-
PDF
- Split View
-
Views
-
Cite
Cite
Blanka Stiburkova, Katerina Pavelcova, Jakub Zavada, Lenka Petru, Pavel Simek, Pavel Cepek, Marketa Pavlikova, Hirotaka Matsuo, Tony R Merriman, Karel Pavelka, Functional non-synonymous variants of ABCG2 and gout risk, Rheumatology, Volume 56, Issue 11, November 2017, Pages 1982–1992, https://doi.org/10.1093/rheumatology/kex295
Close - Share Icon Share
Abstract
Common dysfunctional variants of ATP binding cassette subfamily G member 2 (Junior blood group) (ABCG2), a high-capacity urate transporter gene, that result in decreased urate excretion are major causes of hyperuricemia and gout. In the present study, our objective was to determine the frequency and effect on gout of common and rare non-synonymous and other functional allelic variants in the ABCG2 gene.
The main cohort recruited from the Czech Republic consisted of 145 gout patients; 115 normouricaemic controls were used for comparison. We amplified, directly sequenced and analysed 15 ABCG2 exons. The associations between genetic variants and clinical phenotype were analysed using the t-test, Fisher’s exact test and a logistic and linear regression approach. Data from a New Zealand Polynesian sample set and the UK Biobank were included for the p.V12M analysis.
In the ABCG2 gene, 18 intronic (one dysfunctional splicing) and 11 exonic variants were detected: 9 were non-synonymous (2 common, 7 rare including 1 novel), namely p.V12M, p.Q141K, p.R147W, p.T153M, p.F373C, p.T434M, p.S476P, p.D620N and p.K360del. The p.Q141K (rs2231142) variant had a significantly higher minor allele frequency (0.23) in the gout patients compared with the European-origin population (0.09) and was significantly more common among gout patients than among normouricaemic controls (odds ratio = 3.26, P < 0.0001). Patients with non-synonymous allelic variants had an earlier onset of gout (42 vs 48 years, P = 0.0143) and a greater likelihood of a familial history of gout (41% vs 27%, odds ratio = 1.96, P = 0.053). In a meta-analysis p.V12M exerted a protective effect from gout (P < 0.0001).
Genetic variants of ABCG2, common and rare, increased the risk of gout. Non-synonymous allelic variants of ABCG2 had a significant effect on earlier onset of gout and the presence of a familial gout history. ABCG2 should thus be considered a common and significant risk factor for gout.
Rheumatology key messages
Dysfunctional genetic variants of ABCG2, common and rare, markedly increased the risk of gout.
Dysfunctional genetic variants of ABCG2 associate with an earlier onset of gout.
ABCG2 should be considered a common and significant risk factor for gout.
Introduction
Over the past decade, genome-wide association studies and meta-analyses have revealed over 30 common sequence variants influencing hyperuricaemia or gout, mostly in urate transporters [1]. Recently, novel gout risk loci HIST1H2BF-HIST1H4E, NIPAL1 and FAM35A were identified [2]. However, detailed knowledge of the degree to which genetic variants predict serum uric acid (SUA) concentrations remains limited.
Uric acid (UA) is the end product of purine metabolism in humans. Transport mechanisms for UA are localized mainly in the proximal tubules of the kidneys, where UA is extensively filtered and reabsorbed with some (∼10%) excreted [3]. The intestine can also excrete UA; it is estimated that up to one-third of UA may be excreted into the gut and this fraction may increase in patients with chronic renal failure [4]. SUA concentrations are highly heritable (proportion 0.38–0.63 [5–8]), consistent with a significant genetic component.
Genes that influence the level of SUA via renal UA excretion primarily encode urate transporters such as URAT1 (SLC22A12) and GLUT9 (SLC2A9). The heritable secretion component of urate homeostasis is principally mediated by the product of the ATP-binding cassette, subfamily G, member 2 (ABCG2/BCRP) gene. It is expressed on the plasma membranes of a variety of tissues, including the placenta, pharynx, bladder, brain and kidney, where it mediates the efflux of xenobiotics [9, 10]. Recent studies suggest that ABCG2 also plays an important role in intestinal excretion [11, 12]. Decreased UA excretion caused by ABCG2 dysfunction is a common mechanism of hyperuricaemia. The polymorphism rs2231142, allelic variant p.Q141K, results in a 53% reduction in UA transport with at least 10% of all gout cases in people of European ancestry attributable to this variant [13–16]. Moreover, a significant association between rs2231142 and an increased risk of a poor response to allopurinol has been described [17–19]. The aim of the present study was to determine the effect of common and rare non-synonymous and other functional allelic variants in the ABCG2 gene in patients with gout, and investigate the relationship between rs2231142 and the response to allopurinol.
Methods
Subjects
The main cohort of 145 subjects with gout was selected from patients of the Institute of Rheumatology, Prague, the Czech Republic. The control group of 115 normouricaemic subjects was selected from the personnel of the Institute of Rheumatology. Gouty arthritis was diagnosed according to the 1977 American Rheumatism Association preliminary criteria [20]. Patients suffering from secondary gout and other purine metabolic disorders associated with pathological concentrations of SUA were excluded. For each patient, a family history of gout, age of disease onset, and details of their gout treatment were recorded.
Excess production of UA associated with purine metabolic disorders was excluded through investigation of purine metabolites. For this reason, two separate measurements were performed: one set of samples was taken while patients were receiving allopurinol/febuxostat treatment, and the second set of samples was taken 72 h after temporary suspension or before initiation of allopurinol/febuxostat treatment.
To specifically study the p.V12M (rs2231137) variant, a New Zealand sample set of Māori and Pacific (Polynesian) ancestry (929 cases and 861 controls [21]) was typed for the surrogate rs4148153 using the Illumina CoreExome platform (Illumina, Inc., San Diego, CA, USA). It was necessary to use the surrogate because rs2231137 was not included on the CoreExome platform and imputation was not possible owing to the lack of a reference haplotype sample set from the Polynesian population. Data for rs2231137 were extracted from the publicly available UK Biobank (2432 cases and 102 989 controls).
The study was conducted in accordance with the Declaration of Helsinki. Before entering the study, each patient signed an informed consent regarding biological sample collection, storage, and genetic testing. All tests were performed in accordance with standards set by the institutional ethics committees, which approved the project in Prague (no.6181/2015) and New Zealand (no. MEC 05/130/10). UK Biobank data were accessed under approval no. 12611.
Clinical and biochemical investigations
Biochemical analytes were measured using a Beckman Coulter AU system (Beckman Coulter, Brea, CA, USA). High performance liquid chromatography determination of hypoxanthine, xanthine and oxypurinol in urine were performed on an Alliance 2695 and a 2998 photodiode array detector (Waters, Milford, MA, USA) as described previously [22].
