Abstract
Long non-coding RNAs (lncRNAs) are implicated in various cellular and pathological processes. Two lncRNAs, myocardial infarction-associated transcript (MIAT) and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), may be involved in the pathogenesis of coronary artery disease (CAD). Here, we aimed to determine the relative circulating levels of MIAT and MALAT1 in 110 stable CAD patients and 117 controls and to correlate their levels with the clinical and laboratory data. Peripheral blood expression levels were quantified by Real-Time qPCR. The median MIAT expression level in CAD patients was significantly 12-fold higher than controls (p<0.001). Otherwise, the median MALAT1 expression level was comparable in patient and control groups. Both lncRNAs showed significantly higher relative expression levels in patients with positive history of previous cardiac ischemic events, and MIAT showed significantly higher expression in diabetic CAD patients. The area under the curve of MIAT (0.888 ± 0.02 with sensitivity 95.5% and specificity 72.7%), was significantly larger than that of MALAT1 (0.601 ± 0.04 with sensitivity 50% and specificity 63.6%) for detecting the presence of significant CAD. The current findings suggest that lncRNA MIAT could have a diagnostic significance in CAD patients. MALAT1 levels, however, are not sufficiently reliable to have much clinical use in our cases.
Keywords:
CAD; lncRNA; MALAT1; MIAT; Real-time qPCR
Introduction
In the past years, the discovery of long non-coding RNAs (lncRNAs) has piqued global interest. These RNAs are non-coding sequences of more than 200 nucleotides that are able to regulate gene expression through affecting transcription, post-transcriptional events, as well as translation (Rinn and Chang, 2012Rinn JL and Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81:145-166; Huang and Zhang, 2014Huang B and Zhang R (2014) Regulatory non-coding RNAs: Revolutionizing the RNA world. Mol Biol Rep 41:3915-3923.). In turn, lncRNAs can influence various aspects of cell biology and disease development (Li et al., 2013Li X, Wu Z, Fu X and Han W (2013) Long noncoding RNAs: Insights from biological features and functions to diseases. Med Res Rev 33:517-553.; Boon et al., 2016Boon RA, Jaé N, Holdt L and Dimmeler S (2016) Long noncoding RNAs: From clinical genetics to therapeutic targets? J Am Coll Cardiol 67:1214-1226.). Several studies have detected and characterized the expression of lncRNAs in cardiac diseases and identified several types that control hypertrophy, cardiomyocyte apoptosis and mitochondrial function (Archer et al., 2015Archer K, Broskova Z, Bayoumi AS, Teoh JP, Davila A, Tang Y, Su H and Kim IM (2015) Long non-coding RNAs as master regulators in cardiovascular diseases. Int J Mol Sci 16:2651-23667.; Uchida and Dimmeler, 2015Uchida S and Dimmeler S (2015) Long noncoding RNAs in cardiovascular diseases. Circ Res 116:737-750.). One of the early identified myocardial infarct (MI)-related lncRNAs is the myocardial infarction-associated transcript (MIAT), encoded by the five exon MIAT gene on chromosome 22q12.1 (Ishii et al., 2006Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M et al. (2006) Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 51:1087-1099.). In vitro functional analysis has revealed an MI susceptibility variant in exon 5 of MIAT, associated with an increased transcriptional level of the gene (Ishii et al., 2006Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M et al. (2006) Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 51:1087-1099.), suggesting a possible role of MIAT in the pathogenesis of MI. In addition, MIAT was shown to be involved in vascular endothelial dysfunction (Yan et al., 2015Yan B, Yao J, Liu J-Y, Li XM, Wang XQ, Li YJ, Tao ZF, Song YC, Chen Q and Jiang Q (2015) LncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res 116:1143-1156.), which suggests a role in the pathogenesis of atherosclerosis.
Another lncRNA with a possible relation to the pathogenesis of coronary artery disease (CAD) is MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), coded by the MALAT1 gene on chromosome 11q13.1.(Michalik et al., 2014Michalik KM, You X, Manavski Y, Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S et al. (2014) Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 114:1389-1397.). In the vascular system, this endothelial-expressed lncRNA can regulate vessel growth and function (Michalik et al., 2014Michalik KM, You X, Manavski Y, Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S et al. (2014) Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 114:1389-1397.) and was proven to have a role in the regulation of endothelial cell proliferation (Liu et al., 2014Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ, Yan B and Jiang Q (2014) Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis 5:e1506.), as well as in the promotion of skeletal muscle differentiation (Watts et al., 2013Watts R, Johnsen VL, Shearer J and Hittel DS (2013) Myostatin-induced inhibition of the long noncoding RNA MALAT1 is associated with decreased myogenesis. Am J Physiol Cell Physiol 304:C995-1001.). Interestingly, levels of MALAT1 have been shown to rise considerably with hypoxia (Michalik et al., 2014Michalik KM, You X, Manavski Y, Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S et al. (2014) Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 114:1389-1397.). This might again point to a possible role of MALAT1 in CAD, in which there is local hypoxia in the ischemic regions.
