ITS1F [70] have subsequently been designed to be fungal specific, they can be used for amplification based on pure culture as well as directly form clinical specimens, as they will avoid co-amplification of human DNA. The general PCR amplification conditions are given for each of the primer pairs in Table 2 [67–72]. All PCR products were sequenced in both the forward and reverse directions. Bidirectional sequences were assembled and edited using Sequencher® [73]. Trace files were manually checked and ambiguous bases were corrected based on the forward and reverse sequences taking into account the PHRED scores received with the sequence trace files.
Primers and amplification conditions used to amplify ITS sequences maintained in the ISHAM-ITS reference database.
| Primers . | Amplification conditions . |
|---|---|
| SR6R (5′ AAGTATAAGTCGTAACAAGG 3′) and LR1 (5′ GGTTGGTTTCTTTTCCT 3′)(68) | 97ºC for 3 min; 30 cycles of denaturation (94ºC for 35 s), annealing (50ºC for 45 s), and extension (72ºC for 45 s); and a final extension step at 72ºC for 7 min |
| ITS1 (5′ TCCGTAGGTGAACCTGCGG 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′)(67) | 94ºC for 3 min; 35 cycles of denaturation (94ºC for 60 s), annealing (56ºC for 60 s), and extension (72ºC for 2 min); and a final extension step at 72ºC for 7 min |
| ITS5 (5′ GGAAGTAAAAGTCGTAACAAGG 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′)(67) | 94ºC for 5 min.; 35 cycles of denaturation (94ºC for 30 s), annealing (55ºC for 1 min), and extension (72ºC 1 min and 20 s); and a final extension step at 72ºC for 7 min |
| ITS5 (5′ GGAAGTAAAAGTCGTAACAAGG 3′) and NL4b (5′ GGATTCTCACCCTCTATGAC 3′)(67,71) | 94ºC for 5 min.; 35 cycles of denaturation (94ºC for 30 s), annealing (53ºC for 1 min), and extension (72ºC 1 min and 30 s); and a final extension step at 72ºC for 7 min |
| V9D (5′ TTAAGTCCCTGCCCTTTGTA 3′) and LS266 (5′ GCATTCCCAAACAACTCGACTC 3′)(69) | 95ºC for 10 min; 30 cycles of denaturation (94ºC for 30 s), annealing (58ºC for 30 s), and extension (72ºC for 30 s); and a final extension step at 72ºC for 10 min |
| V9G (5′ TTACGTCCCTGCCCTTTGTA 3′) and LS266 (5′ GCATTCCCAAACAACTCGACTC 3′)(69) | 94ºC for 5 min; 35 cycles of denaturation (94ºC for 60 min), annealing (56ºC for 30 s), and extension (72ºC for 2 min); and a final extension step at 72ºC for 10 min |
| ITS1F (5′ CTTGGTCATTTAGAGGAAGTAA 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′)(67,70) | 95ºC for 5 min; 30 cycles of denaturation (95ºC for 30 s), annealing (58ºC for 30 s), and extension (72ºC for 1 min); and a final extension (72ºC for 10 min). |
| ITS1 (5′ TCCGTAGGTGAACCTGCGG 3′) and IT2 (5′ CCTCCGCTTATTGATATGCTTAGG 3′)(67,72) | 94ºC for 3 min; 35 cycles of denaturation (94ºC for 45 s), annealing (52ºC for 45 s), and extension (72ºC for 60 s); and a final extension at 72ºC for 7 min |
| ITS3 (5′ GCATCGATGAAGAACGCAGC 3′) and LS266 (5′ GCATTCCCAAACAACTCGACTC 3′)(67,69) | 95ºC for 10 min; 30 cycles of denaturation (94ºC for 30 s), annealing (58ºC for 30 s), and extension (72ºC for 30 s); and a final extension step at 72ºC for 10 min |
| Primers . | Amplification conditions . |
|---|---|
| SR6R (5′ AAGTATAAGTCGTAACAAGG 3′) and LR1 (5′ GGTTGGTTTCTTTTCCT 3′)(68) | 97ºC for 3 min; 30 cycles of denaturation (94ºC for 35 s), annealing (50ºC for 45 s), and extension (72ºC for 45 s); and a final extension step at 72ºC for 7 min |
| ITS1 (5′ TCCGTAGGTGAACCTGCGG 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′)(67) | 94ºC for 3 min; 35 cycles of denaturation (94ºC for 60 s), annealing (56ºC for 60 s), and extension (72ºC for 2 min); and a final extension step at 72ºC for 7 min |
| ITS5 (5′ GGAAGTAAAAGTCGTAACAAGG 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′)(67) | 94ºC for 5 min.; 35 cycles of denaturation (94ºC for 30 s), annealing (55ºC for 1 min), and extension (72ºC 1 min and 20 s); and a final extension step at 72ºC for 7 min |
| ITS5 (5′ GGAAGTAAAAGTCGTAACAAGG 3′) and NL4b (5′ GGATTCTCACCCTCTATGAC 3′)(67,71) | 94ºC for 5 min.; 35 cycles of denaturation (94ºC for 30 s), annealing (53ºC for 1 min), and extension (72ºC 1 min and 30 s); and a final extension step at 72ºC for 7 min |
| V9D (5′ TTAAGTCCCTGCCCTTTGTA 3′) and LS266 (5′ GCATTCCCAAACAACTCGACTC 3′)(69) | 95ºC for 10 min; 30 cycles of denaturation (94ºC for 30 s), annealing (58ºC for 30 s), and extension (72ºC for 30 s); and a final extension step at 72ºC for 10 min |
| V9G (5′ TTACGTCCCTGCCCTTTGTA 3′) and LS266 (5′ GCATTCCCAAACAACTCGACTC 3′)(69) | 94ºC for 5 min; 35 cycles of denaturation (94ºC for 60 min), annealing (56ºC for 30 s), and extension (72ºC for 2 min); and a final extension step at 72ºC for 10 min |
| ITS1F (5′ CTTGGTCATTTAGAGGAAGTAA 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′)(67,70) | 95ºC for 5 min; 30 cycles of denaturation (95ºC for 30 s), annealing (58ºC for 30 s), and extension (72ºC for 1 min); and a final extension (72ºC for 10 min). |
| ITS1 (5′ TCCGTAGGTGAACCTGCGG 3′) and IT2 (5′ CCTCCGCTTATTGATATGCTTAGG 3′)(67,72) | 94ºC for 3 min; 35 cycles of denaturation (94ºC for 45 s), annealing (52ºC for 45 s), and extension (72ºC for 60 s); and a final extension at 72ºC for 7 min |
| ITS3 (5′ GCATCGATGAAGAACGCAGC 3′) and LS266 (5′ GCATTCCCAAACAACTCGACTC 3′)(67,69) | 95ºC for 10 min; 30 cycles of denaturation (94ºC for 30 s), annealing (58ºC for 30 s), and extension (72ºC for 30 s); and a final extension step at 72ºC for 10 min |
Primers and amplification conditions used to amplify ITS sequences maintained in the ISHAM-ITS reference database.
| Primers . | Amplification conditions . |
|---|---|
| SR6R (5′ AAGTATAAGTCGTAACAAGG 3′) and LR1 (5′ GGTTGGTTTCTTTTCCT 3′)(68) | 97ºC for 3 min; 30 cycles of denaturation (94ºC for 35 s), annealing (50ºC for 45 s), and extension (72ºC for 45 s); and a final extension step at 72ºC for 7 min |
| ITS1 (5′ TCCGTAGGTGAACCTGCGG 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′)(67) | 94ºC for 3 min; 35 cycles of denaturation (94ºC for 60 s), annealing (56ºC for 60 s), and extension (72ºC for 2 min); and a final extension step at 72ºC for 7 min |
| ITS5 (5′ GGAAGTAAAAGTCGTAACAAGG 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′)(67) | 94ºC for 5 min.; 35 cycles of denaturation (94ºC for 30 s), annealing (55ºC for 1 min), and extension (72ºC 1 min and 20 s); and a final extension step at 72ºC for 7 min |
| ITS5 (5′ GGAAGTAAAAGTCGTAACAAGG 3′) and NL4b (5′ GGATTCTCACCCTCTATGAC 3′)(67,71) | 94ºC for 5 min.; 35 cycles of denaturation (94ºC for 30 s), annealing (53ºC for 1 min), and extension (72ºC 1 min and 30 s); and a final extension step at 72ºC for 7 min |
| V9D (5′ TTAAGTCCCTGCCCTTTGTA 3′) and LS266 (5′ GCATTCCCAAACAACTCGACTC 3′)(69) | 95ºC for 10 min; 30 cycles of denaturation (94ºC for 30 s), annealing (58ºC for 30 s), and extension (72ºC for 30 s); and a final extension step at 72ºC for 10 min |
| V9G (5′ TTACGTCCCTGCCCTTTGTA 3′) and LS266 (5′ GCATTCCCAAACAACTCGACTC 3′)(69) | 94ºC for 5 min; 35 cycles of denaturation (94ºC for 60 min), annealing (56ºC for 30 s), and extension (72ºC for 2 min); and a final extension step at 72ºC for 10 min |
| ITS1F (5′ CTTGGTCATTTAGAGGAAGTAA 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′)(67,70) | 95ºC for 5 min; 30 cycles of denaturation (95ºC for 30 s), annealing (58ºC for 30 s), and extension (72ºC for 1 min); and a final extension (72ºC for 10 min). |
| ITS1 (5′ TCCGTAGGTGAACCTGCGG 3′) and IT2 (5′ CCTCCGCTTATTGATATGCTTAGG 3′)(67,72) | 94ºC for 3 min; 35 cycles of denaturation (94ºC for 45 s), annealing (52ºC for 45 s), and extension (72ºC for 60 s); and a final extension at 72ºC for 7 min |
| ITS3 (5′ GCATCGATGAAGAACGCAGC 3′) and LS266 (5′ GCATTCCCAAACAACTCGACTC 3′)(67,69) | 95ºC for 10 min; 30 cycles of denaturation (94ºC for 30 s), annealing (58ºC for 30 s), and extension (72ºC for 30 s); and a final extension step at 72ºC for 10 min |
| Primers . | Amplification conditions . |
|---|---|
| SR6R (5′ AAGTATAAGTCGTAACAAGG 3′) and LR1 (5′ GGTTGGTTTCTTTTCCT 3′)(68) | 97ºC for 3 min; 30 cycles of denaturation (94ºC for 35 s), annealing (50ºC for 45 s), and extension (72ºC for 45 s); and a final extension step at 72ºC for 7 min |
| ITS1 (5′ TCCGTAGGTGAACCTGCGG 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′)(67) | 94ºC for 3 min; 35 cycles of denaturation (94ºC for 60 s), annealing (56ºC for 60 s), and extension (72ºC for 2 min); and a final extension step at 72ºC for 7 min |
| ITS5 (5′ GGAAGTAAAAGTCGTAACAAGG 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′)(67) | 94ºC for 5 min.; 35 cycles of denaturation (94ºC for 30 s), annealing (55ºC for 1 min), and extension (72ºC 1 min and 20 s); and a final extension step at 72ºC for 7 min |
| ITS5 (5′ GGAAGTAAAAGTCGTAACAAGG 3′) and NL4b (5′ GGATTCTCACCCTCTATGAC 3′)(67,71) | 94ºC for 5 min.; 35 cycles of denaturation (94ºC for 30 s), annealing (53ºC for 1 min), and extension (72ºC 1 min and 30 s); and a final extension step at 72ºC for 7 min |
| V9D (5′ TTAAGTCCCTGCCCTTTGTA 3′) and LS266 (5′ GCATTCCCAAACAACTCGACTC 3′)(69) | 95ºC for 10 min; 30 cycles of denaturation (94ºC for 30 s), annealing (58ºC for 30 s), and extension (72ºC for 30 s); and a final extension step at 72ºC for 10 min |
| V9G (5′ TTACGTCCCTGCCCTTTGTA 3′) and LS266 (5′ GCATTCCCAAACAACTCGACTC 3′)(69) | 94ºC for 5 min; 35 cycles of denaturation (94ºC for 60 min), annealing (56ºC for 30 s), and extension (72ºC for 2 min); and a final extension step at 72ºC for 10 min |
| ITS1F (5′ CTTGGTCATTTAGAGGAAGTAA 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′)(67,70) | 95ºC for 5 min; 30 cycles of denaturation (95ºC for 30 s), annealing (58ºC for 30 s), and extension (72ºC for 1 min); and a final extension (72ºC for 10 min). |
| ITS1 (5′ TCCGTAGGTGAACCTGCGG 3′) and IT2 (5′ CCTCCGCTTATTGATATGCTTAGG 3′)(67,72) | 94ºC for 3 min; 35 cycles of denaturation (94ºC for 45 s), annealing (52ºC for 45 s), and extension (72ºC for 60 s); and a final extension at 72ºC for 7 min |
| ITS3 (5′ GCATCGATGAAGAACGCAGC 3′) and LS266 (5′ GCATTCCCAAACAACTCGACTC 3′)(67,69) | 95ºC for 10 min; 30 cycles of denaturation (94ºC for 30 s), annealing (58ºC for 30 s), and extension (72ºC for 30 s); and a final extension step at 72ºC for 10 min |
Data analysis
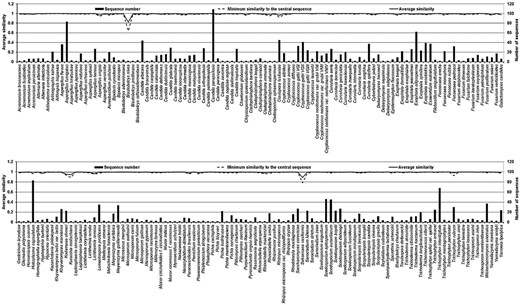
The length, continuity and annotation of the ITS sequences were checked using ITSx 1.0.7. [74] and membership in one species was verified by centrality analysis [75] using the software BioloMICS ver. 7.5.44 [76]. Briefly, sequences of each species were aligned to find the “central sequence”, which is the one having the highest average similarity to other members of the group. Questionable sequences that were very divergent from their central sequence, therefore doubtful as clear members of a species, were removed from further analyses. The sequences for each taxon were aligned using the program CLUSTALW [77] that is part of the software MEGA ver. 5.2.2 [78]. Resulting multiple alignments were then checked visually and edited when needed. For further analyses, the sequences were truncated at conserved sites to obtain equal 3′- and 5′-endings.
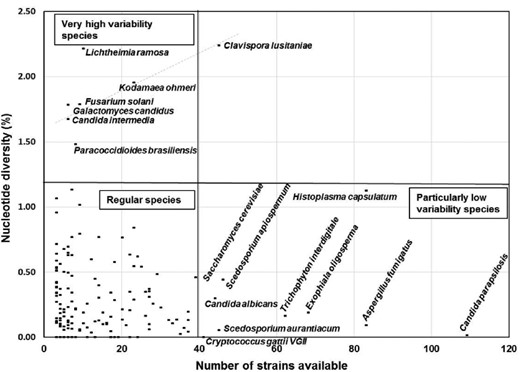
The intraspecies diversity was estimated by calculating the average nucleotide diversity (π), which gives the proportion of nucleotide differences in all haplotypes in the studied sample, the number of segregating polymorphic sites (S), and the proportion of polymorphic sites on base pair basis in a sample (Theta, Θ) of each species with sequences from more than two strains, using the software DnaSP ver. 5.10.01 [79].
For interspecies analyses, all taxa were subjected to pairwise sequence divergence calculations using the Kimura 2-parameter distance model (K2P) [80] using MEGA ver. 5.2.2. [78]. This model provides the best metric when genetic distances are low [81].
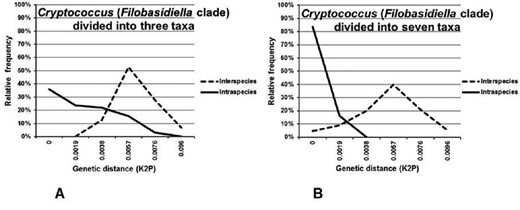
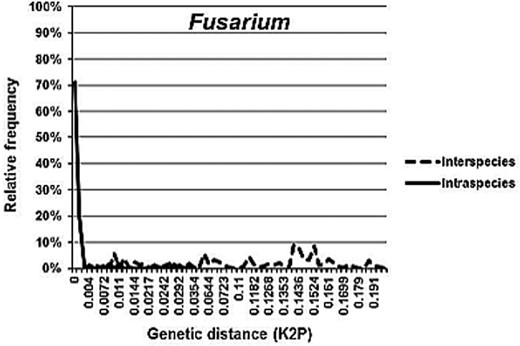
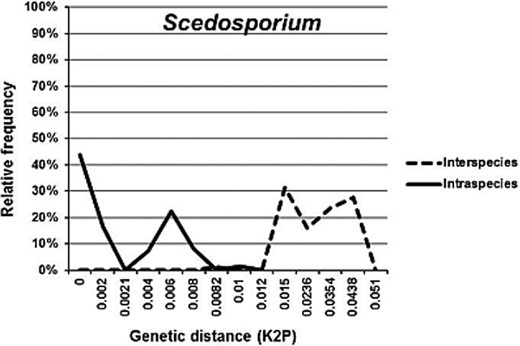
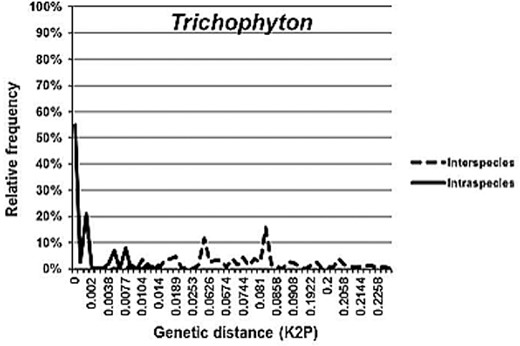
Barcoding gaps were evaluated by comparing the distribution of interspecies to intraspecies divergence within taxa sharing the same phylogenetic lineage [10]. In total, 17 barcoding gap analyses (of genera and phylogenetic clades), including two variants of the analysis for Cryptococcus neoformans/Cryptococcus gattii and Arthrodermataceae/Trichophyton, were performed (Table 3).
