-
PDF
- Split View
-
Views
-
Cite
Cite
P.J. Volders, K. Verheggen, G. Menschaert, K. Vandepoele, L. Martens, J. Vandesompele, P. Mestdagh, An update on LNCipedia: a database for annotated human lncRNA sequences, Nucleic Acids Research, Volume 43, Issue 8, 30 April 2015, Pages 4363–4364, https://doi.org/10.1093/nar/gkv295
Close - Share Icon Share
Nucleic Acids Res. 2015 Jan;43(Database issue):D174–80. doi: 10.1093/nar/gku1060.
LNCipedia collects long non-coding RNA sequences and annotation from different sources. In version 3.0, over 90,000 new transcripts were added to the database. 6917 of these transcripts were obtained from RefSeq by filtering for accession prefix (NR_) and size (200bp). This filtering strategy however, does not confine to long non-coding RNAs and also yields transcripts associated with protein coding genes. Transcripts with incomplete open reading frames that are subject to nonsense-mediated mRNA decay for instance are also annotated with accession prefix NR_. These transcripts are generally not considered as true lncRNAs and typically exhibit a high coding potential score when assessed by PhyloCSF. The authors therefore chose to exclude these transcripts from the database and confine their analysis to the RefSeq subset with keyword biomol_ncrna_lncrna as suggested by RefSeq's Dr. Kimm D. Pruit. This change is reflected in LNCipedia.org update 3.1 and this corrigendum serves to elucidate the discrepancies in the article caused by this update.
LNCipedia.org has been updated to version 3.1. As a consequence, the following results need to be adjusted (new values are shown in bold, original values are in italics).
- Under Protein-coding potential
‘When applying our pre-computed cutoff, these transcripts add up to about 25% (26%) of the collection’
- Under HIGH-CONFIDENCE SET
‘3406 (4127) lncRNA transcripts containing at least one TIS are thus withdrawn’
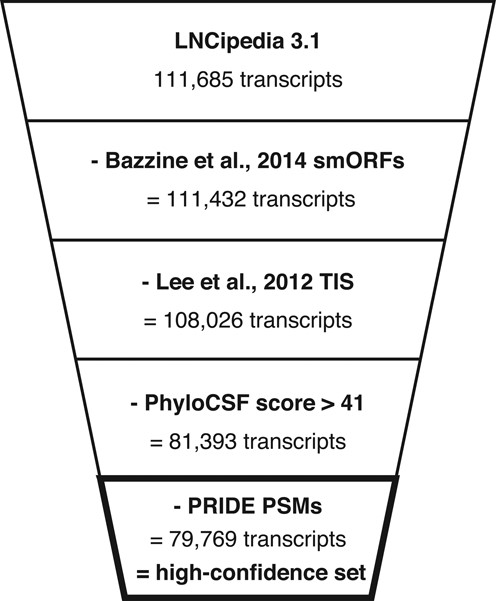
‘As such, 26 633 (27 293) transcripts with a PhyloCSF score higher than 41 are discarded. Finally, the 1624 (2040) PSM containing transcripts from the PRIDE reprocessing pipeline are excluded as well. The resulting set of 79 769 (80 216) transcripts (71% of LNCipedia 3.1) representing 47 877 (48 028) genes (77% (76%)) is referred to as “high-confidence set” and is available for download on the LNCipedia website’
- Table 1
| Source . | Version . | Number of transcripts . |
|---|---|---|
| Ensembl (52) | 75 | 23 498 |
| Refseq (43) | December (March) 2014 | 4774 (6917) |
| Nielsen et al., 2014 (45) | 7656 | |
| Hangauer et al., 2013 (46) | 5339 | |
| NONCODE (44) | 4 | 93 164 |
| LNCipedia (41) | 1.0 | 21 504 |
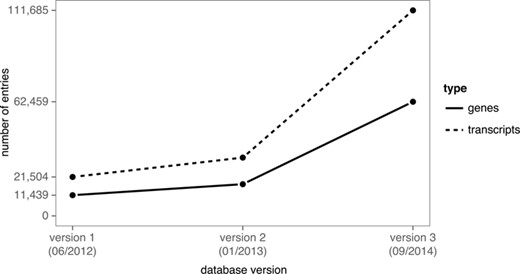
| Total number of unique transcripts | 111 685 (113 513) |
| Source . | Version . | Number of transcripts . |
|---|---|---|
| Ensembl (52) | 75 | 23 498 |
| Refseq (43) | December (March) 2014 | 4774 (6917) |
| Nielsen et al., 2014 (45) | 7656 | |
| Hangauer et al., 2013 (46) | 5339 | |
| NONCODE (44) | 4 | 93 164 |
| LNCipedia (41) | 1.0 | 21 504 |
| Total number of unique transcripts | 111 685 (113 513) |
| Source . | Version . | Number of transcripts . |
|---|---|---|
| Ensembl (52) | 75 | 23 498 |
| Refseq (43) | December (March) 2014 | 4774 (6917) |
| Nielsen et al., 2014 (45) | 7656 | |
| Hangauer et al., 2013 (46) | 5339 | |
| NONCODE (44) | 4 | 93 164 |
| LNCipedia (41) | 1.0 | 21 504 |
| Total number of unique transcripts | 111 685 (113 513) |
| Source . | Version . | Number of transcripts . |
|---|---|---|
| Ensembl (52) | 75 | 23 498 |
| Refseq (43) | December (March) 2014 | 4774 (6917) |
| Nielsen et al., 2014 (45) | 7656 | |
| Hangauer et al., 2013 (46) | 5339 | |
| NONCODE (44) | 4 | 93 164 |
| LNCipedia (41) | 1.0 | 21 504 |
| Total number of unique transcripts | 111 685 (113 513) |
‘Overview of data sources contributing to lncRNA content in LNCipedia 3.1 (3.0). In the case of RefSeq, only entries with property “biomol_ncrna_lncrna” were considered’
The following Figures 1 and 4 should be considered in place of the original figures:
LNCipedia has grown substantially since its first release. The first version (41) was based on sequences and annotation from three different sources and was made available to the public in 2012. For the 2013 release of LNCipedia (unpublished), no additional sources were used, but the different sources were updated to the most recent version. For version 3.1 (3.0) of LNCipedia, both new sources were added and existing sources were updated.
Transcripts with a likely coding potential are removed in the definition of a high-confidence set. Transcripts containing small open reading frames (25), translation initiation sites (24), PhyloCSF score greater than 41 or PSMs with an identification confidence higher than 90% are excluded.
The conclusions of the article are not affected and remain valid. The Authors apologise to Readers for these errors and inconvenience caused.




Comments