PCR amplification of ABCG2 and sequence analysis
Genomic DNA of the Czech data set was extracted from EDTA whole blood using a QIAmp DNA Mini Kit (Qiagen, GmbH., Hilden, Germany). All protein-coding exons (2–16) were amplified using PCR and purified using a PCR DNA Fragments Extraction Kit (Geneaid, New Taipei City, Taiwan). DNA sequencing was performed with a DNA sequencer (Applied Biosystems 3130 Genetic Analyzer; Thermo Fisher Scientific, Waltham, MA, USA). The genotypes of allelic variants in the Czech control cohort were determined using PCR with allele-specific primers. Primer sequences and PCR conditions are available upon request. The reference sequence was defined as version ENST00000237612.7; chromosome 4: 88 090 269–88 158 912 reverse strand (www.ensembl.org). The reference protein sequence was defined as Q9UNQ0 (http://www.uniprot.org/uniprot).
Prediction of the possible impact of finding non-synonymous allelic variants on protein function was determined using PolyPhen, Provean, Mutation Taster, SIFT, Human Splicing Finder and MutPred predictive software.
Statistical analysis
The data were summarized as absolute and relative frequencies, means (s.d.) and/or medians (with interquartile range; IQR), where appropriate. Linear and logistic regression models were used to examine association of allelic variant with SUA and with gout patient/normouricaemic status, respectively. Comparisons of patient characteristics between different groups of patients, according to presence/absence of allelic variants, were performed using Student’s two-sample t test, Wilcoxon’s test, chi-square test or Fisher’s exact test, as appropriate.
To replicate the study of Roberts et al. [17] in the Czech cohort, we divided the gout patients based on their response to allopurinol treatment according to Roberts et al.'s definition (i.e. good responders were defined as having SUA on treatment ⩽357 µmol/l = 6 mg/dl with an allopurinol dose ⩽300 mg, poor responders as having SUA on treatment >357 µmol/l = 6 mg/dl with allopurinol dose >300 mg). We were, however, unable to verify compliance by measuring the allopurinol metabolite oxypurinol as was done by Roberts et al. [17]. Differences in the rs2231142 allele frequency between good and poor responders were tested using Fisher’s exact test and a logistic regression model.
The New Zealand Polynesian and UK Biobank p.V121M association analyses were adjusted by age and sex with the Polynesian sample set additionally adjusted by the number of self-reported Polynesian grandparents with gout. Gout was ascertained in the New Zealand sample set by clinical examination and in the UK Biobank by a combination of urate-lowering therapy and self-reporting of doctor-diagnosed gout [20].
All analyses were performed in the statistical language and environment R, version 3.2.2 (R Foundation for Statistical Computing, Vienna, Austria) with Rmeta used for the p.V12M meta-analysis. The level of statistical significance was set at 0.05.
Results
Subjects
The main demographic and biochemical characteristics of the subjects are summarized in Table 1. Our cohort consisted of 145 individuals with primary gout. In total 48 patients (33%) had a positive family history of gout; of those, 30 patients had first-degree relatives affected, 8 patients had second-degree relatives affected and 10 patients did not provide information about affected relatives.
Main demographic, biochemical and genetic characteristics of the subjects (n = 145)a
| Characteristic . | N (%) . | |
|---|---|---|
| Sex | ||
| Male | 131 (90.3) | |
| Female | 14 (9.7) | |
| Familial occurrence | 48 (33.1) | |
| First degree | 30 (20.7) | |
| Second degree | 8 (5.5) | |
| No information provided | 10 (6.9) | |
| Allopurinol treatment | 116 (80.0) | |
| Febuxostat treatment | 14 (9.7) | |
| At least one non-synonymous variantb | 71 (49.0) | |
| Mean (s.d.) | Range | |
| Age, years | 55.5 (13.5) | 14–90 |
| Age of onset, years | 44.7 (14.9) | 13–84 |
| SUA on treatment, µmol/l(n = 134, M/F: 123/11) | 377.0 (98.1) | 163–725 |
| SUA off treatment, µmol/l(n = 90, M/F: 79/11) | 441.7 (94.3) | 245–683 |
| FEUA on treatment(n = 134, M/F: 123/11) | 3.75 (1.86) | 0.90–11.76 |
| FEUA off treatment(n = 87, M/F: 76/11) | 3.92 (1.50) | 0.75–11.27 |
| BMI, kg/m2 (n = 114) | 29.5 (4.8) | 19.5–43.4 |
| eGFR, ml/min (n = 134) | 86.5 (20.9) | 22.9–127.9 |
| Plasma oxypurinol, µmol/l(n = 96) | 70.5 (48.3) | 4.3–270.4 |
| Median (IQR) | Range | |
| Treatment dose, mgc(n = 130, M/F: 118/12) | 300 (100) | 0–900 |
| Characteristic . | N (%) . | |
|---|---|---|
| Sex | ||
| Male | 131 (90.3) | |
| Female | 14 (9.7) | |
| Familial occurrence | 48 (33.1) | |
| First degree | 30 (20.7) | |
| Second degree | 8 (5.5) | |
| No information provided | 10 (6.9) | |
| Allopurinol treatment | 116 (80.0) | |
| Febuxostat treatment | 14 (9.7) | |
| At least one non-synonymous variantb | 71 (49.0) | |
| Mean (s.d.) | Range | |
| Age, years | 55.5 (13.5) | 14–90 |
| Age of onset, years | 44.7 (14.9) | 13–84 |
| SUA on treatment, µmol/l(n = 134, M/F: 123/11) | 377.0 (98.1) | 163–725 |
| SUA off treatment, µmol/l(n = 90, M/F: 79/11) | 441.7 (94.3) | 245–683 |
| FEUA on treatment(n = 134, M/F: 123/11) | 3.75 (1.86) | 0.90–11.76 |
| FEUA off treatment(n = 87, M/F: 76/11) | 3.92 (1.50) | 0.75–11.27 |
| BMI, kg/m2 (n = 114) | 29.5 (4.8) | 19.5–43.4 |
| eGFR, ml/min (n = 134) | 86.5 (20.9) | 22.9–127.9 |
| Plasma oxypurinol, µmol/l(n = 96) | 70.5 (48.3) | 4.3–270.4 |
| Median (IQR) | Range | |
| Treatment dose, mgc(n = 130, M/F: 118/12) | 300 (100) | 0–900 |
The differences in characteristic between male and female gout patients were mostly non-significant.
For some parameters, there were missing data; in cases where the missing data amounted to 5% or more, the real n is mentioned in parentheses.
Exon non-synonymous allelic variant as described in Table 2.
Febuxostat dose recomputed so that 40 mg febuxostat = 300 mg allopurinol. eGFR: estimated glomerular filtration rate; F: female; FEUA: excretion fraction of uric acid; M: male; SUA: serum uric acid.