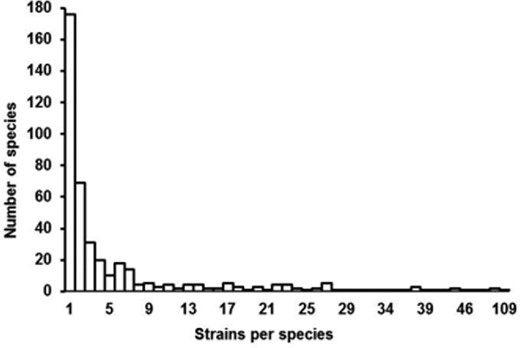
Intraspecies diversity of the 176 fungal species with more than two strains in the ISHAM-ITS reference database.
| . | . | Number of . | . | Number of . | Proportion of . | . |
|---|---|---|---|---|---|---|
| . | Number . | nucleotide . | Nucleotide . | polymorphic . | polymorphic sites . | ITS is sufficient . |
| Species . | of strains . | sites . | diversity (π) . | sites (S) . | in a sample (Θ) . | for identification . |
| Acremonium fusidioides | 3 | 520 | 0.00641 | 5 | 0.00641 | yes |
| Acremonium implicatum | 6 | 498 | 0.00375 | 5 | 0.000887 | yes |
| Acremonium persicinum | 6 | 494 | 0.00067 | 1 | 0.000887 | yes |
| Alternaria alternata | 7 | 475 | 0 | 0 | 0 | yes |
| Alternaria infectoria | 7 | 475 | 0 | 0 | 0 | yes |
| Arthrographis kalrae | 21 | 480 | 0.00091 | 2 | 0.001158 | yes |
| Arthropsis hispanica | 4 | 598 | 0.00251 | 3 | 0.002736 | yes |
| Aspergillus calidoustus | 5 | 482 | 0 | 0 | 0 | yes |
| Aspergillus flavus | 36 | 499 | 0.00071 | 1 | 0.000483 | yes |
| Aspergillus fumigatiaffinis | 4 | 505 | 0 | 0 | 0 | yes |
| Aspergillus fumigatus | 83 | 463 | 0.00094 | 6 | 0.002597 | yes |
| Aspergillus hiratsukae | 3 | 502 | 0.00531 | 4 | 0.005312 | yes |
| Aspergillus nidulans | 17 | 473 | 0.00047 | 1 | 0.000625 | yes |
| Aspergillus niger | 19 | 392 | 0 | 0 | 0 | yes |
| Aspergillus ochraceus | 3 | 491 | 0.00272 | 2 | 0.002716 | yes |
| Aspergillus sydowii | 3 | 480 | 0.00417 | 3 | 0.004167 | yes |
| Aspergillus terreus | 27 | 464 | 0.00061 | 2 | 0.001118 | yes |
| Aspergillus tubingensis | 18 | 425 | 0 | 0 | 0 | yes |
| Aspergillus versicolor | 6 | 433 | 0.00631 | 5 | 0.005057 | yes |
| Aureobasidium pullulans | 20 | 459 | 0.00764 | 15 | 0.009083 | yes |
| Bipolaris cynodontis | 9 | 376 | 0.00059 | 1 | 0.000981 | yes |
| Bipolaris micropus | 3 | 455 | 0.00147 | 1 | 0.001465 | yes |
| Blastobotrys adeninivorans | 4 | 547 | 0.00146 | 2 | 0.001755 | yes |
| Blastobotrys raffinosifermentans | 3 | 517 | 0.00387 | 3 | 0.003868 | yes |
| Candida albicans | 44 | 440 | 0.00298 | 10 | 0.005225 | yes |
| Candida blankii | 7 | 459 | 0 | 0 | 0 | yes |
| Candida carpophila | 3 | 602 | 0.00337 | 4 | 0.003681 | yes |
| Candida catenulata | 13 | 378 | 0.00122 | 1 | 0.000853 | yes |
| Candida deformans | 14 | 320 | 0.0077 | 7 | 0.008244 | yes |
| Candida diddensiae | 3 | 541 | 0 | 0 | 0 | yes |
| Candida dubliniensis | 16 | 451 | 0.00111 | 4 | 0.002673 | yes |
| Candida duobushaemulonis | 4 | 295 | 0 | 0 | 0 | yes |
| Candida glabrata | 29 | 791 | 0.00485 | 22 | 0.007304 | yes |
| Candida haemulonis | 6 | 285 | 0 | 0 | 0 | yes |
| Candida inconspicua | 7 | 413 | 0.0063 | 7 | 0.007423 | yes |
| Candida intermedia | 6 | 299 | 0.01672 | 12 | 0.017577 | yes |
| Candida mesorugosa | 13 | 314 | 0.00449 | 5 | 0.005131 | yes |
| Candida metapsilosis | 14 | 410 | 0.00397 | 4 | 0.003068 | yes |
| Candida orthopsilosis | 28 | 413 | 0.00255 | 5 | 0.005907 | yes |
| Candida palmioleophila | 3 | 632 | 0.00422 | 4 | 0.004219 | yes |
| Candida parapsilosis | 109 | 408 | 0.00014 | 2 | 0.000933 | yes |
| Candida pararugosa | 7 | 412 | 0.01133 | 11 | 0.010898 | yes |
| Candida tropicalis | 27 | 432 | 0.00352 | 13 | 0.007807 | yes |
| Candida zeylanoides | 4 | 579 | 0 | 0 | 0 | yes |
| Cladophialophora bantiana | 3 | 626 | 0 | 0 | 0 | yes |
| Cladophialophora boppii | 4 | 543 | 0.00184 | 2 | 0.002009 | yes |
| Cladophialophora carrionii | 6 | 538 | 0.00372 | 6 | 0.004884 | yes |
| Clavispora lusitaniae | 45 | 293 | 0.02248 | 22 | 0.018258 | no |
| Cryptococcus albidus | 18 | 583 | 0.00577 | 21 | 0.010472 | yes |
| Cryptococcus carnescens | 6 | 485 | 0 | 0 | 0 | yes |
| Cryptococcus diffluens | 3 | 612 | 0.00109 | 1 | 0.001089 | yes |
| Cryptococcus gattii VGI | 33 | 463 | 0.00108 | 1 | 0.000536 | yes |
| Cryptococcus gattii VGII | 41 | 463 | 0 | 0 | 0 | yes |
| Cryptococcus gattii VGIII | 24 | 463 | 0 | 0 | 0 | yes |
| Cryptococcus gattii VGIV | 13 | 463 | 0 | 0 | 0 | yes |
| Cryptococcus laurentii | 6 | 444 | 0.00495 | 4 | 0.003946 | yes |
| Cryptococcus magnus | 6 | 522 | 0 | 0 | 0 | yes |
| Cryptococcus neoformans var. grubii VNI | 22 | 452 | 0 | 0 | 0 | no |
| Cryptococcus neoformans var. grubii VNII | 13 | 460 | 0 | 0 | 0 | no |
| Cryptococcus neoformans var. neoformans VNIV | 17 | 463 | 0 | 0 | 0 | yes |
| Curvularia aeria | 27 | 442 | 0.00311 | 11 | 0.006457 | yes |
| Curvularia borreriae | 4 | 572 | 0.00322 | 3 | 0.002861 | yes |
| Curvularia geniculata | 15 | 503 | 0.00101 | 2 | 0.00125 | yes |
| Curvularia hawaiiensis | 20 | 379 | 0.00136 | 1 | 0.000755 | yes |
| Curvularia inaequalis | 6 | 518 | 0.00129 | 2 | 0.001691 | yes |
| Curvularia lunata | 10 | 467 | 0.00107 | 1 | 0.000788 | yes |
| Curvularia protuberata | 3 | 562 | 0 | 0 | 0 | yes |
| Curvularia sorghina | 4 | 490 | 0.00102 | 1 | 0.001113 | yes |
| Curvularia spicifera | 37 | 367 | 0.00044 | 3 | 0.001958 | yes |
| Curvularia verruculosa | 6 | 524 | 0 | 0 | 0 | yes |
| Cyberlindnera jadinii | 7 | 520 | 0.00769 | 10 | 0.007849 | yes |
| Debaryomyces hansenii | 15 | 540 | 0.00187 | 3 | 0.001709 | yes |
| Epidermophyton floccosum | 5 | 692 | 0.00058 | 1 | 0.000694 | yes |
| Exophiala bergeri | 9 | 495 | 0.01016 | 12 | 0.00892 | yes |
| Exophiala dermatitidis | 22 | 539 | 0.00347 | 9 | 0.004777 | yes |
| Exophiala exophialae | 3 | 538 | 0.00124 | 1 | 0.001239 | yes |
| Exophiala jeanselmei | 26 | 470 | 0.00349 | 10 | 0.005576 | yes |
| Exophiala oligosperma | 62 | 460 | 0.00165 | 3 | 0.001389 | yes |
| Exophiala spinifera | 23 | 501 | 0.00841 | 16 | 0.008653 | yes |
| Exophiala xenobiotica | 39 | 476 | 0.00458 | 18 | 0.008838 | yes |
| Exserohilum rostratum | 37 | 411 | 0.00197 | 10 | 0.00532 | yes |
| Filobasidium uniguttulatum | 4 | 616 | 0.00081 | 1 | 0.000885 | yes |
| Fonsecaea monophora | 22 | 528 | 0.00634 | 17 | 0.008832 | yes |
| Fonsecaea nubica | 3 | 512 | 0.00586 | 6 | 0.006392 | yes |
| Fonsecaea pedrosoi | 32 | 483 | 0.00132 | 5 | 0.00257 | yes |
| Fusarium delphinoides | 3 | 526 | 0 | 0 | 0 | yes |
| Fusarium falciforme | 7 | 458 | 0 | 0 | 0 | no |
| Fusarium keratoplasticum | 8 | 469 | 0.00213 | 6 | 0.004236 | no |
| Fusarium oxysporum | 14 | 455 | 0.00128 | 2 | 0.001382 | yes |
| Fusarium petroliphilum | 6 | 481 | 0.00091 | 1 | 0.00071 | no |
| Fusarium proliferatum | 11 | 451 | 0.00073 | 1 | 0.000757 | yes |
| Fusarium solani | 9 | 466 | 0.01788 | 21 | 0.016581 | no |
| Fusarium verticillioides | 17 | 455 | 0 | 0 | 0 | yes |
| Galactomyces candidus | 6 | 333 | 0.01782 | 10 | 0.013152 | yes |
| Hanseniaspora uvarum | 3 | 633 | 0.00316 | 3 | 0.00316 | yes |
| Histoplasma capsulatum | 83 | 416 | 0.01126 | 38 | 0.018351 | yes |
| Hormographiella aspergillata | 4 | 566 | 0.00088 | 1 | 0.000964 | yes |
| Hyphopichia burtonii | 5 | 359 | 0.00501 | 4 | 0.005348 | yes |
| Hypocrea orientalis | 7 | 438 | 0.00065 | 1 | 0.000932 | yes |
| Kazachstania pintolopesii | 3 | 650 | 0.00513 | 5 | 0.005128 | yes |
| Kluyveromyces lactis var. lactis | 11 | 618 | 0 | 0 | 0 | yes |
| Kluyveromyces marxianus | 26 | 603 | 0.00165 | 5 | 0.002173 | yes |
| Kodamaea ohmeri | 23 | 341 | 0.01954 | 23 | 0.018275 | no |
| Leptosphaeria senegalensis | 3 | 573 | 0.00116 | 1 | 0.001163 | yes |
| Lichtheimia corymbifera | 5 | 650 | 0.00677 | 11 | 0.008123 | yes |
| Lichtheimia ramosa | 10 | 770 | 0.02214 | 55 | 0.025054 | yes |
| Lomentospora prolificans | 35 | 475 | 0.00024 | 2 | 0.001022 | yes |
| Magnusiomyces capitatus | 4 | 365 | 0 | 0 | 0 | yes |
| Medicopsis romeroi | 3 | 467 | 0.00714 | 5 | 0.007138 | yes |
| Meyerozyma caribbica | 17 | 516 | 0.00155 | 3 | 0.001985 | yes |
| Meyerozyma guilliermondii | 34 | 516 | 0.00134 | 3 | 0.001444 | yes |
| Microascus cirrosus | 3 | 502 | 0 | 0 | 0 | yes |
| Microsporum audouinii | 7 | 666 | 0 | 0 | 0 | yes |
| Microsporum canis | 8 | 632 | 0 | 0 | 0 | yes |
| Microsporum fulvum | 6 | 617 | 0.00648 | 10 | 0.007098 | yes |
| Microsporum gypseum | 5 | 619 | 0 | 0 | 0 | yes |
| Microsporum racemosum | 3 | 556 | 0.00959 | 8 | 0.009592 | yes |
| Millerozyma farinosa | 3 | 626 | 0.01065 | 10 | 0.01065 | yes |
| Mucor circinelloides | 9 | 547 | 0.00792 | 11 | 0.007399 | yes |
| Neoscytalidium dimidiatum | 9 | 464 | 0.00048 | 1 | 0.000793 | yes |
| Paracoccidioides brasiliensis | 8 | 468 | 0.0148 | 17 | 0.01401 | yes |
| Penicillium brevicompactum | 3 | 539 | 0 | 0 | 0 | yes |
| Phialemonium atrogriseum | 3 | 524 | 0.00509 | 4 | 0.005089 | yes |
| Pichia kudriavzevii | 22 | 404 | 0.00206 | 4 | 0.002716 | yes |
| Pichia manshurica | 3 | 434 | 0 | 0 | 0 | yes |
| Pichia norvegensis | 14 | 398 | 0.00303 | 4 | 0.003239 | yes |
| Pithomyces chartarum | 7 | 568 | 0.00469 | 8 | 0.006168 | yes |
| Pithomyces sacchari | 6 | 549 | 0.00231 | 3 | 0.002393 | yes |
| Purpureocillium lilacinum | 5 | 501 | 0.0008 | 1 | 0.000958 | yes |
| Rasamsonia aegroticola | 10 | 467 | 0.0019 | 4 | 0.003151 | yes |
| Rhinocladiella similis | 18 | 497 | 0.00285 | 11 | 0.006435 | yes |
| Rhizomucor pusillus | 3 | 586 | 0.00341 | 3 | 0.003413 | yes |
| Rhizopus microsporus | 6 | 587 | 0.00693 | 8 | 0.005969 | yes |
| Rhizopus oryzae | 4 | 538 | 0.00217 | 2 | 0.002028 | yes |
| Rhodotorula mucilaginosa | 16 | 527 | 0.001 | 2 | 0.001144 | yes |
| Saccharomyces cerevisiae | 27 | 664 | 0.00098 | 7 | 0.002735 | yes |
| Sarocladium kiliense | 23 | 483 | 0.00546 | 16 | 0.009208 | yes |
| Sarocladium strictum | 8 | 484 | 0.00221 | 2 | 0.001594 | yes |
| Scedosporium angustum | 3 | 523 | 0.00382 | 3 | 0.003824 | yes |
| Scedosporium apiospermum | 46 | 497 | 0.00442 | 11 | 0.004587 | yes |
| Scedosporium aurantiacum | 45 | 497 | 0.00052 | 4 | 0.001841 | yes |
| Scedosporium boydii | 23 | 480 | 0.00287 | 9 | 0.005021 | yes |
| Scedosporium dehoogii | 27 | 518 | 0.0037 | 6 | 0.003005 | yes |
| Scedosporium ellipsoideum | 5 | 523 | 0.00191 | 2 | 0.001836 | yes |
| Scedosporium minutisporum | 7 | 520 | 0.00275 | 5 | 0.003925 | yes |
| Scopulariopsis brevicaulis | 17 | 459 | 0.00343 | 4 | 0.002578 | yes |
| Scopulariopsis brumptii | 7 | 416 | 0.00343 | 4 | 0.003925 | yes |
| Scopulariopsis cinerea | 5 | 502 | 0.00159 | 2 | 0.001912 | yes |
| Scopulariopsis gracilis | 12 | 533 | 0.00034 | 1 | 0.000621 | yes |
| Scytalidium cuboideum | 4 | 516 | 0.00129 | 1 | 0.001057 | yes |
| Sporothrix schenckii | 11 | 484 | 0.00255 | 4 | 0.002822 | yes |
| Torulaspora delbrueckii | 4 | 711 | 0.00563 | 8 | 0.006137 | yes |
| Trichoderma atroviride | 5 | 567 | 0.00212 | 3 | 0.00254 | yes |
| Trichoderma citrinoviride | 11 | 493 | 0.00074 | 2 | 0.001385 | yes |
| Trichoderma harzianum | 12 | 526 | 0.00599 | 9 | 0.005666 | yes |
| Trichoderma koningiopsis | 3 | 549 | 0 | 0 | 0 | yes |
| Trichoderma longibrachiatum | 20 | 521 | 0.00213 | 6 | 0.003246 | yes |
| Trichophyton ajelloi | 6 | 594 | 0.00112 | 2 | 0.001475 | yes |
| Trichophyton erinacei | 25 | 579 | 0.00541 | 16 | 0.007318 | yes |
| Trichophyton interdigitale | 68 | 525 | 0.00189 | 4 | 0.001591 | yes |
| Trichophyton mentagrophytes = T. quinckeanum | 5 | 603 | 0 | 0 | 0 | yes |
| Trichophyton persicolor | 3 | 601 | 0.00111 | 1 | 0.001109 | yes |
| Trichophyton rubrum | 30 | 540 | 0.00228 | 4 | 0.00187 | yes |
| Trichophyton schoenleinii | 4 | 623 | 0 | 0 | 0 | yes |
| Trichophyton simii | 7 | 608 | 0.00157 | 2 | 0.001343 | yes |
| Trichophyton terrestre | 4 | 615 | 0 | 0 | 0 | yes |
| Trichophyton tonsurans | 6 | 597 | 0.00112 | 2 | 0.001467 | yes |
| Trichophyton verrucosum | 4 | 534 | 0 | 0 | 0 | yes |
| Trichosporon asahii | 7 | 447 | 0.00107 | 1 | 0.000913 | yes |
| Trichosporon dermatis | 4 | 440 | 0 | 0 | 0 | yes |
| Trichosporon inkin | 4 | 539 | 0.00371 | 4 | 0.004048 | yes |
| Trichosporon montevideense | 4 | 528 | 0 | 0 | 0 | yes |
| Wickerhamomyces anomalus | 37 | 522 | 0.00131 | 7 | 0.003212 | yes |
| Yamadazyma mexicana | 3 | 561 | 0.00119 | 1 | 0.001188 | yes |
| Yamadazyma scolyti | 3 | 622 | 0.00536 | 5 | 0.005359 | yes |
| Yarrowia lipolytica | 24 | 347 | 0.0062 | 15 | 0.011576 | yes |
