Abstract
This study aimed to develop a joint population pharmacokinetic model for an antipsychotic agent in development (S33138) and its active metabolite (S35424) produced by reversible metabolism. Because such a model leads to identifiability problems and numerical difficulties, the model building was performed using the FOCE-I and the Stochastic Approximation Expectation Maximization (SAEM) estimation algorithms in NONMEM and MONOLIX, respectively. Four different structural models were compared based on Bayesian information criteria. Models were first written as ordinary differential equations systems and then in closed form (CF) to facilitate further analyses. The impact of polymorphisms on genes coding for the CYP2C19 and CYP2D6 enzymes, respectively involved in the parent drug and the metabolite elimination were investigated using permutation Wald test. The parent drug and metabolite plasma concentrations of 101 patients were analyzed on two occasions after 4 and 8 weeks of treatment at 1, 3, 6, and 24 h following daily oral administration. All configurations led to a two compartment model with back-transformation of the metabolite into the parent drug and a first-pass effect. The elimination clearance of the metabolite through other processes than back-transformation was decreased by 35% [9–53%] in CYP2D6 poor metabolizer. Permutation tests were performed to ensure the robustness of the analysis, using SAEM and CF. In conclusion, we developed a complex joint pharmacokinetic model adequately predicting the impact of CYP2D6 polymorphisms on the parent drug and its metabolite concentrations through the back-transformation mechanism.








Similar content being viewed by others
References
Shargel L, Wu-Pong S, Yu A. Applied biopharmaceutics & pharmacokinetics. 5th ed. USA: McGraw-Hill Medical; 2005.
Younis IR, Malone S, Friedman HS, Schaaf LJ, Petros WP. Enterohepatic recirculation model of irinotecan (CPT-11) and metabolite pharmacokinetics in patients with glioma. Cancer Chemother Pharmacol. 2009;63:517–24.
Panhard X, Goujard C, Legrand M, Taburet AM, Diquet B, Mentré F. Population pharmacokinetic analysis for nelfinavir and its metabolite M8 in virologically controlled HIV-infected patients on HAART. Br J Clin Pharmacol. 2005;60:390–403.
Feng Y, Pollock BG, Coley K, Marder S, Miller D, Kirshner M, et al. Population pharmacokinetic analysis for risperidone using highly sparse sampling measurements from the CATIE study. Br J Clin Pharmacol. 2008;66:629–39.
Lindstrom M, Bates D. Nonlinear mixed effects models for repeated measures data. Biometrics. 1990;46:673–87.
Beal S, Sheiner L, Boeckmann A, Bauer R. NONMEM user's guides (1989–2009). Ellicott City, MD, USA; 2009
Girard P, Mentré F. A comparison of estimation methods in nonlinear mixed effects models using a blind analysis. Pamplona, Spain: 14th Population Approach Group in Europe; 2005. Available from: http://www.page-meeting.org/page/page2005/PAGE2005O08.pdf.
Bertrand J, Comets E, Mentré F. Comparison of model-based tests and selection strategies to detect genetic polymorphisms influencing pharmacokinetic parameters. J Biopharm Stat. 2008;18:1084–102.
Kuhn E, Lavielle M. Maximum likelihood estimation in nonlinear mixed effects models. Comput Stat Data Anal. 2005;49:1020–38.
Lavielle M. MONOLIX (MOdèles NOn LInéaires à effets miXtes). Orsay, France; 2008. http://software.monolix.org/index.php.
Bauer RJ. S-ADAPT/MCPEM User's Guide Version 1.56. Los Angeles, CA, USA; 2008.
Millan MJ, Loiseau F, Dekeyne A, Gobert A, Flik G, Cremers TI, et al. S33138 (N-[4-[2-[(3aS,9bR)-8-cyano-1,3a,4,9b-tetrahydro[1] benzopyrano[3,4-c]pyrrol-2(3 H)-yl)-ethyl]phenyl-acetamide), a preferential dopamine D3 versus D2 receptor antagonist and potential antipsychotic agent: III. Actions in models of therapeutic activity and induction of side effects. J Pharmacol Exp Ther. 2008;324:1212–26.