Main demographic, biochemical and genetic characteristics of the subjects (n = 145)a
| Characteristic . | N (%) . | |
|---|---|---|
| Sex | ||
| Male | 131 (90.3) | |
| Female | 14 (9.7) | |
| Familial occurrence | 48 (33.1) | |
| First degree | 30 (20.7) | |
| Second degree | 8 (5.5) | |
| No information provided | 10 (6.9) | |
| Allopurinol treatment | 116 (80.0) | |
| Febuxostat treatment | 14 (9.7) | |
| At least one non-synonymous variantb | 71 (49.0) | |
| Mean (s.d.) | Range | |
| Age, years | 55.5 (13.5) | 14–90 |
| Age of onset, years | 44.7 (14.9) | 13–84 |
| SUA on treatment, µmol/l(n = 134, M/F: 123/11) | 377.0 (98.1) | 163–725 |
| SUA off treatment, µmol/l(n = 90, M/F: 79/11) | 441.7 (94.3) | 245–683 |
| FEUA on treatment(n = 134, M/F: 123/11) | 3.75 (1.86) | 0.90–11.76 |
| FEUA off treatment(n = 87, M/F: 76/11) | 3.92 (1.50) | 0.75–11.27 |
| BMI, kg/m2 (n = 114) | 29.5 (4.8) | 19.5–43.4 |
| eGFR, ml/min (n = 134) | 86.5 (20.9) | 22.9–127.9 |
| Plasma oxypurinol, µmol/l(n = 96) | 70.5 (48.3) | 4.3–270.4 |
| Median (IQR) | Range | |
| Treatment dose, mgc(n = 130, M/F: 118/12) | 300 (100) | 0–900 |
| Characteristic . | N (%) . | |
|---|---|---|
| Sex | ||
| Male | 131 (90.3) | |
| Female | 14 (9.7) | |
| Familial occurrence | 48 (33.1) | |
| First degree | 30 (20.7) | |
| Second degree | 8 (5.5) | |
| No information provided | 10 (6.9) | |
| Allopurinol treatment | 116 (80.0) | |
| Febuxostat treatment | 14 (9.7) | |
| At least one non-synonymous variantb | 71 (49.0) | |
| Mean (s.d.) | Range | |
| Age, years | 55.5 (13.5) | 14–90 |
| Age of onset, years | 44.7 (14.9) | 13–84 |
| SUA on treatment, µmol/l(n = 134, M/F: 123/11) | 377.0 (98.1) | 163–725 |
| SUA off treatment, µmol/l(n = 90, M/F: 79/11) | 441.7 (94.3) | 245–683 |
| FEUA on treatment(n = 134, M/F: 123/11) | 3.75 (1.86) | 0.90–11.76 |
| FEUA off treatment(n = 87, M/F: 76/11) | 3.92 (1.50) | 0.75–11.27 |
| BMI, kg/m2 (n = 114) | 29.5 (4.8) | 19.5–43.4 |
| eGFR, ml/min (n = 134) | 86.5 (20.9) | 22.9–127.9 |
| Plasma oxypurinol, µmol/l(n = 96) | 70.5 (48.3) | 4.3–270.4 |
| Median (IQR) | Range | |
| Treatment dose, mgc(n = 130, M/F: 118/12) | 300 (100) | 0–900 |
The differences in characteristic between male and female gout patients were mostly non-significant.
For some parameters, there were missing data; in cases where the missing data amounted to 5% or more, the real n is mentioned in parentheses.
Exon non-synonymous allelic variant as described in Table 2.
Febuxostat dose recomputed so that 40 mg febuxostat = 300 mg allopurinol. eGFR: estimated glomerular filtration rate; F: female; FEUA: excretion fraction of uric acid; M: male; SUA: serum uric acid.
Sequencing analysis of ABCG2
In the ABCG2 gene, 18 intronic variants and 11 exonic sequence variants (one not annotated) were detected (Table 2 and supplementary Table S1, available at Rheumatology Online). In the case of c.689 + 1 G > A, related to an individual with severe gouty phenotype, two abnormal ABCG2 splicing variants were identified: r.[532_689del] deletion of exon 6, and r.[532_689del; 944_949del] deletion of exon 6 and deletion of the first six base pairs of exon 9. These deletions, as we published previously, lead to a frameshift, a premature stop codon, a mis-localized ABCG2 signal on the plasma membrane and no urate transport activity in HEK293 cells [24].
Identified non-synonymous ABCG2 allelic variants, genotype distribution and alternative allele frequency in the gout cohort and database in 145 subjects with primary gout
| Reference SNP number . | Amino acid substitution or deletion . | Heterozygotes/ homozygotes . | Allele frequency in study subjects . | Allele frequency in Caucasian population . | In silico prediction software . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| PolyPhen . | SIFT . | PROVEAN . | Mutation taster . | Human splicing finder . | MutPred . | |||||
| rs2231137 | p.V12M | 7/0 | 0.02 | 0.06 | Benign (0.003) | Tolerated (1) | Neutral (0.656) | Polymorphism (21) | Potential alteration | 0.114 |
| rs2231142 | p.Q141K | 55/6 | 0.23 | 0.09 | Benign (0.035) | Tolerated (0.19) | Neutral (−1.588) | Polymorphism (53) | No impact | 0.214 |
| rs372192400 | p.R147W | 1/0 | 0.003 | 0.0001 | Probably damaging (0.999) | Damaging (0) | Deleterious (−7.146) | Disease causing (101) | Potential alteration | 0.804 (deleterious) |
| rs753759474 | p.T153M | 1/0 | 0.003 | N/A | Benign (0.268) | Damaging (0.04) | Neutral (−2.415) | Polymorphism (81) | Potential alteration | 0.387 |
| rs750972998 | p.K360del | 1/0 | 0.003 | 0.007 | N/A | N/A | Neutral (0.9) | Polymorphism (N/A) | Potential alteration | NS |
| rs752626614 | p.F373C | 1/0 | 0.003 | N/A | Probably damaging (0.988) | Damaging (0) | Deleterious (−7,828) | Disease causing (205) | No impact | 0.627 |
| rs769734146 | p.T434M | 1/0 | 0.003 | N/A | Probably damaging (0.223) | Tolerated (0.02) | Deleterious (−3.369) | Disease causing (81) | Potential alteration | 0.482 |
| N/A | p.S476P | 1/0 | 0.003 | N/A | Probably damaging (0.979) | Tolerated (0.06) | Deleterious (−3.16) | Disease causing (N/A) | No impact | 0.702 |
| rs34783571 | p.D620N | 2/0 | 0.007 | 0.004 | Probably damaging (0.028) | Tolerated (0.07) | Deleterious (−3.331) | Disease causing (23) | No impact | 0.158 |
| N/A | r.[532_689del], r.[532_689del; 944_949del] | 1/0 | 0.003 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Reference SNP number . | Amino acid substitution or deletion . | Heterozygotes/ homozygotes . | Allele frequency in study subjects . | Allele frequency in Caucasian population . | In silico prediction software . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| PolyPhen . | SIFT . | PROVEAN . | Mutation taster . | Human splicing finder . | MutPred . | |||||
| rs2231137 | p.V12M | 7/0 | 0.02 | 0.06 | Benign (0.003) | Tolerated (1) | Neutral (0.656) | Polymorphism (21) | Potential alteration | 0.114 |
| rs2231142 | p.Q141K | 55/6 | 0.23 | 0.09 | Benign (0.035) | Tolerated (0.19) | Neutral (−1.588) | Polymorphism (53) | No impact | 0.214 |
| rs372192400 | p.R147W | 1/0 | 0.003 | 0.0001 | Probably damaging (0.999) | Damaging (0) | Deleterious (−7.146) | Disease causing (101) | Potential alteration | 0.804 (deleterious) |
| rs753759474 | p.T153M | 1/0 | 0.003 | N/A | Benign (0.268) | Damaging (0.04) | Neutral (−2.415) | Polymorphism (81) | Potential alteration | 0.387 |
| rs750972998 | p.K360del | 1/0 | 0.003 | 0.007 | N/A | N/A | Neutral (0.9) | Polymorphism (N/A) | Potential alteration | NS |
| rs752626614 | p.F373C | 1/0 | 0.003 | N/A | Probably damaging (0.988) | Damaging (0) | Deleterious (−7,828) | Disease causing (205) | No impact | 0.627 |
| rs769734146 | p.T434M | 1/0 | 0.003 | N/A | Probably damaging (0.223) | Tolerated (0.02) | Deleterious (−3.369) | Disease causing (81) | Potential alteration | 0.482 |
| N/A | p.S476P | 1/0 | 0.003 | N/A | Probably damaging (0.979) | Tolerated (0.06) | Deleterious (−3.16) | Disease causing (N/A) | No impact | 0.702 |
| rs34783571 | p.D620N | 2/0 | 0.007 | 0.004 | Probably damaging (0.028) | Tolerated (0.07) | Deleterious (−3.331) | Disease causing (23) | No impact | 0.158 |
| N/A | r.[532_689del], r.[532_689del; 944_949del] | 1/0 | 0.003 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
N/A: not available; SNP: single nucleotide polymorphism.