| . | . | Number of . | . | Number of . | Proportion of . | . |
|---|---|---|---|---|---|---|
| . | Number . | nucleotide . | Nucleotide . | polymorphic . | polymorphic sites . | ITS is sufficient . |
| Species . | of strains . | sites . | diversity (π) . | sites (S) . | in a sample (Θ) . | for identification . |
| Acremonium fusidioides | 3 | 520 | 0.00641 | 5 | 0.00641 | yes |
| Acremonium implicatum | 6 | 498 | 0.00375 | 5 | 0.000887 | yes |
| Acremonium persicinum | 6 | 494 | 0.00067 | 1 | 0.000887 | yes |
| Alternaria alternata | 7 | 475 | 0 | 0 | 0 | yes |
| Alternaria infectoria | 7 | 475 | 0 | 0 | 0 | yes |
| Arthrographis kalrae | 21 | 480 | 0.00091 | 2 | 0.001158 | yes |
| Arthropsis hispanica | 4 | 598 | 0.00251 | 3 | 0.002736 | yes |
| Aspergillus calidoustus | 5 | 482 | 0 | 0 | 0 | yes |
| Aspergillus flavus | 36 | 499 | 0.00071 | 1 | 0.000483 | yes |
| Aspergillus fumigatiaffinis | 4 | 505 | 0 | 0 | 0 | yes |
| Aspergillus fumigatus | 83 | 463 | 0.00094 | 6 | 0.002597 | yes |
| Aspergillus hiratsukae | 3 | 502 | 0.00531 | 4 | 0.005312 | yes |
| Aspergillus nidulans | 17 | 473 | 0.00047 | 1 | 0.000625 | yes |
| Aspergillus niger | 19 | 392 | 0 | 0 | 0 | yes |
| Aspergillus ochraceus | 3 | 491 | 0.00272 | 2 | 0.002716 | yes |
| Aspergillus sydowii | 3 | 480 | 0.00417 | 3 | 0.004167 | yes |
| Aspergillus terreus | 27 | 464 | 0.00061 | 2 | 0.001118 | yes |
| Aspergillus tubingensis | 18 | 425 | 0 | 0 | 0 | yes |
| Aspergillus versicolor | 6 | 433 | 0.00631 | 5 | 0.005057 | yes |
| Aureobasidium pullulans | 20 | 459 | 0.00764 | 15 | 0.009083 | yes |
| Bipolaris cynodontis | 9 | 376 | 0.00059 | 1 | 0.000981 | yes |
| Bipolaris micropus | 3 | 455 | 0.00147 | 1 | 0.001465 | yes |
| Blastobotrys adeninivorans | 4 | 547 | 0.00146 | 2 | 0.001755 | yes |
| Blastobotrys raffinosifermentans | 3 | 517 | 0.00387 | 3 | 0.003868 | yes |
| Candida albicans | 44 | 440 | 0.00298 | 10 | 0.005225 | yes |
| Candida blankii | 7 | 459 | 0 | 0 | 0 | yes |
| Candida carpophila | 3 | 602 | 0.00337 | 4 | 0.003681 | yes |
| Candida catenulata | 13 | 378 | 0.00122 | 1 | 0.000853 | yes |
| Candida deformans | 14 | 320 | 0.0077 | 7 | 0.008244 | yes |
| Candida diddensiae | 3 | 541 | 0 | 0 | 0 | yes |
| Candida dubliniensis | 16 | 451 | 0.00111 | 4 | 0.002673 | yes |
| Candida duobushaemulonis | 4 | 295 | 0 | 0 | 0 | yes |
| Candida glabrata | 29 | 791 | 0.00485 | 22 | 0.007304 | yes |
| Candida haemulonis | 6 | 285 | 0 | 0 | 0 | yes |
| Candida inconspicua | 7 | 413 | 0.0063 | 7 | 0.007423 | yes |
| Candida intermedia | 6 | 299 | 0.01672 | 12 | 0.017577 | yes |
| Candida mesorugosa | 13 | 314 | 0.00449 | 5 | 0.005131 | yes |
| Candida metapsilosis | 14 | 410 | 0.00397 | 4 | 0.003068 | yes |
| Candida orthopsilosis | 28 | 413 | 0.00255 | 5 | 0.005907 | yes |
| Candida palmioleophila | 3 | 632 | 0.00422 | 4 | 0.004219 | yes |
| Candida parapsilosis | 109 | 408 | 0.00014 | 2 | 0.000933 | yes |
| Candida pararugosa | 7 | 412 | 0.01133 | 11 | 0.010898 | yes |
| Candida tropicalis | 27 | 432 | 0.00352 | 13 | 0.007807 | yes |
| Candida zeylanoides | 4 | 579 | 0 | 0 | 0 | yes |
| Cladophialophora bantiana | 3 | 626 | 0 | 0 | 0 | yes |
| Cladophialophora boppii | 4 | 543 | 0.00184 | 2 | 0.002009 | yes |
| Cladophialophora carrionii | 6 | 538 | 0.00372 | 6 | 0.004884 | yes |
| Clavispora lusitaniae | 45 | 293 | 0.02248 | 22 | 0.018258 | no |
| Cryptococcus albidus | 18 | 583 | 0.00577 | 21 | 0.010472 | yes |
| Cryptococcus carnescens | 6 | 485 | 0 | 0 | 0 | yes |
| Cryptococcus diffluens | 3 | 612 | 0.00109 | 1 | 0.001089 | yes |
| Cryptococcus gattii VGI | 33 | 463 | 0.00108 | 1 | 0.000536 | yes |
| Cryptococcus gattii VGII | 41 | 463 | 0 | 0 | 0 | yes |
| Cryptococcus gattii VGIII | 24 | 463 | 0 | 0 | 0 | yes |
| Cryptococcus gattii VGIV | 13 | 463 | 0 | 0 | 0 | yes |
| Cryptococcus laurentii | 6 | 444 | 0.00495 | 4 | 0.003946 | yes |
| Cryptococcus magnus | 6 | 522 | 0 | 0 | 0 | yes |
| Cryptococcus neoformans var. grubii VNI | 22 | 452 | 0 | 0 | 0 | no |
| Cryptococcus neoformans var. grubii VNII | 13 | 460 | 0 | 0 | 0 | no |
| Cryptococcus neoformans var. neoformans VNIV | 17 | 463 | 0 | 0 | 0 | yes |
| Curvularia aeria | 27 | 442 | 0.00311 | 11 | 0.006457 | yes |
| Curvularia borreriae | 4 | 572 | 0.00322 | 3 | 0.002861 | yes |
| Curvularia geniculata | 15 | 503 | 0.00101 | 2 | 0.00125 | yes |
| Curvularia hawaiiensis | 20 | 379 | 0.00136 | 1 | 0.000755 | yes |
| Curvularia inaequalis | 6 | 518 | 0.00129 | 2 | 0.001691 | yes |
| Curvularia lunata | 10 | 467 | 0.00107 | 1 | 0.000788 | yes |
| Curvularia protuberata | 3 | 562 | 0 | 0 | 0 | yes |
| Curvularia sorghina | 4 | 490 | 0.00102 | 1 | 0.001113 | yes |
| Curvularia spicifera | 37 | 367 | 0.00044 | 3 | 0.001958 | yes |
| Curvularia verruculosa | 6 | 524 | 0 | 0 | 0 | yes |
| Cyberlindnera jadinii | 7 | 520 | 0.00769 | 10 | 0.007849 | yes |
| Debaryomyces hansenii | 15 | 540 | 0.00187 | 3 | 0.001709 | yes |
| Epidermophyton floccosum | 5 | 692 | 0.00058 | 1 | 0.000694 | yes |
| Exophiala bergeri | 9 | 495 | 0.01016 | 12 | 0.00892 | yes |
| Exophiala dermatitidis | 22 | 539 | 0.00347 | 9 | 0.004777 | yes |
| Exophiala exophialae | 3 | 538 | 0.00124 | 1 | 0.001239 | yes |
| Exophiala jeanselmei | 26 | 470 | 0.00349 | 10 | 0.005576 | yes |
| Exophiala oligosperma | 62 | 460 | 0.00165 | 3 | 0.001389 | yes |
| Exophiala spinifera | 23 | 501 | 0.00841 | 16 | 0.008653 | yes |
| Exophiala xenobiotica | 39 | 476 | 0.00458 | 18 | 0.008838 | yes |
| Exserohilum rostratum | 37 | 411 | 0.00197 | 10 | 0.00532 | yes |
| Filobasidium uniguttulatum | 4 | 616 | 0.00081 | 1 | 0.000885 | yes |
| Fonsecaea monophora | 22 | 528 | 0.00634 | 17 | 0.008832 | yes |
| Fonsecaea nubica | 3 | 512 | 0.00586 | 6 | 0.006392 | yes |
| Fonsecaea pedrosoi | 32 | 483 | 0.00132 | 5 | 0.00257 | yes |
| Fusarium delphinoides | 3 | 526 | 0 | 0 | 0 | yes |
| Fusarium falciforme | 7 | 458 | 0 | 0 | 0 | no |
| Fusarium keratoplasticum | 8 | 469 | 0.00213 | 6 | 0.004236 | no |
| Fusarium oxysporum | 14 | 455 | 0.00128 | 2 | 0.001382 | yes |
| Fusarium petroliphilum | 6 | 481 | 0.00091 | 1 | 0.00071 | no |
| Fusarium proliferatum | 11 | 451 | 0.00073 | 1 | 0.000757 | yes |
| Fusarium solani | 9 | 466 | 0.01788 | 21 | 0.016581 | no |
| Fusarium verticillioides | 17 | 455 | 0 | 0 | 0 | yes |
| Galactomyces candidus | 6 | 333 | 0.01782 | 10 | 0.013152 | yes |
| Hanseniaspora uvarum | 3 | 633 | 0.00316 | 3 | 0.00316 | yes |
| Histoplasma capsulatum | 83 | 416 | 0.01126 | 38 | 0.018351 | yes |
| Hormographiella aspergillata | 4 | 566 | 0.00088 | 1 | 0.000964 | yes |
| Hyphopichia burtonii | 5 | 359 | 0.00501 | 4 | 0.005348 | yes |
| Hypocrea orientalis | 7 | 438 | 0.00065 | 1 | 0.000932 | yes |
| Kazachstania pintolopesii | 3 | 650 | 0.00513 | 5 | 0.005128 | yes |
| Kluyveromyces lactis var. lactis | 11 | 618 | 0 | 0 | 0 | yes |
| Kluyveromyces marxianus | 26 | 603 | 0.00165 | 5 | 0.002173 | yes |
| Kodamaea ohmeri | 23 | 341 | 0.01954 | 23 | 0.018275 | no |
| Leptosphaeria senegalensis | 3 | 573 | 0.00116 | 1 | 0.001163 | yes |
| Lichtheimia corymbifera | 5 | 650 | 0.00677 | 11 | 0.008123 | yes |
| Lichtheimia ramosa | 10 | 770 | 0.02214 | 55 | 0.025054 | yes |
| Lomentospora prolificans | 35 | 475 | 0.00024 | 2 | 0.001022 | yes |
| Magnusiomyces capitatus | 4 | 365 | 0 | 0 | 0 | yes |
| Medicopsis romeroi | 3 | 467 | 0.00714 | 5 | 0.007138 | yes |
| Meyerozyma caribbica | 17 | 516 | 0.00155 | 3 | 0.001985 | yes |
| Meyerozyma guilliermondii | 34 | 516 | 0.00134 | 3 | 0.001444 | yes |
| Microascus cirrosus | 3 | 502 | 0 | 0 | 0 | yes |
| Microsporum audouinii | 7 | 666 | 0 | 0 | 0 | yes |
| Microsporum canis | 8 | 632 | 0 | 0 | 0 | yes |
| Microsporum fulvum | 6 | 617 | 0.00648 | 10 | 0.007098 | yes |
| Microsporum gypseum | 5 | 619 | 0 | 0 | 0 | yes |
| Microsporum racemosum | 3 | 556 | 0.00959 | 8 | 0.009592 | yes |
| Millerozyma farinosa | 3 | 626 | 0.01065 | 10 | 0.01065 | yes |
| Mucor circinelloides | 9 | 547 | 0.00792 | 11 | 0.007399 | yes |
| Neoscytalidium dimidiatum | 9 | 464 | 0.00048 | 1 | 0.000793 | yes |
| Paracoccidioides brasiliensis | 8 | 468 | 0.0148 | 17 | 0.01401 | yes |
| Penicillium brevicompactum | 3 | 539 | 0 | 0 | 0 | yes |
| Phialemonium atrogriseum | 3 | 524 | 0.00509 | 4 | 0.005089 | yes |
| Pichia kudriavzevii | 22 | 404 | 0.00206 | 4 | 0.002716 | yes |
| Pichia manshurica | 3 | 434 | 0 | 0 | 0 | yes |
| Pichia norvegensis | 14 | 398 | 0.00303 | 4 | 0.003239 | yes |
| Pithomyces chartarum | 7 | 568 | 0.00469 | 8 | 0.006168 | yes |
| Pithomyces sacchari | 6 | 549 | 0.00231 | 3 | 0.002393 | yes |
| Purpureocillium lilacinum | 5 | 501 | 0.0008 | 1 | 0.000958 | yes |
| Rasamsonia aegroticola | 10 | 467 | 0.0019 | 4 | 0.003151 | yes |
| Rhinocladiella similis | 18 | 497 | 0.00285 | 11 | 0.006435 | yes |
| Rhizomucor pusillus | 3 | 586 | 0.00341 | 3 | 0.003413 | yes |
| Rhizopus microsporus | 6 | 587 | 0.00693 | 8 | 0.005969 | yes |
| Rhizopus oryzae | 4 | 538 | 0.00217 | 2 | 0.002028 | yes |
| Rhodotorula mucilaginosa | 16 | 527 | 0.001 | 2 | 0.001144 | yes |
| Saccharomyces cerevisiae | 27 | 664 | 0.00098 | 7 | 0.002735 | yes |
| Sarocladium kiliense | 23 | 483 | 0.00546 | 16 | 0.009208 | yes |
| Sarocladium strictum | 8 | 484 | 0.00221 | 2 | 0.001594 | yes |
| Scedosporium angustum | 3 | 523 | 0.00382 | 3 | 0.003824 | yes |
| Scedosporium apiospermum | 46 | 497 | 0.00442 | 11 | 0.004587 | yes |
| Scedosporium aurantiacum | 45 | 497 | 0.00052 | 4 | 0.001841 | yes |
| Scedosporium boydii | 23 | 480 | 0.00287 | 9 | 0.005021 | yes |
| Scedosporium dehoogii | 27 | 518 | 0.0037 | 6 | 0.003005 | yes |
| Scedosporium ellipsoideum | 5 | 523 | 0.00191 | 2 | 0.001836 | yes |
| Scedosporium minutisporum | 7 | 520 | 0.00275 | 5 | 0.003925 | yes |
| Scopulariopsis brevicaulis | 17 | 459 | 0.00343 | 4 | 0.002578 | yes |
| Scopulariopsis brumptii | 7 | 416 | 0.00343 | 4 | 0.003925 | yes |
| Scopulariopsis cinerea | 5 | 502 | 0.00159 | 2 | 0.001912 | yes |
| Scopulariopsis gracilis | 12 | 533 | 0.00034 | 1 | 0.000621 | yes |
| Scytalidium cuboideum | 4 | 516 | 0.00129 | 1 | 0.001057 | yes |
| Sporothrix schenckii | 11 | 484 | 0.00255 | 4 | 0.002822 | yes |
| Torulaspora delbrueckii | 4 | 711 | 0.00563 | 8 | 0.006137 | yes |
| Trichoderma atroviride | 5 | 567 | 0.00212 | 3 | 0.00254 | yes |
| Trichoderma citrinoviride | 11 | 493 | 0.00074 | 2 | 0.001385 | yes |
| Trichoderma harzianum | 12 | 526 | 0.00599 | 9 | 0.005666 | yes |
| Trichoderma koningiopsis | 3 | 549 | 0 | 0 | 0 | yes |
| Trichoderma longibrachiatum | 20 | 521 | 0.00213 | 6 | 0.003246 | yes |
| Trichophyton ajelloi | 6 | 594 | 0.00112 | 2 | 0.001475 | yes |
| Trichophyton erinacei | 25 | 579 | 0.00541 | 16 | 0.007318 | yes |
| Trichophyton interdigitale | 68 | 525 | 0.00189 | 4 | 0.001591 | yes |
| Trichophyton mentagrophytes = T. quinckeanum | 5 | 603 | 0 | 0 | 0 | yes |
| Trichophyton persicolor | 3 | 601 | 0.00111 | 1 | 0.001109 | yes |
| Trichophyton rubrum | 30 | 540 | 0.00228 | 4 | 0.00187 | yes |
| Trichophyton schoenleinii | 4 | 623 | 0 | 0 | 0 | yes |
| Trichophyton simii | 7 | 608 | 0.00157 | 2 | 0.001343 | yes |
| Trichophyton terrestre | 4 | 615 | 0 | 0 | 0 | yes |
| Trichophyton tonsurans | 6 | 597 | 0.00112 | 2 | 0.001467 | yes |
| Trichophyton verrucosum | 4 | 534 | 0 | 0 | 0 | yes |
| Trichosporon asahii | 7 | 447 | 0.00107 | 1 | 0.000913 | yes |
| Trichosporon dermatis | 4 | 440 | 0 | 0 | 0 | yes |
| Trichosporon inkin | 4 | 539 | 0.00371 | 4 | 0.004048 | yes |
| Trichosporon montevideense | 4 | 528 | 0 | 0 | 0 | yes |
| Wickerhamomyces anomalus | 37 | 522 | 0.00131 | 7 | 0.003212 | yes |
| Yamadazyma mexicana | 3 | 561 | 0.00119 | 1 | 0.001188 | yes |
| Yamadazyma scolyti | 3 | 622 | 0.00536 | 5 | 0.005359 | yes |
| Yarrowia lipolytica | 24 | 347 | 0.0062 | 15 | 0.011576 | yes |
Intraspecies diversity of the 176 fungal species with more than two strains in the ISHAM-ITS reference database.