There are still several gaps in the existing knowledge. While MIAT variants have already been linked to MI (Ishii et al., 2006Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M et al. (2006) Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 51:1087-1099.), only a few controversial studies are available to date on the expression level of MIAT in peripheral blood of CAD patients (Vausort et al., 2014Vausort M, Wagner DR and Devaux Y (2014) Long noncoding RNAs in patients with acute myocardial infarction. Circ Res 115:668-677.; Liao et al., 2016Liao J, He Q, Li M, Chen Y, Liu Y and Wang J (2016) LncRNA MIAT: Myocardial infarction associated and more. Gene 578:158-161.). In addition, MALAT1 does have a role in endothelial cell function, but whether it plays a causative role in CAD is unknown. This study aims to determine the expression level of MIAT and MALAT1 in CAD patients and to correlate these levels with the clinical and laboratory data of the patients.
Subjects and Methods
Study participants and protocol
This observational, case-controlled study included 110 nonconsanguineous stable CAD patients and 117 age- and sex-matched controls. Patients were consecutively recruited from the Cardiology Department, Suez Canal University (SCU) Hospital, Egypt, during the period between October 2015 and January 2016. Diagnosis of CAD was based on detailed history taking, clinical examination, resting electrocardiography and echocardiography, and further confirmed by coronary angiography (≥ 70% stenosis of at least one major coronary vessel caused by atherosclerosis) (Fawzy et al., 2017aFawzy MS, Toraih EA, Aly NM, Fakhr-Eldeen A, Badran DI and Hussein MH (2017a) Atherosclerotic and thrombotic genetic and environmental determinants in Egyptian coronary artery disease patients: A pilot study. BMC Cardiovasc Disord 17:26., 2018Fawzy MS, Toraih EA, Hamed EO, Hussein MH and Ismail HM (2018) Association of MIR-499a expression and seed region variant (rs3746444) with cardiovascular disease in Egyptian patients. Acta Cardiol 73:131-140.). Patients with congenital heart disease, valvular heart disease or non-atherosclerotic CAD (e.g. vasculitis, fibromuscular dysplasia, etc.), were excluded. Controls had no past history of cardiovascular problems and had a normal resting ECG. Selective coronary angiography was not performed for controls following our institutional ethical guidelines that do not permit to apply invasive procedures for controls in research. The study was conducted in accordance with the guidelines in the Declaration of Helsinki and approved by the Medical Research Ethics Committee of Faculty of Medicine, Suez Canal University (approval no. 2714). All participants enrolled in this study provided a written informed consent.
As part of risk assessment, the body mass index (BMI) of all participants was calculated. Hypertension was defined as repeated blood pressure ≥ 140/90 mmHg or regular use of anti-hypertensive drugs. Diabetes was defined as receiving hypoglycemic drugs, blood sugar > 11.1 mmol/Lon admission, or fasting hyperglycemia ≥ 7 mmol/Lin two determinants. Dyslipidemia was defined as serum triglycerides (TG) ≥ 1.7 mmol/L, total cholesterol (TC) > 2.3 mmol/L or high-density lipoprotein-cholesterol (HDL-c) < 1.0 mmol/L in males and < 1.3 mmol/L in females, or the intake of lipid-lowering drugs (Jellinger et al., 2017Jellinger PS, Handelsman Y, Rosenblit PD, Bloomgarden ZT, Fonseca VA, Garber AJ, Grunberger G, Guerin CK, Bell DSH, Mechanick JI et al. (2017) American Association of Clinical Endocrinologists and American College of Endocrinology guidelines for management of dyslipidemia and prevention of atherosclerosis. Endocr Pract 23:479-497.). Other risk factors, such as smoking and family history (FH) of premature CAD, first degree male relatives <55 years or females <65 years were documented. The Framingham Risk Score was used to estimate the 10-year cardiovascular risk in participants. Low risk was defined as a score less than 10%, intermediate risk between 10 and 20%, and high risk over 20% (Ford et al., 2004Ford ES, Giles WH and Mokdad AH (2004) The distribution of 10-Year risk for coronary heart disease among US adults: Findings from the National Health and Nutrition Examination Survey III. J Am Coll Cardiol 43:1791-1796.). Thrombolysis In Myocardial Infarction (TIMI) score Risk Index (TRI) was used to provide mortality estimates in CAD patients in terms of mild, intermediate, and high risk (Antman et al., 2000Antman EM, Cohen M, Bernink PJ, McCabe CH, Horacek T, Papuchis G, Mautner B, Corbalan R, Radley D and Braunwald E (2000) The TIMI risk score for unstable angina/non-ST elevation MI: A method for prognostication and therapeutic decision making. JAMA 284:835-842.).