Identified non-synonymous ABCG2 allelic variants, genotype distribution and alternative allele frequency in the gout cohort and database in 145 subjects with primary gout
| Reference SNP number . | Amino acid substitution or deletion . | Heterozygotes/ homozygotes . | Allele frequency in study subjects . | Allele frequency in Caucasian population . | In silico prediction software . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| PolyPhen . | SIFT . | PROVEAN . | Mutation taster . | Human splicing finder . | MutPred . | |||||
| rs2231137 | p.V12M | 7/0 | 0.02 | 0.06 | Benign (0.003) | Tolerated (1) | Neutral (0.656) | Polymorphism (21) | Potential alteration | 0.114 |
| rs2231142 | p.Q141K | 55/6 | 0.23 | 0.09 | Benign (0.035) | Tolerated (0.19) | Neutral (−1.588) | Polymorphism (53) | No impact | 0.214 |
| rs372192400 | p.R147W | 1/0 | 0.003 | 0.0001 | Probably damaging (0.999) | Damaging (0) | Deleterious (−7.146) | Disease causing (101) | Potential alteration | 0.804 (deleterious) |
| rs753759474 | p.T153M | 1/0 | 0.003 | N/A | Benign (0.268) | Damaging (0.04) | Neutral (−2.415) | Polymorphism (81) | Potential alteration | 0.387 |
| rs750972998 | p.K360del | 1/0 | 0.003 | 0.007 | N/A | N/A | Neutral (0.9) | Polymorphism (N/A) | Potential alteration | NS |
| rs752626614 | p.F373C | 1/0 | 0.003 | N/A | Probably damaging (0.988) | Damaging (0) | Deleterious (−7,828) | Disease causing (205) | No impact | 0.627 |
| rs769734146 | p.T434M | 1/0 | 0.003 | N/A | Probably damaging (0.223) | Tolerated (0.02) | Deleterious (−3.369) | Disease causing (81) | Potential alteration | 0.482 |
| N/A | p.S476P | 1/0 | 0.003 | N/A | Probably damaging (0.979) | Tolerated (0.06) | Deleterious (−3.16) | Disease causing (N/A) | No impact | 0.702 |
| rs34783571 | p.D620N | 2/0 | 0.007 | 0.004 | Probably damaging (0.028) | Tolerated (0.07) | Deleterious (−3.331) | Disease causing (23) | No impact | 0.158 |
| N/A | r.[532_689del], r.[532_689del; 944_949del] | 1/0 | 0.003 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Reference SNP number . | Amino acid substitution or deletion . | Heterozygotes/ homozygotes . | Allele frequency in study subjects . | Allele frequency in Caucasian population . | In silico prediction software . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| PolyPhen . | SIFT . | PROVEAN . | Mutation taster . | Human splicing finder . | MutPred . | |||||
| rs2231137 | p.V12M | 7/0 | 0.02 | 0.06 | Benign (0.003) | Tolerated (1) | Neutral (0.656) | Polymorphism (21) | Potential alteration | 0.114 |
| rs2231142 | p.Q141K | 55/6 | 0.23 | 0.09 | Benign (0.035) | Tolerated (0.19) | Neutral (−1.588) | Polymorphism (53) | No impact | 0.214 |
| rs372192400 | p.R147W | 1/0 | 0.003 | 0.0001 | Probably damaging (0.999) | Damaging (0) | Deleterious (−7.146) | Disease causing (101) | Potential alteration | 0.804 (deleterious) |
| rs753759474 | p.T153M | 1/0 | 0.003 | N/A | Benign (0.268) | Damaging (0.04) | Neutral (−2.415) | Polymorphism (81) | Potential alteration | 0.387 |
| rs750972998 | p.K360del | 1/0 | 0.003 | 0.007 | N/A | N/A | Neutral (0.9) | Polymorphism (N/A) | Potential alteration | NS |
| rs752626614 | p.F373C | 1/0 | 0.003 | N/A | Probably damaging (0.988) | Damaging (0) | Deleterious (−7,828) | Disease causing (205) | No impact | 0.627 |
| rs769734146 | p.T434M | 1/0 | 0.003 | N/A | Probably damaging (0.223) | Tolerated (0.02) | Deleterious (−3.369) | Disease causing (81) | Potential alteration | 0.482 |
| N/A | p.S476P | 1/0 | 0.003 | N/A | Probably damaging (0.979) | Tolerated (0.06) | Deleterious (−3.16) | Disease causing (N/A) | No impact | 0.702 |
| rs34783571 | p.D620N | 2/0 | 0.007 | 0.004 | Probably damaging (0.028) | Tolerated (0.07) | Deleterious (−3.331) | Disease causing (23) | No impact | 0.158 |
| N/A | r.[532_689del], r.[532_689del; 944_949del] | 1/0 | 0.003 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
N/A: not available; SNP: single nucleotide polymorphism.