| . | . | Number of . | . | Number of . | Proportion of . | . |
|---|---|---|---|---|---|---|
| . | Number . | nucleotide . | Nucleotide . | polymorphic . | polymorphic sites . | ITS is sufficient . |
| Species . | of strains . | sites . | diversity (π) . | sites (S) . | in a sample (Θ) . | for identification . |
| Acremonium fusidioides | 3 | 520 | 0.00641 | 5 | 0.00641 | yes |
| Acremonium implicatum | 6 | 498 | 0.00375 | 5 | 0.000887 | yes |
| Acremonium persicinum | 6 | 494 | 0.00067 | 1 | 0.000887 | yes |
| Alternaria alternata | 7 | 475 | 0 | 0 | 0 | yes |
| Alternaria infectoria | 7 | 475 | 0 | 0 | 0 | yes |
| Arthrographis kalrae | 21 | 480 | 0.00091 | 2 | 0.001158 | yes |
| Arthropsis hispanica | 4 | 598 | 0.00251 | 3 | 0.002736 | yes |
| Aspergillus calidoustus | 5 | 482 | 0 | 0 | 0 | yes |
| Aspergillus flavus | 36 | 499 | 0.00071 | 1 | 0.000483 | yes |
| Aspergillus fumigatiaffinis | 4 | 505 | 0 | 0 | 0 | yes |
| Aspergillus fumigatus | 83 | 463 | 0.00094 | 6 | 0.002597 | yes |
| Aspergillus hiratsukae | 3 | 502 | 0.00531 | 4 | 0.005312 | yes |
| Aspergillus nidulans | 17 | 473 | 0.00047 | 1 | 0.000625 | yes |
| Aspergillus niger | 19 | 392 | 0 | 0 | 0 | yes |
| Aspergillus ochraceus | 3 | 491 | 0.00272 | 2 | 0.002716 | yes |
| Aspergillus sydowii | 3 | 480 | 0.00417 | 3 | 0.004167 | yes |
| Aspergillus terreus | 27 | 464 | 0.00061 | 2 | 0.001118 | yes |
| Aspergillus tubingensis | 18 | 425 | 0 | 0 | 0 | yes |
| Aspergillus versicolor | 6 | 433 | 0.00631 | 5 | 0.005057 | yes |
| Aureobasidium pullulans | 20 | 459 | 0.00764 | 15 | 0.009083 | yes |
| Bipolaris cynodontis | 9 | 376 | 0.00059 | 1 | 0.000981 | yes |
| Bipolaris micropus | 3 | 455 | 0.00147 | 1 | 0.001465 | yes |
| Blastobotrys adeninivorans | 4 | 547 | 0.00146 | 2 | 0.001755 | yes |
| Blastobotrys raffinosifermentans | 3 | 517 | 0.00387 | 3 | 0.003868 | yes |
| Candida albicans | 44 | 440 | 0.00298 | 10 | 0.005225 | yes |
| Candida blankii | 7 | 459 | 0 | 0 | 0 | yes |
| Candida carpophila | 3 | 602 | 0.00337 | 4 | 0.003681 | yes |
| Candida catenulata | 13 | 378 | 0.00122 | 1 | 0.000853 | yes |
| Candida deformans | 14 | 320 | 0.0077 | 7 | 0.008244 | yes |
| Candida diddensiae | 3 | 541 | 0 | 0 | 0 | yes |
| Candida dubliniensis | 16 | 451 | 0.00111 | 4 | 0.002673 | yes |
| Candida duobushaemulonis | 4 | 295 | 0 | 0 | 0 | yes |
| Candida glabrata | 29 | 791 | 0.00485 | 22 | 0.007304 | yes |
| Candida haemulonis | 6 | 285 | 0 | 0 | 0 | yes |
| Candida inconspicua | 7 | 413 | 0.0063 | 7 | 0.007423 | yes |
| Candida intermedia | 6 | 299 | 0.01672 | 12 | 0.017577 | yes |
| Candida mesorugosa | 13 | 314 | 0.00449 | 5 | 0.005131 | yes |
| Candida metapsilosis | 14 | 410 | 0.00397 | 4 | 0.003068 | yes |
| Candida orthopsilosis | 28 | 413 | 0.00255 | 5 | 0.005907 | yes |
| Candida palmioleophila | 3 | 632 | 0.00422 | 4 | 0.004219 | yes |
| Candida parapsilosis | 109 | 408 | 0.00014 | 2 | 0.000933 | yes |
| Candida pararugosa | 7 | 412 | 0.01133 | 11 | 0.010898 | yes |
| Candida tropicalis | 27 | 432 | 0.00352 | 13 | 0.007807 | yes |
| Candida zeylanoides | 4 | 579 | 0 | 0 | 0 | yes |
| Cladophialophora bantiana | 3 | 626 | 0 | 0 | 0 | yes |
| Cladophialophora boppii | 4 | 543 | 0.00184 | 2 | 0.002009 | yes |
| Cladophialophora carrionii | 6 | 538 | 0.00372 | 6 | 0.004884 | yes |
| Clavispora lusitaniae | 45 | 293 | 0.02248 | 22 | 0.018258 | no |
| Cryptococcus albidus | 18 | 583 | 0.00577 | 21 | 0.010472 | yes |
| Cryptococcus carnescens | 6 | 485 | 0 | 0 | 0 | yes |
| Cryptococcus diffluens | 3 | 612 | 0.00109 | 1 | 0.001089 | yes |
| Cryptococcus gattii VGI | 33 | 463 | 0.00108 | 1 | 0.000536 | yes |
| Cryptococcus gattii VGII | 41 | 463 | 0 | 0 | 0 | yes |
| Cryptococcus gattii VGIII | 24 | 463 | 0 | 0 | 0 | yes |
| Cryptococcus gattii VGIV | 13 | 463 | 0 | 0 | 0 | yes |
| Cryptococcus laurentii | 6 | 444 | 0.00495 | 4 | 0.003946 | yes |
| Cryptococcus magnus | 6 | 522 | 0 | 0 | 0 | yes |
| Cryptococcus neoformans var. grubii VNI | 22 | 452 | 0 | 0 | 0 | no |
| Cryptococcus neoformans var. grubii VNII | 13 | 460 | 0 | 0 | 0 | no |
| Cryptococcus neoformans var. neoformans VNIV | 17 | 463 | 0 | 0 | 0 | yes |
| Curvularia aeria | 27 | 442 | 0.00311 | 11 | 0.006457 | yes |
| Curvularia borreriae | 4 | 572 | 0.00322 | 3 | 0.002861 | yes |
| Curvularia geniculata | 15 | 503 | 0.00101 | 2 | 0.00125 | yes |
| Curvularia hawaiiensis | 20 | 379 | 0.00136 | 1 | 0.000755 | yes |
| Curvularia inaequalis | 6 | 518 | 0.00129 | 2 | 0.001691 | yes |
| Curvularia lunata | 10 | 467 | 0.00107 | 1 | 0.000788 | yes |
| Curvularia protuberata | 3 | 562 | 0 | 0 | 0 | yes |
| Curvularia sorghina | 4 | 490 | 0.00102 | 1 | 0.001113 | yes |
| Curvularia spicifera | 37 | 367 | 0.00044 | 3 | 0.001958 | yes |
| Curvularia verruculosa | 6 | 524 | 0 | 0 | 0 | yes |
| Cyberlindnera jadinii | 7 | 520 | 0.00769 | 10 | 0.007849 | yes |
| Debaryomyces hansenii | 15 | 540 | 0.00187 | 3 | 0.001709 | yes |
| Epidermophyton floccosum | 5 | 692 | 0.00058 | 1 | 0.000694 | yes |
| Exophiala bergeri | 9 | 495 | 0.01016 | 12 | 0.00892 | yes |
| Exophiala dermatitidis | 22 | 539 | 0.00347 | 9 | 0.004777 | yes |
| Exophiala exophialae | 3 | 538 | 0.00124 | 1 | 0.001239 | yes |
| Exophiala jeanselmei | 26 | 470 | 0.00349 | 10 | 0.005576 | yes |
| Exophiala oligosperma | 62 | 460 | 0.00165 | 3 | 0.001389 | yes |
| Exophiala spinifera | 23 | 501 | 0.00841 | 16 | 0.008653 | yes |
| Exophiala xenobiotica | 39 | 476 | 0.00458 | 18 | 0.008838 | yes |
| Exserohilum rostratum | 37 | 411 | 0.00197 | 10 | 0.00532 | yes |
| Filobasidium uniguttulatum | 4 | 616 | 0.00081 | 1 | 0.000885 | yes |
| Fonsecaea monophora | 22 | 528 | 0.00634 | 17 | 0.008832 | yes |
| Fonsecaea nubica | 3 | 512 | 0.00586 | 6 | 0.006392 | yes |
| Fonsecaea pedrosoi | 32 | 483 | 0.00132 | 5 | 0.00257 | yes |
| Fusarium delphinoides | 3 | 526 | 0 | 0 | 0 | yes |
| Fusarium falciforme | 7 | 458 | 0 | 0 | 0 | no |
| Fusarium keratoplasticum | 8 | 469 | 0.00213 | 6 | 0.004236 | no |
| Fusarium oxysporum | 14 | 455 | 0.00128 | 2 | 0.001382 | yes |
| Fusarium petroliphilum | 6 | 481 | 0.00091 | 1 | 0.00071 | no |
| Fusarium proliferatum | 11 | 451 | 0.00073 | 1 | 0.000757 | yes |
| Fusarium solani | 9 | 466 | 0.01788 | 21 | 0.016581 | no |
| Fusarium verticillioides | 17 | 455 | 0 | 0 | 0 | yes |
| Galactomyces candidus | 6 | 333 | 0.01782 | 10 | 0.013152 | yes |
| Hanseniaspora uvarum | 3 | 633 | 0.00316 | 3 | 0.00316 | yes |
| Histoplasma capsulatum | 83 | 416 | 0.01126 | 38 | 0.018351 | yes |
| Hormographiella aspergillata | 4 | 566 | 0.00088 | 1 | 0.000964 | yes |
| Hyphopichia burtonii | 5 | 359 | 0.00501 | 4 | 0.005348 | yes |
| Hypocrea orientalis | 7 | 438 | 0.00065 | 1 | 0.000932 | yes |
| Kazachstania pintolopesii | 3 | 650 | 0.00513 | 5 | 0.005128 | yes |
| Kluyveromyces lactis var. lactis | 11 | 618 | 0 | 0 | 0 | yes |
| Kluyveromyces marxianus | 26 | 603 | 0.00165 | 5 | 0.002173 | yes |
| Kodamaea ohmeri | 23 | 341 | 0.01954 | 23 | 0.018275 | no |
| Leptosphaeria senegalensis | 3 | 573 | 0.00116 | 1 | 0.001163 | yes |
| Lichtheimia corymbifera | 5 | 650 | 0.00677 | 11 | 0.008123 | yes |
| Lichtheimia ramosa | 10 | 770 | 0.02214 | 55 | 0.025054 | yes |
| Lomentospora prolificans | 35 | 475 | 0.00024 | 2 | 0.001022 | yes |
| Magnusiomyces capitatus | 4 | 365 | 0 | 0 | 0 | yes |
| Medicopsis romeroi | 3 | 467 | 0.00714 | 5 | 0.007138 | yes |
| Meyerozyma caribbica | 17 | 516 | 0.00155 | 3 | 0.001985 | yes |
| Meyerozyma guilliermondii | 34 | 516 | 0.00134 | 3 | 0.001444 | yes |
| Microascus cirrosus | 3 | 502 | 0 | 0 | 0 | yes |
| Microsporum audouinii | 7 | 666 | 0 | 0 | 0 | yes |
| Microsporum canis | 8 | 632 | 0 | 0 | 0 | yes |
| Microsporum fulvum | 6 | 617 | 0.00648 | 10 | 0.007098 | yes |
| Microsporum gypseum | 5 | 619 | 0 | 0 | 0 | yes |
| Microsporum racemosum | 3 | 556 | 0.00959 | 8 | 0.009592 | yes |
| Millerozyma farinosa | 3 | 626 | 0.01065 | 10 | 0.01065 | yes |
| Mucor circinelloides | 9 | 547 | 0.00792 | 11 | 0.007399 | yes |
| Neoscytalidium dimidiatum | 9 | 464 | 0.00048 | 1 | 0.000793 | yes |
| Paracoccidioides brasiliensis | 8 | 468 | 0.0148 | 17 | 0.01401 | yes |
| Penicillium brevicompactum | 3 | 539 | 0 | 0 | 0 | yes |
| Phialemonium atrogriseum | 3 | 524 | 0.00509 | 4 | 0.005089 | yes |
| Pichia kudriavzevii | 22 | 404 | 0.00206 | 4 | 0.002716 | yes |
| Pichia manshurica | 3 | 434 | 0 | 0 | 0 | yes |
| Pichia norvegensis | 14 | 398 | 0.00303 | 4 | 0.003239 | yes |
| Pithomyces chartarum | 7 | 568 | 0.00469 | 8 | 0.006168 | yes |
| Pithomyces sacchari | 6 | 549 | 0.00231 | 3 | 0.002393 | yes |
| Purpureocillium lilacinum | 5 | 501 | 0.0008 | 1 | 0.000958 | yes |
| Rasamsonia aegroticola | 10 | 467 | 0.0019 | 4 | 0.003151 | yes |
| Rhinocladiella similis | 18 | 497 | 0.00285 | 11 | 0.006435 | yes |
| Rhizomucor pusillus | 3 | 586 | 0.00341 | 3 | 0.003413 | yes |
| Rhizopus microsporus | 6 | 587 | 0.00693 | 8 | 0.005969 | yes |
| Rhizopus oryzae | 4 | 538 | 0.00217 | 2 | 0.002028 | yes |
| Rhodotorula mucilaginosa | 16 | 527 | 0.001 | 2 | 0.001144 | yes |
| Saccharomyces cerevisiae | 27 | 664 | 0.00098 | 7 | 0.002735 | yes |
| Sarocladium kiliense | 23 | 483 | 0.00546 | 16 | 0.009208 | yes |
| Sarocladium strictum | 8 | 484 | 0.00221 | 2 | 0.001594 | yes |
| Scedosporium angustum | 3 | 523 | 0.00382 | 3 | 0.003824 | yes |
| Scedosporium apiospermum | 46 | 497 | 0.00442 | 11 | 0.004587 | yes |
| Scedosporium aurantiacum | 45 | 497 | 0.00052 | 4 | 0.001841 | yes |
| Scedosporium boydii | 23 | 480 | 0.00287 | 9 | 0.005021 | yes |
| Scedosporium dehoogii | 27 | 518 | 0.0037 | 6 | 0.003005 | yes |
| Scedosporium ellipsoideum | 5 | 523 | 0.00191 | 2 | 0.001836 | yes |
| Scedosporium minutisporum | 7 | 520 | 0.00275 | 5 | 0.003925 | yes |
| Scopulariopsis brevicaulis | 17 | 459 | 0.00343 | 4 | 0.002578 | yes |
| Scopulariopsis brumptii | 7 | 416 | 0.00343 | 4 | 0.003925 | yes |
| Scopulariopsis cinerea | 5 | 502 | 0.00159 | 2 | 0.001912 | yes |
| Scopulariopsis gracilis | 12 | 533 | 0.00034 | 1 | 0.000621 | yes |
| Scytalidium cuboideum | 4 | 516 | 0.00129 | 1 | 0.001057 | yes |
| Sporothrix schenckii | 11 | 484 | 0.00255 | 4 | 0.002822 | yes |
| Torulaspora delbrueckii | 4 | 711 | 0.00563 | 8 | 0.006137 | yes |
| Trichoderma atroviride | 5 | 567 | 0.00212 | 3 | 0.00254 | yes |
| Trichoderma citrinoviride | 11 | 493 | 0.00074 | 2 | 0.001385 | yes |
| Trichoderma harzianum | 12 | 526 | 0.00599 | 9 | 0.005666 | yes |
| Trichoderma koningiopsis | 3 | 549 | 0 | 0 | 0 | yes |
| Trichoderma longibrachiatum | 20 | 521 | 0.00213 | 6 | 0.003246 | yes |
| Trichophyton ajelloi | 6 | 594 | 0.00112 | 2 | 0.001475 | yes |
| Trichophyton erinacei | 25 | 579 | 0.00541 | 16 | 0.007318 | yes |
| Trichophyton interdigitale | 68 | 525 | 0.00189 | 4 | 0.001591 | yes |
| Trichophyton mentagrophytes = T. quinckeanum | 5 | 603 | 0 | 0 | 0 | yes |
| Trichophyton persicolor | 3 | 601 | 0.00111 | 1 | 0.001109 | yes |
| Trichophyton rubrum | 30 | 540 | 0.00228 | 4 | 0.00187 | yes |
| Trichophyton schoenleinii | 4 | 623 | 0 | 0 | 0 | yes |
| Trichophyton simii | 7 | 608 | 0.00157 | 2 | 0.001343 | yes |
| Trichophyton terrestre | 4 | 615 | 0 | 0 | 0 | yes |
| Trichophyton tonsurans | 6 | 597 | 0.00112 | 2 | 0.001467 | yes |
| Trichophyton verrucosum | 4 | 534 | 0 | 0 | 0 | yes |
| Trichosporon asahii | 7 | 447 | 0.00107 | 1 | 0.000913 | yes |
| Trichosporon dermatis | 4 | 440 | 0 | 0 | 0 | yes |
| Trichosporon inkin | 4 | 539 | 0.00371 | 4 | 0.004048 | yes |
| Trichosporon montevideense | 4 | 528 | 0 | 0 | 0 | yes |
| Wickerhamomyces anomalus | 37 | 522 | 0.00131 | 7 | 0.003212 | yes |
| Yamadazyma mexicana | 3 | 561 | 0.00119 | 1 | 0.001188 | yes |
| Yamadazyma scolyti | 3 | 622 | 0.00536 | 5 | 0.005359 | yes |
| Yarrowia lipolytica | 24 | 347 | 0.0062 | 15 | 0.011576 | yes |
| . | . | Number of . | . | Number of . | Proportion of . | . |