A two-dimensional conventional echocardiographic study was performed for all patients using the standard views to exclude the presence of structural heart disease using a commercially available system (General Electric Healthcare Company, Vivid seven Dimensions Vingmed and Horten- Norway) with the 2.5-MHz phased array probe. All echocardiographic data were interpreted by two independent experts in echocardiography.
Selective coronary angiography was performed for all patients from the right femoral approach using the standard modified Seldinger technique (Schussler, 2011Schussler JM (2011) Effectiveness and safety of transradial artery access for cardiac catheterization.Proc (Bayl Univ Med Cent) 24:205-209.). CAD extent and severity were assessed according to our hospital protocol using the Gensini score (Saha et al., 2015Saha T, Khalequzzaman M, Akanda MAK, Saha S, Tushar AZ, Ahmed R, Saha GK and Ullah M (2015) Association of GRACE risk score with angiographic severity of coronary artery disease in patients with ST elevation myocardial infarction. Cardiovasc J 8:30-34.). The score is calculated based on the vessel affected, the location of the lesion, and the degree of stenosisl. This was determined and interpreted by two independent angiographers who were blinded to the clinical data.
Specimen collection and laboratory investigations
Six milliliters of fasting blood samples were withdrawn from the median cubital vein of participants under aseptic conditions in sodium citrate, plain, and EDTA-Na2vacutainers. The citrated and clotted blood tubes were centrifuged at 2500 rpm for 20 minutes. Citrated plasma was immediately used to measure the prothrombin time (PT) using an automated analyzer (Sysmex CA-1500, USA), and the serum was kept at -20 °C in aliquots until the time of laboratory assessment. Fasting blood glucose(FBG), serum TC, HDL-c and TG were determined by the enzymatic method using a Hitachi 912 automated chemistry analyzer (Roche Diagnostics Co, Mannheim, Germany). Serum low density lipoprotein-cholesterol (LDL-c) was calculated by Friedewald’s equation, as all participants had TG values less than 4.5 mmol/L (Friedewald et al., 1972Friedewald WT, Levy RI and Fredrickson DS (1972) Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 18:499-502.). The EDTA tubes were used for subsequent genetic analysis.
RNA extraction and reverse transcription (RT)
The total RNA was isolated freshly from whole blood using ABIOpureTM Total RNA (Alliance Bio, USA, Catalog no. M541RP50-B) following the manufacturer’s protocol. RNA concentration and purity at the absorbance ratio 260/280 nm were determined by NanoDrop ND-1000 spectrophotometer (NanoDrop Tech., Inc. Wilmington, USA) followed by agarose gel electrophoresis to check for RNA integrity. The range of the extracted RNA was 20-65 ng/μL and the total RNA concentration in every sample was checked to be the same for the subsequent reverse transcription (RT) step. High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, P/N 4368814) was used. For each 20 μL RT reaction, a 10 μL RNA sample was combined with 10 μL of RT reaction mix containing 2 μL of 10x Buffer, 0.8 μL of 25x dNTP (100 mM), 2 μL of 10x random primers, 1 μL of MultiScribe Reverse Transcriptase, 1 μL of RNase inhibitor, and 3.2 μL of nuclease-free water. RT was carried out in T-Professional Basic, Biometra PCR System (Biometra, Göttingen, Germany) at 25 °C for 10 min, followed by 37 °C for 120 min, and finally 85 °C for 5 min followed by a final hold step at 4 °C (Toraih et al., 2018Toraih EA, Ellawindy A, Fala SY, Al Ageeli E, Gouda NS, Fawzy MS and Hosny S (2018) Oncogenic long noncoding RNA MALAT1 and HCV-related hepatocellular carcinoma. Biomed Pharmacother 102:653-669.). All reactions included two types of controls: non-template control (NTC) and no-reverse transcriptase control. The cDNA products were subjected immediately to quantification by real-time PCR without storage.