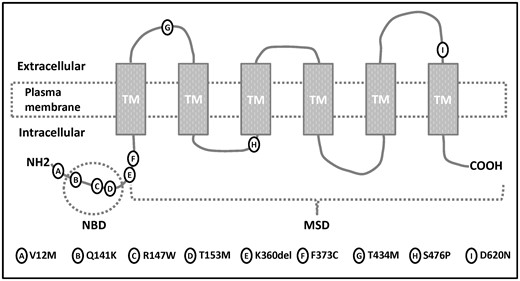
Of the exonic variants, nine were non-synonymous: p.V12M (rs2231137), p.Q141K (rs2231142), p.R147W (rs372192400), p.T153M (rs753759474), p.F373C (rs752626614), p.T434M (rs769734146), p.S476P (not annotated), p.D620N (rs34783571) and a three base deletion p.K360del (rs750972998). Heterozygous p.V12M was detected in seven individuals. Heterozygous variants p.R147W, p.T153M, p.F373C, p.T434M, p.K360del and p.S476P were detected once, and variant p.D620N was detected twice. In silico, all seven allelic variants with unknown and/or having a rare minor allele frequency (i.e. <0.01) were predicted as probably damaging. The p.Q141K variant was present with a significantly higher minor allele frequency (MAF; 0.23, 55 heterozygotes/6 homozygotes) in the Czech cohort of gout patients compared with the European-origin population (MAF = 0.09) and the worldwide population (MAF = 0.12). In total, 71 patients (49.0%) harboured at least one of these nine non-synonymous variants. Of those, 11 harboured two non-synonymous variants: six had two copies of p.Q141K, five had one copy of p.Q141K and one of the other identified non-synonymous variants. There was no patient with three or more copies of identified non-synonymous variants. The identified variants, their genotype distribution, alternative allele frequency in the Czech gout cohort and database source are shown in Table 2 and supplementary Table S1, available at Rheumatology Online. Positions of identified non-synonymous allelic variants in the membrane topology model of ABCG2 are showed in Fig. 1.
Position of identified allelic variants in membrane topology model of ABCG2
Association between allelic variants in ABCG2, presence of gout and levels of hyperuricaemia
The results from our association analyses are shown in supplementary Table S1, available at Rheumatology Online. A univariate association of SUA, measured on allopurinol/febuxostat treatment, with allelic variants showed the minor alleles of rs2231138 and rs2231165 to be potentially positively associated with increased SUA concentration (P = 0.038 and P = 0.022, respectively). Similarly, the minor alleles of rs2231156 and rs2231165 showed a positive association with increased SUA, off-treatment (P = 0.041 and P = 0.043, respectively). These variants showed associations significant at the 0.05 level; however, they were not statistically significant with the Bonferroni correction for multiple comparisons applied. The comparison of the gout group vs normouricaemic group showed a strong association between rs2231142 and gout/normouricaemic status: the variant frequency in gout patients was almost triple the frequency in normouricaemic controls (23% vs 8%, odds ratio (OR) = 3.26, 95% CI: 1.96, 5.36, P < 0.0001).
Association between allelic variants in ABCG2 and response to allopurinol
There were 42 (29%) good responders and 9 (6%) poor responders. The rest of the cohort (94 patients, 65%) had either higher SUA with lower doses of allopurinol (50 patients) or lower SUA when treated with higher doses of allopurinol (9 patients); 35 patients had missing data for genotype, dose or UA concentration, or received treatment other than allopurinol. Characteristics of good and poor responders are presented in Table 3.
Baseline demographics, frequency of rs2231142 and its association with allopurinol response among patients with gout
| Variable . | Good responder (n = 42) . | Poor responder (n = 9) . | P-valuea . |
|---|---|---|---|
| Familial occurrence, n (%) | 9 (21.4) | 3 (33.3) | 1.0000b |
| Age, mean (range), years | 61.0 (25-90) | 64.3 (50-74) | 0.4333 |
| Male, n (%) | 36 (85.7) | 7 (100) | 0.5749b |
| BMI, mean (s.e.), kg/m2 | 28.4 (4.5) | 33.0 (4.0) | 0.0245 |
| eGFR, mean (s.e.), ml/min | 87.1 (18.2) | 72.0 (12.0) | 0.0214 |
| Serum urate, mean (s.e.), µmol/l | 300.3 (47.0) | 421.8 (76.6) | <0.0001 |
| Allopurinol dose, mean (range), mg/day | 233.3 (100–300) | 588.9 (400–800) | <0.0001 |
| Plasma oxypurinol, mean (s.e.), µmol/l | 76.7 (45.2) | 90.4 (29.6) | 0.4178 |
| Off serum urate, mean (s.e.), µmol/l | 405.3 (64.8) | 439.1 (86.4) | 0.2654 |
| rs2231142 | |||
| GG, N (relative frequency) | 29 (0.69) | 5 (0.56) | 0.4589b |
| GT, N (relative frequency) | 13 (0.34) | 4 (0.44) | |
| TT, N (relative frequency) | 0 (0.00) | 0 (0.00) | |
| MAF, N (relative frequency) | 13 (0.15) | 4 (0.22) | 0.4589b |
| Non-synonymous allele variants | |||
| None, N (relative frequency) | 28 (0.67) | 3 (0.33) | 0.1289b |
| At least one, N (relative frequency) | 14 (0.33) | 6 (0.67) | |
| Association of ABCG2 SNP rs2231142 with allopurinol response in patients with gout/hyperuricaemia | P-valuec | ||
| Unadjusted OR (95% CI) | 1.78 (0.41, 7.75) | 0.4395 | |
| Adjusted on age, BMI (95% CI) | 2.97 (0.35, 25.06) | 0.3168 | |
| Adjusted on GFR (95% CI) | 1.54 (0.32, 7.29) | 0.5894 | |
| Adjusted on off serum urate (95% CI) | 2.86 (0.49, 16.64) | 0.2415 | |
| Association of any identified non-synonymous variant with allopurinol response in patients with gout/hyperuricaemia | P-valuec | ||
| Unadjusted OR (95% CI) | 4.00 (0.87, 18.42) | 0.0752 | |
| Variable . | Good responder (n = 42) . | Poor responder (n = 9) . | P-valuea . |
|---|---|---|---|
| Familial occurrence, n (%) | 9 (21.4) | 3 (33.3) | 1.0000b |
| Age, mean (range), years | 61.0 (25-90) | 64.3 (50-74) | 0.4333 |
| Male, n (%) | 36 (85.7) | 7 (100) | 0.5749b |
| BMI, mean (s.e.), kg/m2 | 28.4 (4.5) | 33.0 (4.0) | 0.0245 |
| eGFR, mean (s.e.), ml/min | 87.1 (18.2) | 72.0 (12.0) | 0.0214 |
| Serum urate, mean (s.e.), µmol/l | 300.3 (47.0) | 421.8 (76.6) | <0.0001 |
| Allopurinol dose, mean (range), mg/day | 233.3 (100–300) | 588.9 (400–800) | <0.0001 |
| Plasma oxypurinol, mean (s.e.), µmol/l | 76.7 (45.2) | 90.4 (29.6) | 0.4178 |
| Off serum urate, mean (s.e.), µmol/l | 405.3 (64.8) | 439.1 (86.4) | 0.2654 |
| rs2231142 | |||
| GG, N (relative frequency) | 29 (0.69) | 5 (0.56) | 0.4589b |
| GT, N (relative frequency) | 13 (0.34) | 4 (0.44) | |
| TT, N (relative frequency) | 0 (0.00) | 0 (0.00) | |
| MAF, N (relative frequency) | 13 (0.15) | 4 (0.22) | 0.4589b |
| Non-synonymous allele variants | |||
| None, N (relative frequency) | 28 (0.67) | 3 (0.33) | 0.1289b |
| At least one, N (relative frequency) | 14 (0.33) | 6 (0.67) | |
| Association of ABCG2 SNP rs2231142 with allopurinol response in patients with gout/hyperuricaemia | P-valuec | ||
| Unadjusted OR (95% CI) | 1.78 (0.41, 7.75) | 0.4395 | |
| Adjusted on age, BMI (95% CI) | 2.97 (0.35, 25.06) | 0.3168 | |
| Adjusted on GFR (95% CI) | 1.54 (0.32, 7.29) | 0.5894 | |
| Adjusted on off serum urate (95% CI) | 2.86 (0.49, 16.64) | 0.2415 | |
| Association of any identified non-synonymous variant with allopurinol response in patients with gout/hyperuricaemia | P-valuec | ||
| Unadjusted OR (95% CI) | 4.00 (0.87, 18.42) | 0.0752 | |
If not stated otherwise, good vs poor responders are compared using two-sample t-test.