|---|---|---|---|---|---|---|
| . | Number . | nucleotide . | Nucleotide . | polymorphic . | polymorphic sites . | ITS is sufficient . |
| Species . | of strains . | sites . | diversity (π) . | sites (S) . | in a sample (Θ) . | for identification . |
| Acremonium fusidioides | 3 | 520 | 0.00641 | 5 | 0.00641 | yes |
| Acremonium implicatum | 6 | 498 | 0.00375 | 5 | 0.000887 | yes |
| Acremonium persicinum | 6 | 494 | 0.00067 | 1 | 0.000887 | yes |
| Alternaria alternata | 7 | 475 | 0 | 0 | 0 | yes |
| Alternaria infectoria | 7 | 475 | 0 | 0 | 0 | yes |
| Arthrographis kalrae | 21 | 480 | 0.00091 | 2 | 0.001158 | yes |
| Arthropsis hispanica | 4 | 598 | 0.00251 | 3 | 0.002736 | yes |
| Aspergillus calidoustus | 5 | 482 | 0 | 0 | 0 | yes |
| Aspergillus flavus | 36 | 499 | 0.00071 | 1 | 0.000483 | yes |
| Aspergillus fumigatiaffinis | 4 | 505 | 0 | 0 | 0 | yes |
| Aspergillus fumigatus | 83 | 463 | 0.00094 | 6 | 0.002597 | yes |
| Aspergillus hiratsukae | 3 | 502 | 0.00531 | 4 | 0.005312 | yes |
| Aspergillus nidulans | 17 | 473 | 0.00047 | 1 | 0.000625 | yes |
| Aspergillus niger | 19 | 392 | 0 | 0 | 0 | yes |
| Aspergillus ochraceus | 3 | 491 | 0.00272 | 2 | 0.002716 | yes |
| Aspergillus sydowii | 3 | 480 | 0.00417 | 3 | 0.004167 | yes |
| Aspergillus terreus | 27 | 464 | 0.00061 | 2 | 0.001118 | yes |
| Aspergillus tubingensis | 18 | 425 | 0 | 0 | 0 | yes |
| Aspergillus versicolor | 6 | 433 | 0.00631 | 5 | 0.005057 | yes |
| Aureobasidium pullulans | 20 | 459 | 0.00764 | 15 | 0.009083 | yes |
| Bipolaris cynodontis | 9 | 376 | 0.00059 | 1 | 0.000981 | yes |
| Bipolaris micropus | 3 | 455 | 0.00147 | 1 | 0.001465 | yes |
| Blastobotrys adeninivorans | 4 | 547 | 0.00146 | 2 | 0.001755 | yes |
| Blastobotrys raffinosifermentans | 3 | 517 | 0.00387 | 3 | 0.003868 | yes |
| Candida albicans | 44 | 440 | 0.00298 | 10 | 0.005225 | yes |
| Candida blankii | 7 | 459 | 0 | 0 | 0 | yes |
| Candida carpophila | 3 | 602 | 0.00337 | 4 | 0.003681 | yes |
| Candida catenulata | 13 | 378 | 0.00122 | 1 | 0.000853 | yes |
| Candida deformans | 14 | 320 | 0.0077 | 7 | 0.008244 | yes |
| Candida diddensiae | 3 | 541 | 0 | 0 | 0 | yes |
| Candida dubliniensis | 16 | 451 | 0.00111 | 4 | 0.002673 | yes |
| Candida duobushaemulonis | 4 | 295 | 0 | 0 | 0 | yes |
| Candida glabrata | 29 | 791 | 0.00485 | 22 | 0.007304 | yes |
| Candida haemulonis | 6 | 285 | 0 | 0 | 0 | yes |
| Candida inconspicua | 7 | 413 | 0.0063 | 7 | 0.007423 | yes |
| Candida intermedia | 6 | 299 | 0.01672 | 12 | 0.017577 | yes |
| Candida mesorugosa | 13 | 314 | 0.00449 | 5 | 0.005131 | yes |
| Candida metapsilosis | 14 | 410 | 0.00397 | 4 | 0.003068 | yes |
| Candida orthopsilosis | 28 | 413 | 0.00255 | 5 | 0.005907 | yes |
| Candida palmioleophila | 3 | 632 | 0.00422 | 4 | 0.004219 | yes |
| Candida parapsilosis | 109 | 408 | 0.00014 | 2 | 0.000933 | yes |
| Candida pararugosa | 7 | 412 | 0.01133 | 11 | 0.010898 | yes |
| Candida tropicalis | 27 | 432 | 0.00352 | 13 | 0.007807 | yes |
| Candida zeylanoides | 4 | 579 | 0 | 0 | 0 | yes |
| Cladophialophora bantiana | 3 | 626 | 0 | 0 | 0 | yes |
| Cladophialophora boppii | 4 | 543 | 0.00184 | 2 | 0.002009 | yes |
| Cladophialophora carrionii | 6 | 538 | 0.00372 | 6 | 0.004884 | yes |
| Clavispora lusitaniae | 45 | 293 | 0.02248 | 22 | 0.018258 | no |
| Cryptococcus albidus | 18 | 583 | 0.00577 | 21 | 0.010472 | yes |
| Cryptococcus carnescens | 6 | 485 | 0 | 0 | 0 | yes |
| Cryptococcus diffluens | 3 | 612 | 0.00109 | 1 | 0.001089 | yes |
| Cryptococcus gattii VGI | 33 | 463 | 0.00108 | 1 | 0.000536 | yes |
| Cryptococcus gattii VGII | 41 | 463 | 0 | 0 | 0 | yes |
| Cryptococcus gattii VGIII | 24 | 463 | 0 | 0 | 0 | yes |
| Cryptococcus gattii VGIV | 13 | 463 | 0 | 0 | 0 | yes |
| Cryptococcus laurentii | 6 | 444 | 0.00495 | 4 | 0.003946 | yes |
| Cryptococcus magnus | 6 | 522 | 0 | 0 | 0 | yes |
| Cryptococcus neoformans var. grubii VNI | 22 | 452 | 0 | 0 | 0 | no |
| Cryptococcus neoformans var. grubii VNII | 13 | 460 | 0 | 0 | 0 | no |
| Cryptococcus neoformans var. neoformans VNIV | 17 | 463 | 0 | 0 | 0 | yes |
| Curvularia aeria | 27 | 442 | 0.00311 | 11 | 0.006457 | yes |
| Curvularia borreriae | 4 | 572 | 0.00322 | 3 | 0.002861 | yes |
| Curvularia geniculata | 15 | 503 | 0.00101 | 2 | 0.00125 | yes |
| Curvularia hawaiiensis | 20 | 379 | 0.00136 | 1 | 0.000755 | yes |
| Curvularia inaequalis | 6 | 518 | 0.00129 | 2 | 0.001691 | yes |
| Curvularia lunata | 10 | 467 | 0.00107 | 1 | 0.000788 | yes |
| Curvularia protuberata | 3 | 562 | 0 | 0 | 0 | yes |
| Curvularia sorghina | 4 | 490 | 0.00102 | 1 | 0.001113 | yes |
| Curvularia spicifera | 37 | 367 | 0.00044 | 3 | 0.001958 | yes |
| Curvularia verruculosa | 6 | 524 | 0 | 0 | 0 | yes |
| Cyberlindnera jadinii | 7 | 520 | 0.00769 | 10 | 0.007849 | yes |
| Debaryomyces hansenii | 15 | 540 | 0.00187 | 3 | 0.001709 | yes |
| Epidermophyton floccosum | 5 | 692 | 0.00058 | 1 | 0.000694 | yes |
| Exophiala bergeri | 9 | 495 | 0.01016 | 12 | 0.00892 | yes |
| Exophiala dermatitidis | 22 | 539 | 0.00347 | 9 | 0.004777 | yes |
| Exophiala exophialae | 3 | 538 | 0.00124 | 1 | 0.001239 | yes |
| Exophiala jeanselmei | 26 | 470 | 0.00349 | 10 | 0.005576 | yes |
| Exophiala oligosperma | 62 | 460 | 0.00165 | 3 | 0.001389 | yes |
| Exophiala spinifera | 23 | 501 | 0.00841 | 16 | 0.008653 | yes |
| Exophiala xenobiotica | 39 | 476 | 0.00458 | 18 | 0.008838 | yes |
| Exserohilum rostratum | 37 | 411 | 0.00197 | 10 | 0.00532 | yes |
| Filobasidium uniguttulatum | 4 | 616 | 0.00081 | 1 | 0.000885 | yes |
| Fonsecaea monophora | 22 | 528 | 0.00634 | 17 | 0.008832 | yes |
| Fonsecaea nubica | 3 | 512 | 0.00586 | 6 | 0.006392 | yes |
| Fonsecaea pedrosoi | 32 | 483 | 0.00132 | 5 | 0.00257 | yes |
| Fusarium delphinoides | 3 | 526 | 0 | 0 | 0 | yes |
| Fusarium falciforme | 7 | 458 | 0 | 0 | 0 | no |
| Fusarium keratoplasticum | 8 | 469 | 0.00213 | 6 | 0.004236 | no |
| Fusarium oxysporum | 14 | 455 | 0.00128 | 2 | 0.001382 | yes |
| Fusarium petroliphilum | 6 | 481 | 0.00091 | 1 | 0.00071 | no |
| Fusarium proliferatum | 11 | 451 | 0.00073 | 1 | 0.000757 | yes |
| Fusarium solani | 9 | 466 | 0.01788 | 21 | 0.016581 | no |
| Fusarium verticillioides | 17 | 455 | 0 | 0 | 0 | yes |
| Galactomyces candidus | 6 | 333 | 0.01782 | 10 | 0.013152 | yes |
| Hanseniaspora uvarum | 3 | 633 | 0.00316 | 3 | 0.00316 | yes |
| Histoplasma capsulatum | 83 | 416 | 0.01126 | 38 | 0.018351 | yes |
| Hormographiella aspergillata | 4 | 566 | 0.00088 | 1 | 0.000964 | yes |
| Hyphopichia burtonii | 5 | 359 | 0.00501 | 4 | 0.005348 | yes |
| Hypocrea orientalis | 7 | 438 | 0.00065 | 1 | 0.000932 | yes |
| Kazachstania pintolopesii | 3 | 650 | 0.00513 | 5 | 0.005128 | yes |
| Kluyveromyces lactis var. lactis | 11 | 618 | 0 | 0 | 0 | yes |
| Kluyveromyces marxianus | 26 | 603 | 0.00165 | 5 | 0.002173 | yes |
| Kodamaea ohmeri | 23 | 341 | 0.01954 | 23 | 0.018275 | no |
| Leptosphaeria senegalensis | 3 | 573 | 0.00116 | 1 | 0.001163 | yes |
| Lichtheimia corymbifera | 5 | 650 | 0.00677 | 11 | 0.008123 | yes |
| Lichtheimia ramosa | 10 | 770 | 0.02214 | 55 | 0.025054 | yes |
| Lomentospora prolificans | 35 | 475 | 0.00024 | 2 | 0.001022 | yes |
| Magnusiomyces capitatus | 4 | 365 | 0 | 0 | 0 | yes |
| Medicopsis romeroi | 3 | 467 | 0.00714 | 5 | 0.007138 | yes |
| Meyerozyma caribbica | 17 | 516 | 0.00155 | 3 | 0.001985 | yes |
| Meyerozyma guilliermondii | 34 | 516 | 0.00134 | 3 | 0.001444 | yes |
| Microascus cirrosus | 3 | 502 | 0 | 0 | 0 | yes |
| Microsporum audouinii | 7 | 666 | 0 | 0 | 0 | yes |
| Microsporum canis | 8 | 632 | 0 | 0 | 0 | yes |
| Microsporum fulvum | 6 | 617 | 0.00648 | 10 | 0.007098 | yes |
| Microsporum gypseum | 5 | 619 | 0 | 0 | 0 | yes |
| Microsporum racemosum | 3 | 556 | 0.00959 | 8 | 0.009592 | yes |
| Millerozyma farinosa | 3 | 626 | 0.01065 | 10 | 0.01065 | yes |
| Mucor circinelloides | 9 | 547 | 0.00792 | 11 | 0.007399 | yes |
| Neoscytalidium dimidiatum | 9 | 464 | 0.00048 | 1 | 0.000793 | yes |
| Paracoccidioides brasiliensis | 8 | 468 | 0.0148 | 17 | 0.01401 | yes |
| Penicillium brevicompactum | 3 | 539 | 0 | 0 | 0 | yes |
| Phialemonium atrogriseum | 3 | 524 | 0.00509 | 4 | 0.005089 | yes |
| Pichia kudriavzevii | 22 | 404 | 0.00206 | 4 | 0.002716 | yes |
| Pichia manshurica | 3 | 434 | 0 | 0 | 0 | yes |
| Pichia norvegensis | 14 | 398 | 0.00303 | 4 | 0.003239 | yes |
| Pithomyces chartarum | 7 | 568 | 0.00469 | 8 | 0.006168 | yes |
| Pithomyces sacchari | 6 | 549 | 0.00231 | 3 | 0.002393 | yes |
| Purpureocillium lilacinum | 5 | 501 | 0.0008 | 1 | 0.000958 | yes |
| Rasamsonia aegroticola | 10 | 467 | 0.0019 | 4 | 0.003151 | yes |
| Rhinocladiella similis | 18 | 497 | 0.00285 | 11 | 0.006435 | yes |
| Rhizomucor pusillus | 3 | 586 | 0.00341 | 3 | 0.003413 | yes |
| Rhizopus microsporus | 6 | 587 | 0.00693 | 8 | 0.005969 | yes |
| Rhizopus oryzae | 4 | 538 | 0.00217 | 2 | 0.002028 | yes |
| Rhodotorula mucilaginosa | 16 | 527 | 0.001 | 2 | 0.001144 | yes |
| Saccharomyces cerevisiae | 27 | 664 | 0.00098 | 7 | 0.002735 | yes |
| Sarocladium kiliense | 23 | 483 | 0.00546 | 16 | 0.009208 | yes |
| Sarocladium strictum | 8 | 484 | 0.00221 | 2 | 0.001594 | yes |
| Scedosporium angustum | 3 | 523 | 0.00382 | 3 | 0.003824 | yes |
| Scedosporium apiospermum | 46 | 497 | 0.00442 | 11 | 0.004587 | yes |
| Scedosporium aurantiacum | 45 | 497 | 0.00052 | 4 | 0.001841 | yes |
| Scedosporium boydii | 23 | 480 | 0.00287 | 9 | 0.005021 | yes |
| Scedosporium dehoogii | 27 | 518 | 0.0037 | 6 | 0.003005 | yes |
| Scedosporium ellipsoideum | 5 | 523 | 0.00191 | 2 | 0.001836 | yes |
| Scedosporium minutisporum | 7 | 520 | 0.00275 | 5 | 0.003925 | yes |
| Scopulariopsis brevicaulis | 17 | 459 | 0.00343 | 4 | 0.002578 | yes |
| Scopulariopsis brumptii | 7 | 416 | 0.00343 | 4 | 0.003925 | yes |
| Scopulariopsis cinerea | 5 | 502 | 0.00159 | 2 | 0.001912 | yes |
| Scopulariopsis gracilis | 12 | 533 | 0.00034 | 1 | 0.000621 | yes |
| Scytalidium cuboideum | 4 | 516 | 0.00129 | 1 | 0.001057 | yes |
| Sporothrix schenckii | 11 | 484 | 0.00255 | 4 | 0.002822 | yes |
| Torulaspora delbrueckii | 4 | 711 | 0.00563 | 8 | 0.006137 | yes |
| Trichoderma atroviride | 5 | 567 | 0.00212 | 3 | 0.00254 | yes |
| Trichoderma citrinoviride | 11 | 493 | 0.00074 | 2 | 0.001385 | yes |
| Trichoderma harzianum | 12 | 526 | 0.00599 | 9 | 0.005666 | yes |
| Trichoderma koningiopsis | 3 | 549 | 0 | 0 | 0 | yes |
| Trichoderma longibrachiatum | 20 | 521 | 0.00213 | 6 | 0.003246 | yes |
| Trichophyton ajelloi | 6 | 594 | 0.00112 | 2 | 0.001475 | yes |
| Trichophyton erinacei | 25 | 579 | 0.00541 | 16 | 0.007318 | yes |
| Trichophyton interdigitale | 68 | 525 | 0.00189 | 4 | 0.001591 | yes |
| Trichophyton mentagrophytes = T. quinckeanum | 5 | 603 | 0 | 0 | 0 | yes |
| Trichophyton persicolor | 3 | 601 | 0.00111 | 1 | 0.001109 | yes |
| Trichophyton rubrum | 30 | 540 | 0.00228 | 4 | 0.00187 | yes |
| Trichophyton schoenleinii | 4 | 623 | 0 | 0 | 0 | yes |
| Trichophyton simii | 7 | 608 | 0.00157 | 2 | 0.001343 | yes |
| Trichophyton terrestre | 4 | 615 | 0 | 0 | 0 | yes |
| Trichophyton tonsurans | 6 | 597 | 0.00112 | 2 | 0.001467 | yes |
| Trichophyton verrucosum | 4 | 534 | 0 | 0 | 0 | yes |
| Trichosporon asahii | 7 | 447 | 0.00107 | 1 | 0.000913 | yes |
| Trichosporon dermatis | 4 | 440 | 0 | 0 | 0 | yes |
| Trichosporon inkin | 4 | 539 | 0.00371 | 4 | 0.004048 | yes |
| Trichosporon montevideense | 4 | 528 | 0 | 0 | 0 | yes |
| Wickerhamomyces anomalus | 37 | 522 | 0.00131 | 7 | 0.003212 | yes |
| Yamadazyma mexicana | 3 | 561 | 0.00119 | 1 | 0.001188 | yes |
| Yamadazyma scolyti | 3 | 622 | 0.00536 | 5 | 0.005359 | yes |
| Yarrowia lipolytica | 24 | 347 | 0.0062 | 15 | 0.011576 | yes |
Sequence data were stored in BioloMICS ver. 7.5.44 [76] and statistical analyses were carried out in the statistical environment R [82].