Zhou S, Liu J, Chowbay B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab Rev. 2009;41:89–295.
Bertrand J, Comets E, Laffont C, Chenel M, Mentré F. Pharmacogenetics and population pharmacokinetics: impact of the design on three tests using the SAEM algorithm. J Pharmacokinet Pharmacodyn. 2009;36:317–39.
Manly B. Randomization, bootstrap and Monte Carlo methods in biology. 2nd ed. London: Chapman & Hall; 1998.
Brendel K, Dartois C, Comets E, Lemenuel-Diot A, Laveille C, Tranchand B, et al. Are population pharmacokinetic and/or pharmacodynamic models adequately evaluated? A survey of the literature from 2002 to 2004. Clin Pharmacokinet. 2007;46:221–34.
Hamrén B, Ericsson H, Samuelsson O, Karlsson M. Mechanistic modelling of tesaglitazar pharmacokinetic data in subjects with various degrees of renal function-evidence of interconversion. Br J Clin Pharmacol. 2008;65:855–63.
Cheng H, Jusko W. Pharmacokinetics of reversible metabolic systems. Biopharm Drug Dispos. 1993;14:721–66.
Gibaldi M, Perrier D. Pharmacokinetics. 2nd ed. New York: Marcel Dekker; 1982.
Vaughan D, Mallard D, Trainor A, Mitchard M. General pharmacokinetic equations for linear mammillary models with drug absorption into peripheral compartments. Europ J Clin Pharmacol. 1975;8:141–8.
Schwartz G. Estimating the dimension of a model. Ann Stat. 1978;6:461–4.
Faes C, Molenberghs G, Aerts M, Verbeke G, Kenward MG. The effective sample size and an alternative small-sample degrees-of-freedom method. Am Stat. 2009;63:389–99.
Visser I, Ray S, Jang W, Berger J. Effective sample size and the Bayes factor. Research Triangle Park, NC: workshop on latent variable modeling in social sciences, SAMSI; 2006.
Stram DO, Lee JW. Variance components testing in the longitudinal mixed effects model. Biometrics. 1994;50(4):1171–1177. Available from: http://www.jstor.org/stable/2533455.
Chou W, Yan F, Robbins-Weilert D, Ryder T, Liu W, Perbost C, et al. Comparison of two CYP2D6 genotyping methods and assessment of genotype-phenotype relationships. Clin Chem. 2003;49:542–51.
Ozdemir V, Kalow W, Tang B, Paterson A, Walker S, Endrenyi L, et al. Evaluation of the genetic component of variability in CYP3A4 activity: a repeated drug administration method. Pharmacogenetics. 2000;10:373–88.
Karlsson M, Holford N. A Tutorial on visual predictive checks. Marseille, France: 17th Population Approach Group in Europe; 2008. Available from: http://www.page-meeting.org/pdf\_assets/8694-Karlsson\_Holford\_VPC\_Tutorial\_hires.pdf.
Brendel K, Comets E, Laffont C, Mentré F. Evaluation of different tests based on observations for external model evaluation of population analyses. J Pharmacokinet Pharmacodyn. 2010;37:49–65.
Comets E, Brendel K, Mentré F. Computing normalised prediction distribution errors to evaluate nonlinear mixed-effect models: the npde add-on package for R. Comput Methods Programs Biomed. 2008;90:154–66.
Rowland M, Tozer T. Clinical pharmacokinetics. Concepts and applications. 3rd ed. USA: Lippincot Williams & Wilkins; 1995.
Martin S, Bishop F, Kerr B, Moor M, Moore M, Sheffels P, et al. Pharmacokinetics and metabolism of the novel anticonvulsant agent N-(2,6-dimethylphenyl)-5-methyl-3-isoxazolecarboxamide (D2624) in rats and humans. Drug Metab Dispos. 1997;25:40–6.