Good vs poor responders are compared using Fisher’s exact test.
OR estimates, 95% CIs and P-values come from logistic regression models with poor/good response as dependent variable, presence of ABCG2 SNP rs2231142 as predictor and different covariates as specified in the table. eGFR: estimated glomerular filtration rate; G: guanine; GFR: glomerular filtration rate; MAF: minor allele frequency; T: thymine.
Baseline demographics, frequency of rs2231142 and its association with allopurinol response among patients with gout
| Variable . | Good responder (n = 42) . | Poor responder (n = 9) . | P-valuea . |
|---|---|---|---|
| Familial occurrence, n (%) | 9 (21.4) | 3 (33.3) | 1.0000b |
| Age, mean (range), years | 61.0 (25-90) | 64.3 (50-74) | 0.4333 |
| Male, n (%) | 36 (85.7) | 7 (100) | 0.5749b |
| BMI, mean (s.e.), kg/m2 | 28.4 (4.5) | 33.0 (4.0) | 0.0245 |
| eGFR, mean (s.e.), ml/min | 87.1 (18.2) | 72.0 (12.0) | 0.0214 |
| Serum urate, mean (s.e.), µmol/l | 300.3 (47.0) | 421.8 (76.6) | <0.0001 |
| Allopurinol dose, mean (range), mg/day | 233.3 (100–300) | 588.9 (400–800) | <0.0001 |
| Plasma oxypurinol, mean (s.e.), µmol/l | 76.7 (45.2) | 90.4 (29.6) | 0.4178 |
| Off serum urate, mean (s.e.), µmol/l | 405.3 (64.8) | 439.1 (86.4) | 0.2654 |
| rs2231142 | |||
| GG, N (relative frequency) | 29 (0.69) | 5 (0.56) | 0.4589b |
| GT, N (relative frequency) | 13 (0.34) | 4 (0.44) | |
| TT, N (relative frequency) | 0 (0.00) | 0 (0.00) | |
| MAF, N (relative frequency) | 13 (0.15) | 4 (0.22) | 0.4589b |
| Non-synonymous allele variants | |||
| None, N (relative frequency) | 28 (0.67) | 3 (0.33) | 0.1289b |
| At least one, N (relative frequency) | 14 (0.33) | 6 (0.67) | |
| Association of ABCG2 SNP rs2231142 with allopurinol response in patients with gout/hyperuricaemia | P-valuec | ||
| Unadjusted OR (95% CI) | 1.78 (0.41, 7.75) | 0.4395 | |
| Adjusted on age, BMI (95% CI) | 2.97 (0.35, 25.06) | 0.3168 | |
| Adjusted on GFR (95% CI) | 1.54 (0.32, 7.29) | 0.5894 | |
| Adjusted on off serum urate (95% CI) | 2.86 (0.49, 16.64) | 0.2415 | |
| Association of any identified non-synonymous variant with allopurinol response in patients with gout/hyperuricaemia | P-valuec | ||
| Unadjusted OR (95% CI) | 4.00 (0.87, 18.42) | 0.0752 | |
| Variable . | Good responder (n = 42) . | Poor responder (n = 9) . | P-valuea . |
|---|---|---|---|
| Familial occurrence, n (%) | 9 (21.4) | 3 (33.3) | 1.0000b |
| Age, mean (range), years | 61.0 (25-90) | 64.3 (50-74) | 0.4333 |
| Male, n (%) | 36 (85.7) | 7 (100) | 0.5749b |
| BMI, mean (s.e.), kg/m2 | 28.4 (4.5) | 33.0 (4.0) | 0.0245 |
| eGFR, mean (s.e.), ml/min | 87.1 (18.2) | 72.0 (12.0) | 0.0214 |
| Serum urate, mean (s.e.), µmol/l | 300.3 (47.0) | 421.8 (76.6) | <0.0001 |
| Allopurinol dose, mean (range), mg/day | 233.3 (100–300) | 588.9 (400–800) | <0.0001 |
| Plasma oxypurinol, mean (s.e.), µmol/l | 76.7 (45.2) | 90.4 (29.6) | 0.4178 |
| Off serum urate, mean (s.e.), µmol/l | 405.3 (64.8) | 439.1 (86.4) | 0.2654 |
| rs2231142 | |||
| GG, N (relative frequency) | 29 (0.69) | 5 (0.56) | 0.4589b |
| GT, N (relative frequency) | 13 (0.34) | 4 (0.44) | |
| TT, N (relative frequency) | 0 (0.00) | 0 (0.00) | |
| MAF, N (relative frequency) | 13 (0.15) | 4 (0.22) | 0.4589b |
| Non-synonymous allele variants | |||
| None, N (relative frequency) | 28 (0.67) | 3 (0.33) | 0.1289b |
| At least one, N (relative frequency) | 14 (0.33) | 6 (0.67) | |
| Association of ABCG2 SNP rs2231142 with allopurinol response in patients with gout/hyperuricaemia | P-valuec | ||
| Unadjusted OR (95% CI) | 1.78 (0.41, 7.75) | 0.4395 | |
| Adjusted on age, BMI (95% CI) | 2.97 (0.35, 25.06) | 0.3168 | |
| Adjusted on GFR (95% CI) | 1.54 (0.32, 7.29) | 0.5894 | |
| Adjusted on off serum urate (95% CI) | 2.86 (0.49, 16.64) | 0.2415 | |
| Association of any identified non-synonymous variant with allopurinol response in patients with gout/hyperuricaemia | P-valuec | ||
| Unadjusted OR (95% CI) | 4.00 (0.87, 18.42) | 0.0752 | |
If not stated otherwise, good vs poor responders are compared using two-sample t-test.
Good vs poor responders are compared using Fisher’s exact test.
OR estimates, 95% CIs and P-values come from logistic regression models with poor/good response as dependent variable, presence of ABCG2 SNP rs2231142 as predictor and different covariates as specified in the table. eGFR: estimated glomerular filtration rate; G: guanine; GFR: glomerular filtration rate; MAF: minor allele frequency; T: thymine.