Definitions
Species = a well-defined organism with a proven clinical relevance. Species complex = are organisms which form a cryptic species for which currently no proven evidence of individual medical relevance is known [83].
Results
Establishment of the quality controlled ISHAM-ITS reference database
A quality-controlled ITS reference database for human and animal pathogenic fungi was established as the result of the collaboration between 14 mycology laboratories from three continents. Altogether, the participating laboratories generated complete ITS (ITS1-5.8S-ITS2) sequences representing most of the pathogenic fungi. The number of ITS sequences and species contributed are shown in Table 1. According to the most recent taxonomic nomenclature, many species with different synonyms proved to be identical. Each sequence was associated with the current taxonomic species name, as well as with the most commonly used scientific names, used in a clinical setting. The sequences are freely accessible at http://www.isham.org/, directly from http://its.mycologylab.org/ or as specifically labelled ISHAM-ITS sequences in GenBank and UNITE. Of the 421 fungal species contained in the ISHAM-ITS sequences 71 representing the type culture of the species have also been submitted RTL at NCBI, following the principles laid out in Schoch et al. [57].
Number of sequences
At present, the quality-controlled ISHAM-ITS reference database contains 2800 complete ITS sequences representing 421 human/animal pathogenic fungal species. It contains 176 species represented by one strain, 69 species by two strains, and 176 species by a minimum of three to a maximum of 109 sequences. The distribution of strains per species was hyperbolic, meaning that the species with few strains were more frequent than those with many (Fig. 1).
Distribution of the number of strains per species in the ISHAM-ITS reference database.
Lengths of the ITS
The lengths of complete ITS sequences in the ISHAM-ITS reference database varied between 285 and 791 bp. The distribution of the number of nucleotides per sequence is given in Figure 2. The shortest complete ITS sequences were assigned to Candida haemulonis (285 bp), Clavispora lusitaniae (293 bp), and the longest ones to Candida glabrata (791 bp) and Lichtheimia ramosa (770 bp). The mean nucleotide length of ITS sequences in the database was 503 bp, while the median was 500 bp, indicating that the distribution of the sequence lengths was almost normal, with 0.08 skewness and 0.71 kurtosis (Fig. 2). These two metrics indicate that the population of sequences is centered around the average (skewness close to 0) and displays a more acute peak (kurtosis >0) than expected in a normal distribution. Altogether these metrics indicate that the sequences can be described rather well by a normal distribution very dense around the mean.
Length distribution of ITS sequences in the ISHAM-ITS reference database.
Quality of the database
There were 206 species, including 69 represented by only two strains, whose sequences showed diversity from the “central sequence” of the species. Figure 3 shows the average and the minimum similarity of the sequences to their central sequence as well as the number of the sequences within these species. The minimum similarity to the central sequence was less than 0.95% in the case of seven species, between 0.95–0.98% in 32 species and 0.98–0.998% in 167 species.
Average and the minimum similarity of the sequences to their central sequence as well as the number of the sequences within these species.
The average nucleotide diversity (π) was compared with the number of strains to test the hypothesis that the number of strain influences the variability. The nucleotide diversity and the number of strains did not show significant correlation, indicating that it is unlikely that the number of strains influences the variability. According to these two parameters, 160 out of the 176 species with more than two strains, were placed within a region spanning from 0 to 40 strains per species and from 0 to 1.1% variability within the species (Fig. 4). Six species (Lichtheimia ramosa, Fusarium solani, Kodamaea ohmeri, Galactomyces candidus, Candida intermedia, and Clavispora lusitaniae) showed a high intraspecies variability of up to 2.25% based on the value of π. Nine species (Histoplasma capsulatum, Scedosporium apiospermum, Scedosporium aurantiacum, Cryptococcus gattii VGII, Exophiala oligosperma, Trichophyton interdigitale, Aspergillus fumigatus, Candida parapsilosis, and Candida albicans) were in a region with less than 1.1% intraspecies variability, although the number of strains per species ranged from 40 to 109. Interestingly, this group of taxa with relatively low variability includes some of the more important pathogenic fungi namely A. fumigatus, C. parapsilosis, and C. albicans.
Nucleotide diversity (π) compared to the number of sequences by species in the ISHAM-ITS reference database.
Intraspecies genetic diversity of pathogenic fungal species in the ISHAM-ITS reference database
The two metrics of nucleotide diversity (π and Θ) generated very similar values (Table 3). The nucleotide diversity (π) estimated the proportion of nucleotide differences in all haplotypes and Θ measured the proportion of all segregating sites in a sample, thus being strongly influenced by rare haplotypes. The average nucleotide diversity per species was expressed as a percentage based on the value of π (Fig. 5).
Average nucleotide diversity per species expressed as a percentage based on the value of π of the 176 fungal species with more than three strains in the ISHAM-ITS reference database. The error bars indicate the standard deviation of nucleotide differences.
In the ISHAM-ITS reference database, the average nucleotide diversity was less than 0.5% for 138 species, between 0.5–1.0% in 27 species, 1.01–1.5% in five species (Exophiala bergeri, Millerozyma farinosa, H. capsulatum, Candida pararugosa, and Paracoccidioides brasiliensis), 1.5–2.0% in four species (C. intermedia, G. candidus, F. solani, and K. ohmeri), and more than 2% in two species (Lichtheimia ramosa and C. lusitaniae) (Table 3, Fig. 5).
The distribution of the distances from the “central sequence” of a species was hyperbolic, with the most frequent class, containing 63 species, representing more than one third of the species with more than two strains in the database, showing intraspecies variability ranging from 0 to 0.1%. More than half of the species with more than two strains in the database (97 species) were represented by species with less than 0.4% distance (Fig. 6).
Distribution of average distance of s within species compared to the number of species in the ISHAM-ITS reference database.
The polymorphic site distribution showed a similar result. In 117 species, the number of polymorphic sites was less than five, in 35 species it was between five and ten, in 11 species between 11 and 15, in six species between 16 and 20 and finally more than 20 in seven species. The species with the highest number of segregating sites were Cryptococcus albidus (21 sites), the complex of F. solani (21 sites), C. lusitaniae (22 sites), C. glabrata (22 sites), K. ohmeri (23 sites), H. capsulatum (38 sites), and L. ramosa (55 sites) (Table 3). The value of Θ showed a strong correlation with the average nucleotide diversity and the number of segregating sites. The proportion of rare haplotypes in a given sample was the highest in F. solani, C. lusitaniae, K. ohmeri, H. capsulatum, and L. ramosa (Table 3).
The intraspecies genetic analyses showed that the majority of medically important species had a low variability in ITS regions. Thus ITS sequencing can be used for the identification of most medical relevant fungal species (Table 3). The species with high intraspecies diversity within the ITS region require analysis of additional molecular markers to be reliably identified (see Table 4).
Taxa with high ITS diversity and alternative methods to be used for their reliable identification.
| Taxa . | Proposed alternatives . |
|---|---|
| Clavispora lusitaniae | Morphological identification by mating unknowns with a strain of known mating type(89,90) |
| Fusarium solani species complex (FSSC) | MLST(116); translation elongation factor 1-α (TEF-1α), RNA polymerase II gene (RPB2), secondary metabolite profiles(94) |
| Kodamaea ohmeri | Further taxonomic studies needed |
| Lichtheimia spp. | D1/D2 region, translation elongation factor 1-α (TEF-1α)(102); MALDI-TOF(105) |
| Cryptococcus | AFLP(110); PCR fingerprinting, RFLP of orotidine monophosphate pyrophosphorylase gene (URA5)(111); MLST(114) |
| Scedosporium | β -tubulin (BT2)(122), AFLP(121); LSU(124) |
| Arthrodermataceae | RAPD, PCR fingerprinting, AFLP, microsatellite markers(102) |
| Taxa . | Proposed alternatives . |
|---|---|
| Clavispora lusitaniae | Morphological identification by mating unknowns with a strain of known mating type(89,90) |
| Fusarium solani species complex (FSSC) | MLST(116); translation elongation factor 1-α (TEF-1α), RNA polymerase II gene (RPB2), secondary metabolite profiles(94) |
| Kodamaea ohmeri | Further taxonomic studies needed |
| Lichtheimia spp. | D1/D2 region, translation elongation factor 1-α (TEF-1α)(102); MALDI-TOF(105) |
| Cryptococcus | AFLP(110); PCR fingerprinting, RFLP of orotidine monophosphate pyrophosphorylase gene (URA5)(111); MLST(114) |
| Scedosporium | β -tubulin (BT2)(122), AFLP(121); LSU(124) |
| Arthrodermataceae | RAPD, PCR fingerprinting, AFLP, microsatellite markers(102) |
Taxa with high ITS diversity and alternative methods to be used for their reliable identification.
| Taxa . | Proposed alternatives . |
|---|---|
| Clavispora lusitaniae | Morphological identification by mating unknowns with a strain of known mating type(89,90) |
| Fusarium solani species complex (FSSC) | MLST(116); translation elongation factor 1-α (TEF-1α), RNA polymerase II gene (RPB2), secondary metabolite profiles(94) |
| Kodamaea ohmeri | Further taxonomic studies needed |
| Lichtheimia spp. | D1/D2 region, translation elongation factor 1-α (TEF-1α)(102); MALDI-TOF(105) |
| Cryptococcus | AFLP(110); PCR fingerprinting, RFLP of orotidine monophosphate pyrophosphorylase gene (URA5)(111); MLST(114) |
| Scedosporium | β -tubulin (BT2)(122), AFLP(121); LSU(124) |
| Arthrodermataceae | RAPD, PCR fingerprinting, AFLP, microsatellite markers(102) |
| Taxa . | Proposed alternatives . |
|---|---|
| Clavispora lusitaniae | Morphological identification by mating unknowns with a strain of known mating type(89,90) |
| Fusarium solani species complex (FSSC) | MLST(116); translation elongation factor 1-α (TEF-1α), RNA polymerase II gene (RPB2), secondary metabolite profiles(94) |
| Kodamaea ohmeri | Further taxonomic studies needed |
| Lichtheimia spp. | D1/D2 region, translation elongation factor 1-α (TEF-1α)(102); MALDI-TOF(105) |
| Cryptococcus | AFLP(110); PCR fingerprinting, RFLP of orotidine monophosphate pyrophosphorylase gene (URA5)(111); MLST(114) |
| Scedosporium | β -tubulin (BT2)(122), AFLP(121); LSU(124) |
| Arthrodermataceae | RAPD, PCR fingerprinting, AFLP, microsatellite markers(102) |
Barcoding gap analysis of the species represented in the ISHAM-ITS reference database
For the estimation of the barcoding gap the distribution of the Kimura 2-parameter (K2P) genetic distances within species and between species was calculated. In the ISHAM-ITS reference database, 17 taxonomical groups with more than two species sharing the same phylogenetic clade were identified based on previous data in MycoBank [63,64], Index Fungorum [65], and The Yeasts [66] (Table 5). The barcoding gap analysis was performed in all 17 taxa, including two versions of analysis for C. neoformans/C. gattii and Arthrodermataceae/Trichophyton (see Table 5). The distribution of genetic distances (intra- and interspecies) in each taxon is shown in Figures 7–10 and Supplementary Figures S1–S13. In 13 taxa (phylogenetic clades), a clear barcoding gap (K2P distance) was found (Table 5). The smallest barcoding gap (0.0002) was found in the Microsporum spp., while the largest one was found in the Cladophialophora spp. (0.09). In these cases, the highest intraspecies distances were smaller than the lowest genetic distances between species, creating a barcoding gap. For the remaining four taxa Cryptococcus (Fig. 7), Fusarium (Fig. 8), Scedosporium (Fig. 9), and Trichophyton (Fig. 10), it was not possible to define a clear barcoding gap, meaning that the distributions of genetic distances within and between species overlapped.
A) Distribution of interspecies (broken line) and intraspecies (solid line) pairwise Kimura 2-parameter genetic distances in Cryptococcus (Filobasidiella clade diveded into three taxa) including C. gattii; C. neoformans var. grubii; C. neoformans var. neoformans.B) Distribution of interspecies (broken line) and intraspecies (solid line) pairwise Kimura 2-parameter genetic distances in Cryptococcus (Filobasidiella clade diveded into seven taxa) including C. gattii VGI; C. gattii VGII; C. gattii VGIII; C. gattii VGIV; C. neoformans var. grubii VNI; C. neoformans var. grubii VNII; C. neoformans var. neoformans VNIV.
Distribution of interspecies (broken line) and intraspecies (solid line) pairwise Kimura 2-parameter genetic distances in Fusarium including F. delphinoides; F. falciforme; F. oxysporum; F. proliferatum; F. solani; F. keratoplasticum; F. petroliphilum; F. verticillioides.
Distribution of interspecies (broken line) and intraspecies (solid line) pairwise Kimura 2-parameter genetic distances in Scedosporium including S. angustum; S. apiospermum; S. aurantiacum; S. boydii; S. dehoogii; S. ellipsoideum; S. minutisporum.
Distribution of interspecies (broken line) and intraspecies (solid line) pairwise Kimura 2-parameter genetic distances in Trichophyton including T. ajelloi; T. erinacei; T. interdigitale; T. mentagrophytes (=T. quinckeanum); T. rubrum; T. schoenleinii; T. simii; T. terrestre; T. verrucosum.
Barcoding gap based on Kimura 2-parameter genetic distances in 17 studied phylogenetic clades represented by more than two species, with two variants of analysis for Cryptococcus neoformans/Cryptococcus gattii, and Arthrodermataceae/Trichophyton in the ISHAM-ITS reference database.