Duffull S, Chabaud S, Nony P, Laveille C, Girard P, Aarons L. A pharmacokinetic simulation model for ivabradine in healthy volunteers. Eur J Pharm Sci. 2000;10:285–94.
Murray M. Role of CYP pharmacogenetics and drug-drug interactions in the efficacy and safety of atypical and other antipsychotic agents. J Pharm Pharmacol. 2006;58:871–85.
Kang R, Jung S, Kim K, Lee D, Cho H, Jung B, et al. Effects of CYP2D6 and CYP3A5 genotypes on the plasma concentrations of risperidone and 9-hydroxyrisperidone in Korean schizophrenic patients. J Clin Psychopharmacol. 2009;29:272–7.
Bertilsson L, Lou Y, Du Y, Liu Y, Kuang T, Liao X, et al. Pronounced differences betweennative Chinese and Swedish populations in the polymorphic hydroxylations of debrisoquin andS-mephenytoin. Clin Pharmacol Ther. 1992;51:388–97.
ACKNOWLEDGMENTS
The authors would like to thank Prof. N. Holford for his clever inputs on the choice of the parameter's notation to facilitate the understanding of the different structural models. We would also like to thank the MONOLIX development team for making the MLXTRAN tool available for this work. During this work, Julie Bertrand was supported by a grant from the Institut de Recherches Internationales Servier (France).
Author information
Authors and Affiliations
Corresponding author
APPENDIX
APPENDIX
In all the equations below, C p is the parent drug concentration in plasma and C m is the active metabolite concentration in plasma following a single oral administration of a dose D of the parent drug.
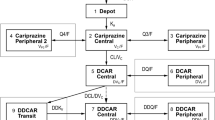
The ordinary differential equation system and the corresponding closed-form solution correspond to the last model on the right-hand side of Fig. 2, with similar absorption rates K a = K ap = K am.
Ordinary Differential Equation System
In the ordinary differential equation system (4), f is the fraction of dose after absorption, D is the dose, F p is the fraction of parent reaching systemic circulation after absorption, K a is the absorption constant for the parent and the metabolite, V p is the volume of distribution for the parent, V m is the volume of distribution for the metabolite, k po is the parent rate constant of elimination by other pathways (=CL po/V p), k pm is the parent rate constant of transformation into the metabolite (=CL pm/V p), k mo is the metabolite rate constant of elimination by other pathways (=CL mo/V m), and k mp is the metabolite rate constant of back-transformation into the parent (=CL mp/V m).
Closed-Form Solutions
In both Eqs. (5) and (6), E 1 is the parent drug total rate constant of elimination (=k po + k pm), E 2 is the metabolite total rate constant of elimination (=k mo + k mp), and λ 1 and λ 2 are the initial and terminal slopes of elimination, respectively defined in Eqs. (7) and (8).
Parameter Identifiability
From the model slopes, the following parameters may be identified: K a, λ 1, and λ 2. From the model intercepts, we can identify the following equations:
From (7) and (8) we can write:
This yields a system of six equations, where V, W, Y, and Z are four reals like λ 1 and λ 2. We can estimate the following parameters: V p/f F p, V m /f (1 − F p), E 1 = k po + k pm, E 2 = k mo + k mp, k pm F p/(1 − F p) and k mp(1 − F p)/F p, with the following solutions:
where \( {T_1} = \left( {\frac{{{\lambda_2}}}{W} - \frac{{{\lambda_1}}}{V}} \right)/\left( {\frac{1}{W} - \frac{1}{V}} \right) \) and \( {T_2} = \left( {\frac{{{\lambda_2}}}{Z} - \frac{{{\lambda_1}}}{Y}} \right)/\left( {\frac{1}{Z} - \frac{1}{Y}} \right) \). Alternatively, the following parameterization may be used instead:
Rights and permissions
About this article
Cite this article
Bertrand, J., Laffont, C.M., Mentré, F. et al. Development of a Complex Parent-Metabolite Joint Population Pharmacokinetic Model. AAPS J 13, 390–404 (2011). https://doi.org/10.1208/s12248-011-9282-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1208/s12248-011-9282-9