The minor allele frequency of rs2231142 was numerically higher in patients who responded poorly to allopurinol therapy (OR = 1.78, 95% CI: 0.41, 7.75; P = 0.440; see Table 3), but the result was not statistically significant. Adjustment for gender, BMI, estimated glomerular filtration rate and SUA concentration without the use of urate-lowering therapy did not change the results, although large confidence intervals suggest that the sample was too small.
The presence of identified non-synonymous allelic variants was two times higher in patients who responded poorly to allopurinol therapy (6 of 9, 67%) compared with good responders (14 of 42, 33%; OR = 4.00, 95% CI: 0.87, 18.42; P = 0.075). The result was not statistically significant; however, this might change with a larger data set.
Association between allelic variants in ABCG2 and age of onset of gout
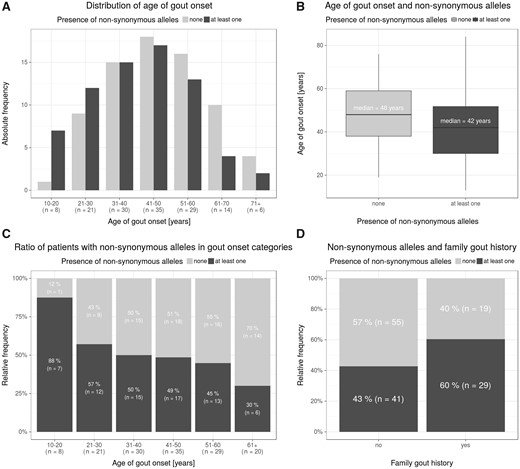
In the Czech cohort, the mean age of gout onset was 44.9 years. Remarkably, we detected a non-synonymous allelic variant in the ABCG2 gene in seven of eight patients (88%) with very early gout onset (between ages 10 and 20 years). Of those, the p.Q141K variant was present in six patients. In the group with early onset between 21 and 30 years, the non-synonymous allelic variants were detected in 12 of 21 patients (57%). On the other hand, these variants were under-represented when the age of onset was over 61 years (6 of 20, 30%). This shows an apparent shift in proportions of patients with non-synonymous alleles who are over-represented in earlier age of onset categories and under-represented in older age of onset categories χ2-test for trend in proportions, P = 0.010). The median age of onset among patients with any non-synonymous allelic variant was 42 years, while among patients without non-synonymous allelic variants it was 48 years (Wilcoxon’s test P = 0.014). Under a dose–response model, the median age of onset among those with two non-synonymous alleles was 31 years (in a linear regression model: β = –4.9 meaning a shift of 4.9 to earlier age of onset with each extra copy of a non-synonynous allele; P = 0.013).
As for family history, patients with non-synonymous variants had familial gout in 29 of 71 cases (40.8%), while patients without non-synonymous variants had familial gout in 19 of 74 cases (25.7%). This association showed borderline significance (OR = 1.96, 95% CI: 0.97, 3.96; Fisher’s test, P = 0.053). The relationship between a family history of gout, onset of gout and allelic variants in ABCG2 is shown in Fig. 2.
Family history, age distribution and allelic variants in the ABCG2 gene
(A) Histogram of age of gout onset among patients with and without any of the nine exon non-synonymous alleles. (B) Box-and-whiskers plot of age of gout onset among patients with and without any of the nine exon non-synonymous alleles. (C) Proportion of patients with and without any of the nine exon non-synonymous alleles in age of gout onset (by decades). (D) Proportion of patients with and without any of the nine exon non-synonymous alleles among patients with and without gout family history.
Association of p.V12M with gout: meta-analysis
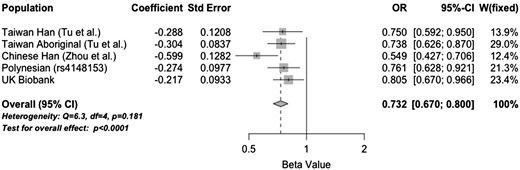
Our data showed that the p.V12M alternative allele was under-represented in the Czech gout cases (frequency = 0.02) compared with the general European population (frequency = 0.06, P = 0.036). While we did not test for an association with gout, these observations are consistent with previous studies showing the p.12M allele to be protective against gout in Han Chinese and Taiwanese Aboriginal sample sets [24]. Therefore we tested p.V12M for association with gout in a NZ Polynesian sample set (using surrogate rs4148153) and in the UK Biobank cohort, and meta-analysed with the Tu et al. [25] and Zhou et al. [26] studies (Fig. 3). There was evidence of a p.V12M association with both the Polynesian (OR = 0.76, P = 0.005) and UK Biobank sample sets (OR = 0.81, P = 0.02), with the p.12 M allele conferring a protective effect. A meta-analysis using published data indicates strong evidence that p.12 M has a gout protective effect (OR = 0.73, P < 0.0001).
Discussion
In this study, we identified a total of 29 sequence variants [one intronic splicing variant, one hitherto unpublished allelic non-synonymous variant, six non-synonymous variants with unknown and/or a very rare MAF (i.e. <0.01) and two non-synonymous common variants] in the ABCG2 gene in a Czech gout cohort. The new findings of this study are the following: identification of novel non-synonymous allelic variant p.S476P; the presence of non-synonymous allelic variants in ABCG2 was significantly higher in the gout cohort compared with the common population; earlier onset of gout was associated with the presence of non-synonymous allelic variants in the ABCG2 transporter.
ABCG2 is highly variable in the human population. Most of the variants are rare, and only two common non-synonymous allelic variants (MAF ⩾ 0.01), p.V12M and p.Q141K, have been identified. A previous study showed that ABCG2 common dysfunction causes hyperuricaemia by two complementary mechanisms: increased urate levels in the blood caused by reduced urate excretion by the kidneys, and renal urate overload caused by reduced urate excretion by the intestines [27]. The functional characterization and impact of most variants, except for p.Q141K, which accounts for approximately one-half the reduction of UA transport, are currently unknown.
The structural model for ABCG2 focuses on the organization and alignment of residues within six transmembrane spanning domains. No identified variant in our cohort was present in transmembrane segments. The variant p.V12M, localized in the short and flexible N-terminal region, had a lower minor allele frequency (MAF = 0.02) in the Czech cohort of gout patients than in the European-origin population (MAF = 0.06). A significant association between p.V12M and gout has been reported in separate samples from different ethnic groups, for example, Taiwanese Han, Taiwan Aborigines [25] and Chinese Han [26]. These findings were extended in this study by a meta-analysis of diverse population groups. The meta-analysis confirmed the protective effect of the p.12 M allele. This variant is genetically independent of p.Q141K (r2 = 0.002 in Europeans) and thus it represents an ABCG2 effect additional to that of p.Q141K. Data from the Japanese population also showed a different haplotype for p.V12M and dysfunctional variants p.Q126X and p.Q141K, which supports the independent protective effect of p.V12M [14]. However, tagging experiments have shown that p.V12M has no measurable effect on the processing or function of the ABCG2 protein [28]. In the Czech cohort, the seven carriers of p.V12M allele had a disease onset at a relatively younger age (range 23–69, median 41), without notable differences in family gout history relative to the whole cohort. Based on the Czech data alone, we cannot support or contradict the possible protective role of p.V12M.