| Taxa . | Barcoding gap . | Species included in the analyses represented with more than two strains by species . |
|---|---|---|
| Acremonium | 0.055 | Acremonium fusidioides; A. implicatum; A. persicinum; Phialemonium atrogriseum; Sarocladium kiliense; S. strictum; |
| Arthrodermataceae | 0.002 | Arthroderma benhamiae; A. fulvum; A. gypseum; A. insingulare; A. otae; A. persicolor; A. simii; A. uncinatum; A. vanbreuseghemii |
| Aspergillus | 0.002 | Aspergillus calidoustus; A. flavus; A. fumigatiaffinis; A. fumigatus; A. hiratsukae; A. nidulans; A. niger; A. ochraceus; A. sydowii; A. terreus; A. tubingensis |
| Cladophialophora | 0.09 | Cladophialophora bantiana; C. boppii; C. carrionii |
| Cryptococcus (Filobasidiella clade divided into three taxa) | – | Cryptococcus gattii; C. neoformans var. grubii; C. neoformans var. neoformans |
| Cryptococcus (Filobasidiella clade divided into seven taxa) | – | Cryptococcus gattii VGI; C. gattii VGII; C. gattii VGIII; C. gattii VGIV; C. neoformans var. grubii VNI; C. neoformans var. grubii VNII; C. neoformans var. neoformans VNIV |
| Curvularia | 0.001 | Curvularia aeria; C. borreriae; C. inaequalis; C. geniculata; C. hawaiiensis; C. inaequalis; C. lunata; C. protuberata; C. spicifera; C. sorghina; C. verruculosa |
| Debaryomycetaceae (Lodderomyces clade) | 0.001 | Candida albicans; C. dubliniensis; C. metapsilosis; C. orthopsilosis; C. parapsilosis; C. tropicalis; Debaryomyces hansenii |
| Exophiala | 0.015 | Exophiala bergeri; E. dermatitidis; E. exophialae; E. jeanselmei; E. oligosperma; E. spinifera; E. xenobiotica |
| Fusarium | – | Fusarium delphinoides; F. falciforme; F. oxysporum; F. proliferatum; F. solani; F. keratoplasticum; F. petroliphilum; F. verticillioides |
| Metschnikowiaceae | 0.0603 | Candida duobushaemulonis; C. haemulonis; C. intermedia; C. lusitaniae; Kodamaea ohmeri |
| Microsporum | 0.0002 | Microsporum audouinii; M. canis; M. fulvum; M. gypseum |
| Pichiaceae | 0.005 | Pichia kudriavzevii; P. norvegensis; P. manshurica |
| Saccharomycetaceae | 0.009 | Kluyveromyces marxianus; K. lactis var. lactis; Saccharomyces cerevisiae; Torulaspora delbrueckii |
| Scedosporium | – | Scedosporium angustum; S. apiospermum; S. aurantiacum; S. boydii; S. dehoogii; S. ellipsoideum; S. minutisporum |
| Scopulariopsis | 0.0034 | Scopulariopsis brevicaulis; S. brumptii; S. cinerea; S. gracilis |
| Trichophyton | – | Trichophyton ajelloi; T. erinacei; T. interdigitale; T. mentagrophytes ( = T. quinckeanum); T. rubrum; T. schoenleinii; T. simii; T. terrestre; T. verrucosum |
| Trichosporon | 0.004 | Trichosporon asahii; T. dermatis; T. inkin; T. montevideense |
| Taxa . | Barcoding gap . | Species included in the analyses represented with more than two strains by species . |
|---|---|---|
| Acremonium | 0.055 | Acremonium fusidioides; A. implicatum; A. persicinum; Phialemonium atrogriseum; Sarocladium kiliense; S. strictum; |
| Arthrodermataceae | 0.002 | Arthroderma benhamiae; A. fulvum; A. gypseum; A. insingulare; A. otae; A. persicolor; A. simii; A. uncinatum; A. vanbreuseghemii |
| Aspergillus | 0.002 | Aspergillus calidoustus; A. flavus; A. fumigatiaffinis; A. fumigatus; A. hiratsukae; A. nidulans; A. niger; A. ochraceus; A. sydowii; A. terreus; A. tubingensis |
| Cladophialophora | 0.09 | Cladophialophora bantiana; C. boppii; C. carrionii |
| Cryptococcus (Filobasidiella clade divided into three taxa) | – | Cryptococcus gattii; C. neoformans var. grubii; C. neoformans var. neoformans |
| Cryptococcus (Filobasidiella clade divided into seven taxa) | – | Cryptococcus gattii VGI; C. gattii VGII; C. gattii VGIII; C. gattii VGIV; C. neoformans var. grubii VNI; C. neoformans var. grubii VNII; C. neoformans var. neoformans VNIV |
| Curvularia | 0.001 | Curvularia aeria; C. borreriae; C. inaequalis; C. geniculata; C. hawaiiensis; C. inaequalis; C. lunata; C. protuberata; C. spicifera; C. sorghina; C. verruculosa |
| Debaryomycetaceae (Lodderomyces clade) | 0.001 | Candida albicans; C. dubliniensis; C. metapsilosis; C. orthopsilosis; C. parapsilosis; C. tropicalis; Debaryomyces hansenii |
| Exophiala | 0.015 | Exophiala bergeri; E. dermatitidis; E. exophialae; E. jeanselmei; E. oligosperma; E. spinifera; E. xenobiotica |
| Fusarium | – | Fusarium delphinoides; F. falciforme; F. oxysporum; F. proliferatum; F. solani; F. keratoplasticum; F. petroliphilum; F. verticillioides |
| Metschnikowiaceae | 0.0603 | Candida duobushaemulonis; C. haemulonis; C. intermedia; C. lusitaniae; Kodamaea ohmeri |
| Microsporum | 0.0002 | Microsporum audouinii; M. canis; M. fulvum; M. gypseum |
| Pichiaceae | 0.005 | Pichia kudriavzevii; P. norvegensis; P. manshurica |
| Saccharomycetaceae | 0.009 | Kluyveromyces marxianus; K. lactis var. lactis; Saccharomyces cerevisiae; Torulaspora delbrueckii |
| Scedosporium | – | Scedosporium angustum; S. apiospermum; S. aurantiacum; S. boydii; S. dehoogii; S. ellipsoideum; S. minutisporum |
| Scopulariopsis | 0.0034 | Scopulariopsis brevicaulis; S. brumptii; S. cinerea; S. gracilis |
| Trichophyton | – | Trichophyton ajelloi; T. erinacei; T. interdigitale; T. mentagrophytes ( = T. quinckeanum); T. rubrum; T. schoenleinii; T. simii; T. terrestre; T. verrucosum |
| Trichosporon | 0.004 | Trichosporon asahii; T. dermatis; T. inkin; T. montevideense |
Barcoding gap based on Kimura 2-parameter genetic distances in 17 studied phylogenetic clades represented by more than two species, with two variants of analysis for Cryptococcus neoformans/Cryptococcus gattii, and Arthrodermataceae/Trichophyton in the ISHAM-ITS reference database.
| Taxa . | Barcoding gap . | Species included in the analyses represented with more than two strains by species . |
|---|---|---|
| Acremonium | 0.055 | Acremonium fusidioides; A. implicatum; A. persicinum; Phialemonium atrogriseum; Sarocladium kiliense; S. strictum; |
| Arthrodermataceae | 0.002 | Arthroderma benhamiae; A. fulvum; A. gypseum; A. insingulare; A. otae; A. persicolor; A. simii; A. uncinatum; A. vanbreuseghemii |
| Aspergillus | 0.002 | Aspergillus calidoustus; A. flavus; A. fumigatiaffinis; A. fumigatus; A. hiratsukae; A. nidulans; A. niger; A. ochraceus; A. sydowii; A. terreus; A. tubingensis |
| Cladophialophora | 0.09 | Cladophialophora bantiana; C. boppii; C. carrionii |
| Cryptococcus (Filobasidiella clade divided into three taxa) | – | Cryptococcus gattii; C. neoformans var. grubii; C. neoformans var. neoformans |
| Cryptococcus (Filobasidiella clade divided into seven taxa) | – | Cryptococcus gattii VGI; C. gattii VGII; C. gattii VGIII; C. gattii VGIV; C. neoformans var. grubii VNI; C. neoformans var. grubii VNII; C. neoformans var. neoformans VNIV |
| Curvularia | 0.001 | Curvularia aeria; C. borreriae; C. inaequalis; C. geniculata; C. hawaiiensis; C. inaequalis; C. lunata; C. protuberata; C. spicifera; C. sorghina; C. verruculosa |
| Debaryomycetaceae (Lodderomyces clade) | 0.001 | Candida albicans; C. dubliniensis; C. metapsilosis; C. orthopsilosis; C. parapsilosis; C. tropicalis; Debaryomyces hansenii |
| Exophiala | 0.015 | Exophiala bergeri; E. dermatitidis; E. exophialae; E. jeanselmei; E. oligosperma; E. spinifera; E. xenobiotica |
| Fusarium | – | Fusarium delphinoides; F. falciforme; F. oxysporum; F. proliferatum; F. solani; F. keratoplasticum; F. petroliphilum; F. verticillioides |
| Metschnikowiaceae | 0.0603 | Candida duobushaemulonis; C. haemulonis; C. intermedia; C. lusitaniae; Kodamaea ohmeri |
| Microsporum | 0.0002 | Microsporum audouinii; M. canis; M. fulvum; M. gypseum |
| Pichiaceae | 0.005 | Pichia kudriavzevii; P. norvegensis; P. manshurica |
| Saccharomycetaceae | 0.009 | Kluyveromyces marxianus; K. lactis var. lactis; Saccharomyces cerevisiae; Torulaspora delbrueckii |
| Scedosporium | – | Scedosporium angustum; S. apiospermum; S. aurantiacum; S. boydii; S. dehoogii; S. ellipsoideum; S. minutisporum |
| Scopulariopsis | 0.0034 | Scopulariopsis brevicaulis; S. brumptii; S. cinerea; S. gracilis |
| Trichophyton | – | Trichophyton ajelloi; T. erinacei; T. interdigitale; T. mentagrophytes ( = T. quinckeanum); T. rubrum; T. schoenleinii; T. simii; T. terrestre; T. verrucosum |
| Trichosporon | 0.004 | Trichosporon asahii; T. dermatis; T. inkin; T. montevideense |
| Taxa . | Barcoding gap . | Species included in the analyses represented with more than two strains by species . |
|---|---|---|
| Acremonium | 0.055 | Acremonium fusidioides; A. implicatum; A. persicinum; Phialemonium atrogriseum; Sarocladium kiliense; S. strictum; |
| Arthrodermataceae | 0.002 | Arthroderma benhamiae; A. fulvum; A. gypseum; A. insingulare; A. otae; A. persicolor; A. simii; A. uncinatum; A. vanbreuseghemii |
| Aspergillus | 0.002 | Aspergillus calidoustus; A. flavus; A. fumigatiaffinis; A. fumigatus; A. hiratsukae; A. nidulans; A. niger; A. ochraceus; A. sydowii; A. terreus; A. tubingensis |
| Cladophialophora | 0.09 | Cladophialophora bantiana; C. boppii; C. carrionii |
| Cryptococcus (Filobasidiella clade divided into three taxa) | – | Cryptococcus gattii; C. neoformans var. grubii; C. neoformans var. neoformans |
| Cryptococcus (Filobasidiella clade divided into seven taxa) | – | Cryptococcus gattii VGI; C. gattii VGII; C. gattii VGIII; C. gattii VGIV; C. neoformans var. grubii VNI; C. neoformans var. grubii VNII; C. neoformans var. neoformans VNIV |
| Curvularia | 0.001 | Curvularia aeria; C. borreriae; C. inaequalis; C. geniculata; C. hawaiiensis; C. inaequalis; C. lunata; C. protuberata; C. spicifera; C. sorghina; C. verruculosa |
| Debaryomycetaceae (Lodderomyces clade) | 0.001 | Candida albicans; C. dubliniensis; C. metapsilosis; C. orthopsilosis; C. parapsilosis; C. tropicalis; Debaryomyces hansenii |
| Exophiala | 0.015 | Exophiala bergeri; E. dermatitidis; E. exophialae; E. jeanselmei; E. oligosperma; E. spinifera; E. xenobiotica |
| Fusarium | – | Fusarium delphinoides; F. falciforme; F. oxysporum; F. proliferatum; F. solani; F. keratoplasticum; F. petroliphilum; F. verticillioides |
| Metschnikowiaceae | 0.0603 | Candida duobushaemulonis; C. haemulonis; C. intermedia; C. lusitaniae; Kodamaea ohmeri |
| Microsporum | 0.0002 | Microsporum audouinii; M. canis; M. fulvum; M. gypseum |
| Pichiaceae | 0.005 | Pichia kudriavzevii; P. norvegensis; P. manshurica |
| Saccharomycetaceae | 0.009 | Kluyveromyces marxianus; K. lactis var. lactis; Saccharomyces cerevisiae; Torulaspora delbrueckii |
| Scedosporium | – | Scedosporium angustum; S. apiospermum; S. aurantiacum; S. boydii; S. dehoogii; S. ellipsoideum; S. minutisporum |
| Scopulariopsis | 0.0034 | Scopulariopsis brevicaulis; S. brumptii; S. cinerea; S. gracilis |
| Trichophyton | – | Trichophyton ajelloi; T. erinacei; T. interdigitale; T. mentagrophytes ( = T. quinckeanum); T. rubrum; T. schoenleinii; T. simii; T. terrestre; T. verrucosum |
| Trichosporon | 0.004 | Trichosporon asahii; T. dermatis; T. inkin; T. montevideense |
Most of the studied taxa could be identified with the ITS barcode, although in some cases a clear discrimination could not be observed. There are two possible reasons for this: either the taxa is insufficiently studied or the ITS region is simply an inappropriate marker for discrimination between biologically consistent groups. Alternative loci and/or molecular methods are required for correct identification of these species (Table 5).
Discussion
ISHAM-ITS reference database
With a significant rise in the diversity of etiological agents of fungal infections in human and animal populations [1,2], rapid and accurate identification of pathogenic fungal species is one of the most important requirements for early and successful clinical treatment. As such, molecular information is expected to become a reliable tool for the identification of fungal species in medical diagnostic laboratories.
DNA barcoding represents a recent attempt to obtain rapid and accurate species identification based on comparative analysis of short but taxonomically significant sequences that has already found broad application in biology. However, the widespread application of fungal barcoding is hindered by a lack of reference databases. We herein report the establishment of the ISHAM-ITS reference database, containing 2800 quality controlled sequences, covering 421 human/animal pathogenic fungal species, which is publicly accessible at http://its.mycologylab.org/ and http://www.isham.org/. The principal roles of this reference database are to provide a reliable source for diagnostic medical and veterinary mycology laboratories, to enable correct identification of the causal agents of fungal infections, rapid diagnosis of mycoses, and early initiation of appropriate antifungal therapy (Fig. 11).
Proposed working flow to identify human and animal pathogenic fungi.
Intraspecies variation
The intraspecies genetic diversity of the ITS region varied between 0 and 2.25% but in 170 species it was less than 1.5%. The data generated in the present study are in agreement with previous studies stating that the genetic diversity of the ITS regions in fungi varies between taxa and that a single cut-off value cannot be established [33,84]. One could hypothesize that highly invasive fungal species show little variability because they are fully adapted to the host environment. However, further analyses are necessary to determine whether or not the variability calculated within the ITS regions is representative of the general genotypic and phenotypic variability within these species. Notably, the intraspecies diversity is more complex, with intragenomic polymorphism of rDNA repeats documented in a number of fungal species [36,85]. Observed intraspecies diversity in medical fungi may partly be due to the intragenomic polymorphism. Although we were not able to address this issue, its impact on the functionality of the database is mitigated because the ITS sequences contained in the ISHAM-ITS reference database are the result of direct sequencing which leads to the amplification of the most abundant sequence in the sample.
Taxa with high intraspecies variation for which identification based solely on the ITS region could be problematic
In the ISHAM-ITS reference database, only six fungal species (C. intermedia, C. lusitaniae, F. solani, G. candidus, K. ohmeri, and L. ramosa) revealed an intraspecies diversity of more than 1.5%.
Clavispora lusitaniae
Among these six species, C. lusitaniae (the teleomorph of Candida lusitaniae) causes approximately 1–2% of episodes of candidemia, including nosocomial outbreaks [86]. The species is exceptionally polymorphic in the ITS region and the D2 domain of the large-subunit rDNA gene, containing more than 30 substitutions [87,88]. In the ISHAM-ITS reference database, the average nucleotide diversity for this species was 2.19%, with 22 polymorphic sites, which may be a problem for identification of strains with sequences that are currently not represented in the database. In this case, correct species identification, may be determined by mating type for sexual reproduction [89,90] (see Table 4). The polyphyletic nature of the genus Clavispora was recently confirmed by multigene sequence analysis [91]. Further taxonomic studies are required for a better delimitation of this species.
Fusarium solani species complex (FSSC)
The second highest intraspecies variation was found amongst Fusarium species, which are primarily saprobes, plant pathogens, often linked with pathological infections, mainly keratitis, in both humans and animals. The F. solani species complex is the most common group of fusaria responsible for human infections, primarily in immunocompromised individuals [92,93]. Before taxonomical reanalysis the ISHAM-ITS database contained ten different Fusarium species including the highly polyphyletic FSSC. Seven of these species showed below 0.5% intraspecies variability suggesting a good taxonomic delimitation which can in turn allow easy identification with the ITS. However, within the FSSC the average nucleotide diversity was 3.76%, indicating that this complex has remained unresolved and contains multiple other cryptic species. According to the latest taxonomic studies [94], F. keratoplasticum, F. petroliphilum, and F. falciforme have been separated from the FSSC as new taxa, reducing the average nucleotide diversity to 1.65% in the ISHAM-ITS reference database. This variation still represents a significantly high degree of sequence diversity, making it necessary to employ different markers for correct identification at the species level (Table 4 and see below).
Galactomyces candidus
G. candidus (anamorph Geotrichum candidum) is a ubiquitous and dimorphic yeast, which occurs commonly on moist substrates rich in nutrients. Occasionally it is found as an opportunistic pathogen in the human respiratory and gastro-intestinal tracts [92,95]. The taxonomic classification of the species was revised in 2004 by de Hoog and Smith [96]. A standardized protocol was proposed for the identification of G. candidus at species and strain level in 2006 [97]. According to a recent study [38], the ITS region, especially the ITS1 region of G. candidus, proved to be highly polymorphic at intraspecies and intragenomic levels. In the ISHAM-ITS database, the species was represented by five strains with 1.78% genetic diversity, mainly in the ITS1 region. Although the 18S-ITS1-5.8S-ITS2-26S as a whole provides an improved phylogenetic resolution for the different phylotypes, use of the ITS region alone is not suitable for rapid identification of the species [38].
Kodamaea ohmeri
Using the ISHAM-ITS reference database, K. ohmeri (syn.: Pichia ohmeri, the teleomorph of Candida guilliermondii var. membranifaciens) has been found to contain high intraspecies diversity. This is an ascosporogenic yeast, mainly used in the food industry for fermentation, but has recently emerged as a fungal pathogen, particularly in immunocompromised patients [98,99]. However, few studies on this species have been done. Recently a number of species have been found with characteristics similar to those of K. ohmeri raising the possibility of cryptic species and the potential misidentification of previously described isolates [100]. Phylogenetic analyses of the ITS sequences contained in the ISHAM-ITS reference database supported two clades not previously identified. Further studies are needed to taxonomically resolve possible cryptic species.
Lichtheimia spp
The next group of fungi with marked ITS intraspecies variation was Lichtheimia species, which causes life-threating rhinocerebral and bronchorespiratory mucormycoses [101]. Multigene sequence analysis (ITS, 28S, EF-1α) of 38 isolates identified morphologically as L. corymbifera revealed a new species, named L. ramosa, which differed in morphology and nucleotide sequences from L. corymbifera [102]. To date, from the five recognized species of the genus Lichtheimia, only three L. corymbifera, L. ornata, and L. ramosa are of clinical relevance [103]. L. ramosa proved to be more polymorphic than L. corymbifera, with more than 2% diversity in the ITS sequences. Similar values for the ITS region of L. ramosa have been reported by Walther et al. in 2013 [104], suggesting that different groups among L. ramosa should be considered as a separate species. If so, the ITS region would be an appropriate marker for identification of these species. In view of the high diversity observed among ITS sequences within Lichtheimia, currently it is recommended to use either a multiple gene approach [102] or MALDI-TOF [105] for a reliable identification (see Table 4).
Barcoding gap analysis
At interspecies level, clear barcoding gaps, ranging from 0.0002 to 0.09, were found in 13 of 17 taxonomical clades, containing at least three species with more than two strains. These included the taxa Acremonium, Arthrodermataceae, Aspergillus, Cladophialophora, Curvularia, Debaryomycetaceae (Lodderomyces clade), Exophiala, Metschnikowiaceae, Microsporum, Pichiaceae, Saccharomycetaceae, Scopulariopsis, and Trichosporon. Thus, the identification of these species based on ITS sequences is reliable, the taxonomy of the groups is well defined and all the species in the current dataset are well delimited. However, four taxa showed no clear barcoding gap: Cryptococcus, Fusarium, Scedosporium, and Trichophyton. The species of these four clades require more insight to fully understand if and why the ITS barcoding fails to dissect this specific group or if these species are not yet well isolated from a taxonomic point of view. Additional molecular methods or genetic markers are required to accurately identify the species in this group (Table 4). The barcoding gap analyses presented herein are based on the current dataset in the ISHAM-ITS database, which may not reflect all known cryptic species of all studied taxa, for example, it is well known that A. fumigatus is species complex, and ITS will only enable an identification to the species complex, with additional sequencing of either β-tubulin [106] and calmodulin [107] being needed to identify the actual species.
Overall, ITS barcoding can be used as a screening system to evaluate and indicate to specialists which species require more attention at the taxonomic level.
Cryptococcus neoformans/C. gattii species complex
The C. neoformans/C. gattii species complex is a good example of how the delimitation of a species can be improved by molecular characterization. Cryptococcosis is a life-threatening systemic mycosis in a broad range of animals and humans. Most cases are due two species belonging to the family Tremellaceae. The causal agent of cryptococcosis was originally considered as one species until four serotypes were identified based on antigenic properties of the polysaccharide capsule [108]. Currently, the etiologic agents of cryptococcosis are divided into two species, C. neoformans (serotypes A, D, and AD) and C. gattii (serotypes B and C) [109]. Molecular genotyping methods have more recently revealed seven major haplotypes among the two species [110–113]. These include three lineages in C. neoformans (VNI/AFLP1, VNII/AFLP1A/1B, and VNIV/AFLP3) and four in C. gattii (VGI/AFLP4, VGII/AFLP6, VGIII/AFLP5 and VGIV/AFLP7) [114]. As with other species complexes, the C. neoformans/C. gattii species complex is a controversial topic and there is no agreement amongst taxonomists regarding the delimitation of the species. This is likely due to the absence of a consensus species definition for fungi. It has been suggested that every molecular type should be considered as a different variety or even as separate species [113]. The ISHAM-ITS reference database contains a large set of ITS sequences representing all seven major haploid molecular types of the C. neoformans/C. gattii species complex. In order to determine the effect of accurate taxonomic recognition, the genetic diversity within and between species was calculated in two different ways: (a) considering only C. neoformans and C. gattii as species and (b) considering the seven major haplotypes as “species.” In the first case, the average intraspecies diversity was 0.35% for C. gattii and 0.19% for C. neoformans. These values are consistent with genetic diversity within species. However, in the barcoding gap analyses the K2P genetic distances overlapped significantly (Table 5, Fig. 7A). In the second analysis based on the seven species assumption, the average genetic diversity among molecular types was 0–0.1%, which was significantly less variation than in the analysis based on the two-species assumption (Table 5, Fig. 7B). However, a clear barcoding gap was still absent, but the overlap was considerably less than in the first set. The only reason for the absence of a barcoding gap was that the VNI and VNII molecular types of C. neoformans could not be separated by ITS sequencing, which confirmed previous findings (43). Alternative methods are therefore needed to fully resolve this species complex. Currently AFLP analysis [110], URA5-RFLP analysis [111], MLMT/SCAR analysis [115] and MLST analysis using the ISHAM consensus MLST scheme for the C. neoformans/C. gattii species complex, which includes the following genetic loci: CAP59, GPD1, LAC1, PLB1, SOD1, URA5, and IGS1 [114] are recommended to separate all major molecular types/potential species in this species complex.
Fusarium solani species complex (FSSC)
The second group of fungi lacking a clear barcoding gap comprised the FSSC. No clear barcoding gap was identified amongst Fusarium species in the ISHAM-ITS database (Fig. 8). The overlap of the K2P genetic distance within and between species was undeniably due to the poorly resolved F. solani species complex. For correct species identification, the following additional genetic loci are recommended: translation elongation factor 1-α (TEF-1α) and the RNA polymerase II gene (RPB2) [94]. An MLST method, including eight protein-coding genes was also developed to identify species in FSSC [116] (Table 4).
Scedosporium
The third group that lacked a barcoding gap was the ascomycetous fungal species of the genus Scedosporium (Microascaceae) (Fig. 9). They are well known emerging pathogens, which are associated with important human diseases [117–119] and animal infections [120]. In this group, important taxonomic changes have been made in recent years using different molecular methodologies [121]. Based on several genetic markers including the ITS region, S. apiospermum and S. boydii have been re-evaluated, resulting in the definition of S. apiospermum (heterothallic teleomorph Pseudallescheria apiosperma), S. boydii (homothallic teleomorph Pseudallescheria boydii), S. dehoogii, S. minutisporum and S. aurantiacum [122,123]. The routine identification of species within the genus Scedosporium is complicated due to a high intraspecies but little constant interspecies variability in morphological characters mixed within the various synanamorphs and teleomorphs [123]. The ITS regions are a widely used molecular marker for the identification of these species, possibly in association with other markers. According to a new molecular study, these species can be reliably identified by ITS sequencing, although the distances between certain species (S. boydii and S. apiospermum) remain very small [121]. The identification of newly described species within the genus, S. ellipsoideum, S. fusoideum, and S. angustum is also questionable if only ITS sequences are used, as they cluster within S. boydii, with limited statistical support [121]. In the ISHAM-ITS reference database, the intraspecies diversity of Scedosporium species was low, indicating that they are all well-delineated taxa. The highest divergence was observed in S. apiospermum, S. boydii, and S. dehoogii. However, at interspecies level, no clear barcoding gap has been found since the smallest interspecies distances (S. boydii – S. apiospermum and S. boydii – S. ellipsoideum) were smaller than the biggest intraspecies distances found in S. apiospermum, S. boydii, and S. dehoogii. As such, to obtain a clear differentiation among all Scedosporium species, the amplification of the large subunit rRNA (LSU) [124], β-tubulin (BT2) [122], or AFLP [121] are recommended (Table 4).
Dermatophytes
The last group of species, which did not show a defined barcoding gap was the dermatophytes (Fig. 10). They comprise a highly polyphyletic group of fungi that attack keratinized tissue of humans and animals, causing dermatophytoses [125]. The anamorphic stages of dermatophyte species belong mainly to the genera Microsporum, Trichophyton, and Epidermophyton, while their teleomorphic stages belonged to Arthroderma [125]. The taxonomy of dermatophyte species has been changed and revised several times [126,127]. The nomenclature has recently become more unsettled because separate names are no longer used for the anamorph/teleomorph stages of fungi [62]. The application of different molecular and biochemical methods has largely contributed to the description, delineation and taxonomical re-evaluation of these species. However, many taxonomic questions still remain unresolved in these taxa. According to a recent phylogenetic study using four genetic markers, including the ITS region, many anamorph species in Trichophyton share the same teleomorph genus Arthroderma [128]. The most recent taxonomy, nomenclature and phylogeny of the family are summarized in a review by Cafarchia et al. [127].
Currently, two opposing concepts exist for the medically well-known species Trichophyton mentagrophytes. In a phylogenetic study of the T. mentagrophytes complex by Gräser et al. [129], three clades containing T. mentagrophytes varieties were recovered. Based on clinical and morphological data, most varieties were reduced to synonym species, whereas two were elevated to species level [126,130,131]. This resulted in three clades assigned to T. erinacei, T. interdigitale and to T. mentagrophytes. The third clade was composed of two strains: CBS 318.56, originally identified as T. mentagrophytes var. mentagrophytes, and CBS 106.67, originally identified as T. mentagrophytes var. quinckeanum. The latter strain was considered incorrectly identified, and CBS 318.56 was designated by Gräser et al. [129] as the neotype for T. mentagrophytes. The choice of this neotype has been under debate ever since, as T. mentagrophytes in this sense are now encountered rarely in clinical surveys that use DNA sequencing for identification. At the same time, an unnamed zoophilic species closely related to T. interdigitale was detected which appeared to be quite common and seemed to fit the original concept of T. mentagrophytes [132,133]. In an article verifying the new dermatophyte taxonomy using mating results and phylogenetic analyses, Kawasaki [128] states that the selected neotype only corresponds to strains of T. mentagrophytes var. quinckeanum, a rather rare dermatophyte causing favus predominantly in rodents. Beguin et al. [72] found that the neotype strain CBS 318.56 was included in a clade consisting exclusively of strains originally identified as T. (mentagrophytes var.) quinckeanum. They also provided arguments on why this epithet should not be disposed of as a nomen nudum. Although part of the medical mycological community disagrees with the current neotype for T. mentagrophytes, no alternative neotype for T. mentagrophytes has been proposed so far.
In the ISHAM-ITS reference database, the three major genera of the dermatophytes are present with a number of species, including six Microsporum, 15 Trichophyton, four Arthroderma and one Epidermophyton species. These species showed a high similarity at the intraspecies level, except T. erinacei, which had still less than 1% ITS sequence variation. To evaluate the interspecies diversity and estimate the existence of a barcoding gap, the distribution of interspecies/intraspecies divergence in the genera Trichophyton and Microsporum was compared. The results indicated that there was a clear, though very small barcoding gap in the genus Microsporum but not in the genus Trichophyton, where the two overlapped. There were species, for example, T. erinacei, where the intraspecies K2P genetic distance exceeded the interspecies K2P distances between two species. The difference in the ITS region was only a few nucleotides, e.g., between T. mentagrophytes (= T. quinckeanum strains) and T. schoenleinii or between T. tonsurans and T. interdigitale. However, evaluation of the former teleomorph stages of the species revealed that there was a clear barcoding gap (Supplementary Fig. S2) in the family Arthrodermataceae, since the different former anamorph species have a common former teleomorph genus. Based on the results of this study and the complex taxonomy of the dermatophytes it is strongly recommended that other molecular or biochemical features, for example, BT2, AFLP, PCR fingerprinting, or microsatellite analysis, be used to accurately identify the closely related species (T. schoenleinii – T. mentagrophytes ( = T. quinckeanum), T. tonsurans – T. interdigitale and T. verrucosum – T. erinacei) of this group [72,134] (Table 4).
Algorithm consideration
The occurrence of taxa without a barcoding gap can be explained by the fact that the algorithms which have long been used by the barcoding community to calculate the genetic distances (K2P) [80] or the algorithm used in BLAST [135] for sequence matching between the query sequence and reference sequences represent different approaches from those commonly used for phylogenetic analyses. Both K2P and BLAST approaches are based on simple sequence similarities. The most commonly applied method for species delimitation using phylogenetic approaches in mycology is the genealogical concordance phylogenetic species recognition (GCPSR), first proposed by Taylor et al. [136]. This relies on the concordant discrimination of characters from three or more unlinked loci. Phylogenetic analysis can be performed using a variety of algorithms relying on complex, computationally intensive evolutionary models based on “phylogenetic signals.” These methods are more robust and require more computational power and expertise. In exchange they give a more reliable summary of the evolutionary relatedness of the members of a specific taxonomic group. A common question often arises in the barcoding community whether a phylogenetic model is necessary for DNA barcode sequence analyses. In this study, we tested the discriminatory power of the official fungal barcode, the ITS regions [28], to identify human and animal pathogenic fungi and showed that it is efficient, using a simple sequence similarity based algorithm, for the identification of an unknown fungal disease agent in the majority of species. However, in sibling/cryptic species with only 1–2 bp differences, identification based only on ITS sequencing may be unreliable. Many articles have been published discriminating species by only one or two polymorphic sites in the ITS region [43,47,121]. However, the majority of these studies used phylogenetic approaches, e.g., maximum likelihood, parsimony or Bayesian analysis [137–139]. It should be noted that, in contrast to phylogenetic methods, the DNA barcoding approach focuses on the use of a universal marker that maximises the number of specimens to be examined, whilst lowering the time spent on processing and analysis. This approach can be simplified in two major indications, namely specimen identification and species discovery [140,141]. The method popularly used in DNA barcoding approaches, K2P genetic distances, does not capture the same level of species distinctiveness with limited genetic variation. [142]. This is especially true when only one marker is used in the barcoding analyses. Specimen identification works best in concert with a well-annotated reference database that incorporates species boundaries delimited with phylogenetic multi-gene analyses. However, due to the paucity of sequence data in many fungi DNA databases barcoding will provide a first sweep of species discovery that should eventually be verified with more robust phylogenetic methods.
A basic step in phylogenetic analysis is the global alignment of all sequences. Beyond causing excessive gap opening and extension when divergent sequences are compared, this approach requires all sequences to be of the same length. It is questionable whether in the hectic practice of diagnostic labs this level of sequence quality and analytical care can be obtained, when the presence of life threatening pathogens has to be determined. Distance based algorithms seem to better fit these situations, maybe with upgrades in terms of taxonomic and bioinformatics conception [75,143,144], and with flexible distance algorithms [76].
The lack of interspecies gaps paves the way to three basic questions: (i) Is this relevant in the diagnostic practice? (ii) Is it due to unresolved taxonomy or to the intrinsic low power of the ITS barcode? and (iii) Are there taxonomic approaches and bioinformatics pipelines to reduce or resolve this problem?
The first question is a trivial one, but as long as the therapies for the unresolved species are similar, the lack of specific gaps is more a biological than a clinical problem. An attentive analysis from this point of view should accompany the purely taxonomic search, in order to pay particular attention to unresolved groups requiring different drug treatments. The second question is more complex. Many fungal species are not easily resolved for an exceeding number of taxonomic questions no matter of the single marker used. More insight on this point is necessary, maybe to develop easy to read indexes describing the ratio between the single marker vs. multiparameter species delimitation. This type of analysis seems to be necessary for further development of molecular markers in order to define their effective “taxonomic resolution power”. The evidence that many species presented a large variability does not impair the validity of ITS as a barcoding gene but suggest that particular attention must be paid in delimiting large species at the taxonomic level. Finally, the third question calls for a more attentive analysis of the species structure and of the algorithms necessary to discriminate them in fungi.
As a result of this study a quality–controlled reference ITS database, containing 2800 strains covering 421 species has been established and is publically accessible at http://its.mycologylab.org/ and http://www.isham.org/. The sequences selected in this study expand the number of medical species represented in the RTL ITS reference database at NCBI. There are several sequences with type information shared between the ISHAM-ITS database and RTL. Curators at NCBI will continuously verify additional single ITS accessions representing species where type information is currently unavailable. After a series of verifications these will serve as “verified” reference sequences [57] until a sequence obtained from type material is available. ISHAM-ITS database records are linked with their appropriate records at NCBI, similarly to the existing link between GenBank records and the UNITE and BOLD databases using Linkout (http://www.ncbi.nlm.nih.gov/projects/linkout/) and db_xref (http://www.ncbi.nlm.nih.gov/genbank/collab/db_xref) links. The results of the analysis of the sequences maintained in the database showed that ITS works well as a barcode for the majority of species. However, it has limitations in resolving species within species complexes and in sibling species delineation, where the difference of only one or a few nucleotide positions exist at the ITS locus. This study does not intend to challenge the current taxonomy of any fungal taxon. The goal was to highlight those taxa for the scientific community where additional genetic markers or molecular algorithms should be used for the reliable species identification.
Call for participation
The database is intended to cover all clinically relevant fungal species. It is open for further sequence submission to cover all medially relevant species with a sufficient number of strains, either via direct submission through the database (http://its.mycologylab.org/) or contacting the curators of the database (Prof. Wieland Meyer, wieland.meyer@sydney.edu.au or Laszlo Irinyi, laszlo.irinyi@sydney.edu.au).
This study was supported by an National Health and Medical Research Council of Australia (NH&MRC) grant [#APP1031952] to W Meyer, S Chen, V Robert, and D Ellis; CNPq [350338/2000-0] and FAPERJ [E-26/103.157/2011] grants to RM Zancopé-Oliveira; CNPq [308011/2010-4] and FAPESP [2007/08575-1] Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) grants to AL Colombo; PEst-OE/BIA/UI4050/2014 from Fundação para a Ciência e Tecnologia (FCT) to C Pais; the Belgian Science Policy Office (Belspo) to BCCM/IHEM; the MEXBOL program of CONACyT-Mexico, [ref. number: 122896] to ML Taylor and [122481] to C Toriello; the Institut Pasteur and Institut de Veille Sanitaire to F Dromer and D Garcia-Hermoso; and the grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and the Fundação de Amparo a Pesquisa do Estado de Goiás (FAPEG) to CM de Almeida Soares and JA Parente Rocha. I Arthur would like to thank G Cherian, A Higgins and the staff of the Molecular Diagnostics Laboratory, Division of Microbiology and Infectious Diseases, PathWest, QEII Medical Centre. F Dromer would like to thank for the technical help of the sequencing facility and specifically that of L Diancourt, A-S Delannoy-Vieillard, J-M Thiberge (Genotyping of Pathogens and Public Health, Institut Pasteur). RM Zancopé-Oliveira would like to thank the Genomic/DNA Sequencing Platform at Fundação Oswaldo Cruz—PDTIS/FIOCRUZ [RPT01A], Brazil for the sequencing. B Robbertse and CL Schoch acknowledge support from the Intramural Research Program of the NIH, National Library of Medicine. T Sorrell's work is funded by the NH&MRC of Australia; she is a Sydney Medical School Foundation Fellow.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and the writing of the paper.
Supplementary material
Supplementary material is available at Medical Mycology online (http://www.mmy.oxfordjournals.org/).