In the Czech cohort, the p.Q141K variant was present with a significantly higher allele frequency, almost triple the frequency in normouricaemic controls (23% vs 8%, P < 0.0001) and higher than in the European-origin population (9%, P < 0.0001). p.Q141K is located in the nucleotide binding domain, the most conserved region in all ABC proteins (where the domain binds and cleaves ATP). The prevalence of this variant is 1–5% in the African, 9% in the Caucasian, and 30% in the Asian population [15]. Our results confirm that this particular alteration in the ABCG2 gene is a common cause of hyperuricaemia. This finding is consistent with previous studies in which rs2231142 has been associated with hyperuricaemia and gout in individuals of European, Han Chinese, Japanese and African American ancestry [14, 15, 29–32]; however, this association was not found in Māori subjects from New Zealand [32].
The rare variants p.R147W (Caucasian MAF = 0.0001) and p.T153M (Caucasian MAF unknown) were identified in our gout cohort in one heterozygous individual each (MAF = 0.003). These variants were localized close to p.Q141K, in the nucleotide-binding domain, and are probably damaging through disruption of ATP binding. The in-frame three nucleotide deletion p.K360del (Caucasian MAF = 0.007) and p.F373C (Caucasian MAF unknown), also identified in one heterozygous individual each, were located in the intracellular membrane-spanning domain. Variant p.T434M (Caucasian MAF unknown), identified in one heterozygous individual, was located in the first extracellular loop between transmembrane domains 1 and 2. The novel variant p.S476P was located in the first intracellular loop between transmembrane domains 2 and 3. Variant p.D620N (Caucasian MAF = 0.0004), identified in two heterozygous individuals, was located in the last extracellular loop between transmembrane domains 5 and 6. This is a specific region with an unusual conformation (consecutive V-shaped helices partially inserted into the membrane) with an experimentally verified N-glycosylation site [33]. These variants have not been characterized yet in ABCG2 functional studies, but the nature of these variants suggests that they may have a functional impact: analysis using PolyPhen software, Provean and Mutation Taster predicted that substitutions p.R147W, p.F373C, p.T434M, p.S476P and p.D620N could possibly be damaging. Analyses using other predictive software such as SIFT, Human Splicing Finder and MutPred were not consistent and emphasize the need for experimental functional characterization.
An association analysis in the Czech cohort showed a relationship between SUA and three single nucleotide polymorphisms within the ABCG2 locus, marked by intronic variants rs2231138, rs2231165 and rs2231156. These variants showed a significant association with the concentration of SUA (P < 0.05) during hyperuricaemic treatment (rs2231138, rs2231165) and after 72 h without treatment (rs2231156, rs2231165). However, the associations were not statistically significant after the Bonferroni correction. A comparison of genotype distribution showed a lower MAF in the gout group vs the Caucasian population for rs2231138 (0.021 vs 0.044) and rs2231165 (0.010 vs 0.021), and a higher MAF in rs2231156 (0.159 vs 0.073). However, the size of the studied groups was not sufficiently large for a detailed analysis of such rare variants with very low MAFs. Furthermore, this finding could have been caused by a linkage disequilibrium with previously reported and more strongly associated common variants that include rs2231142 and intronic rs2622629 [34].
Although we did not find a statistically significant association between rs2231142 and an increased risk of a poor response to allopurinol, the directionality of the ORs of this analysis support the hypothesis that the dysfunctional p.Q141K variant may be associated with a poor response to allopurinol in patients with gout. It is further supported by the increased occurrence of all identified non-synonymous allele variants among poor responders compared with good responders. We can theorize that genotyping rs2231142 may someday be a useful tool for more effective allocation of gout patients relative to therapeutic interventions (e.g. uricosurics and/or febuxostat) as part of the ‘personalized medicine’ concept.
ABCG2 dysfunctional variants have a strong impact on the progression of hyperuricaemia. A study in a cohort of 5005 Japanese participants reported that the ABCG2 population-attributable risk percentage (PAR%) for hyperuricaemia was 29.2%, which is much higher than those of the other typical environmental risks, that is, overweight/obesity (BMI ⩾ 25.0; PAR% = 18.7%), heavy drinking [>196 g/week (male) or >98 g/week (female) of pure alcohol; PAR% = 15.4%] and ageing (⩾60 years old; PAR% = 5.74%) [35].
Gout usually occurs between the fourth and sixth decade of life. In a large cohort study of 23 857 incident gout patients from the UK, the mean age of onset of gout was 61.9 (s.d. 14.5) years [36]. However, the number of patients experiencing onset at a younger age is now increasing [37]. A study of 705 Japanese male gout cases described that 88.2% of early-onset patients (twenties or younger) were positive for mild to severe ABCG2 dysfunctional variants. Severe ABCG2 dysfunction particularly increased the risk of early-onset gout (OR = 22.2, P = 4.66 × 10−6) [38]. Our results confirm that common dysfunction of ABCG2 is a significant cause of familial and/or early-onset gout.
Our study has several strengths. First, our analysis of all ABCG2 exon regions was complete and thorough and we believe that it has not been done before to such an extent in a gout cohort. Second, in the selection of our gout cohort, we controlled for and excluded several potential confounders of secondary hyperuricaemia/gout and other purine metabolic disorders associated with pathological concentrations of SUA. Some limitations of this study should be acknowledged. First, the size of the studied groups was not large enough to rule out that some functional variants may have gone undetected. Second, we studied genetic variants in transcribed regions and exon–intron boundaries only and therefore genetic variants outside these regions would have gone undetected.
Conclusions
In conclusions, our finding of one intronic splicing and one novel, six very rare and two common non-synonymous ABCG2 allelic variants in a sample of 145 gout patients suggests that the ABCG2 gene should be considered a strong and common risk for gout. This finding is supported by the significant effect of these variants, especially the dysfunctional variant p.Q141K, on those with early-onset gout and the presence of a familial gout history. Genotyping the rare variants of ABCG2 in conjunction with its common variants will eventually contribute to evaluation of an individual’s risk for gout, a wider selection of effective treatments and better understanding of why some patients respond poorly to treatment, especially in patients with a family history and/or early gout onset.
Acknowledgements
We would like to thank Tereza Kovacikova (Institute of Inherited Metabolic Disorders, Czech Republic) for her assistance with high performance liquid chromatography determination of purine metabolites). We are grateful to all the patients who took part in this study as well as our colleagues at the Institute of Rheumatology for their help in recruiting patients for the study.
Author contributions: All authors were involved in drafting the manuscript or revising it critically for content. All the authors approved the final version for publication.
Study conception and design: B.S.; clinical observation: J.Z. and K.P.; acquisition of data: B.S., Kat.P., P.S., P.C., M.P. and T.R.M.; analysis and interpretation of data: B.S., Kat.P., M.P., L.P., J.Z., M.H. and T.R.M.
Funding: Supported by a grant from the Czech Republic Ministry of Health AZV 15-26693 A.
Disclosure statement: The authors have declared no conflicts of interest.
Supplementary data
Supplementary data are available at Rheumatology Online.



Comments