Gene expression analysis by quantitative Real-Time Polymerase Chain Reaction
Expression of MIAT, MALAT1 and GAPDH (as an endogenous control) were quantified using pre-designed TaqMan® assays (Applied Biosystems, ID Hs00402814_m1, Hs00273907_s1, and Hs402869, respectively) and Taqman® Universal PCR master mix II (Applied Biosystems, P/N 4440043). The detailed description of each assay is available at (http://www.thermofisher.com). The PCR reactions were carried out in accordance with the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (Bustin et al., 2009Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL et al. (2009) The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622.) in a final volume of 20 μL, including 1.33 μL RT product, 10 μL TaqMan®Universal PCR Master Mix, 1 μL TaqMan® assays. All reactions included three types of control: two controls for RT reactions and NTC for real time PCR (that contains nuclease free water instead of cDNA), to confirm the absence of non-specific PCR product generation. The PCR assays were performed in a StepOne Real-Time PCR System (Applied Biosystems) as follows: 95 °C for 10 minu followed by 40 cycles of 92 °C for 15 s and 60 °C for 1 min (Fawzy et al., 2017bFawzy MS, Toraih EA and Abdallah HY (2017b) Long noncoding RNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1): A molecular predictor of poor survival in glioblastoma multiforme in Egyptian patients. Egypt J Med Hum Genet 18:231-239.). Ten randomly selected study control samples were re-evaluated for the expression of the three studied genes in separate runs to test the reproducibility of the qPCR which showed very close Cq value results and low standard deviations. Serial dilutions by 1/10 to 1/100,000 of cDNA from a pool of CAD patients and three RT-PCR controls were run to test the sensitivity of the studied long non-coding RNAs.
Statistical analysis
Appropriate sample size and power calculations were carried out using Quanto software package version 1.2.4 (http://biostats.usc.edu/software). Chi-square, Fisher’s exact, Student’s t, and Mann-Whitney U tests were used when appropriate. Logistic regression was performed to adjust for smoking status, lipid profile, platelet count, and white blood cell parameters (as MIAT was reported to be positively associated with the percentage of lymphocytes and negatively associated with neutrophils and platelets) (Sun et al., 2018Sun C, Huang L, Li Z, Leng K, Xu Y, Jiang X and Cui Y (2018) Long non-coding RNA MIAT in development and disease: A new player in an old game. J Biomed Sci 25:23.). Receiver operating characteristic (ROC) curves were generated to obtain the best cutoff values of both lncRNAs for discriminating CAD patients from controls. A two-tailed p<0.05 was considered statistically significant. The fold change of lncRNAs expression was calculated using the Livak and Schmittgen (2001)Livak KJ and Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) method. Methods 25:402–408. method based on the quantitative cycle (Cq) values with the following equation: relative quantity = 2-DDCq, where ΔΔCq = (Cq LncRNA – Cq GAPDH) CAD - (Cq LncRNA – Cq GAPDH) Controls. Data were managed using the Statistical Package for the Social Sciences (SPSS) for Windows (version 20.0).
Results
Baseline characteristics of the study population
The demographic, basic laboratory, and cardiac assessment parameters of CAD patients and controls are shown in Table 1.
Cardiac catheterization showed that nearly half the patients had at least a single vessel disease (defined as luminal narrowing of > 70%); almost all of which have affection of the left anterior descending artery, while 10% and 23% of patients showed a two and three-vessel disease respectively. About a fifth of the patients had no discernible obstructive disease in the coronary angiogram. Less than 10% of the patients were classified as intermediate/high TRI, which denotes an in-hospital mortality risk > 10%. Variables for which statistically significant differences were found between the groups were considered for logistic regression correction.
Expression profile of MIAT and MALAT1
The endogenous control GAPDH was found uniformly expressed in all samples (26.9 ± 3.4 in controls, 27.924.58 in patients, p=0.229). In contrast, mean Cq values of MIAT and MALAT1 varied significantly between the studied groups (p=0.005 and p=0.007, respectively). All samples exhibited up-regulation of MIAT with a relative expression ratio > 1.0. Nearly 27% of the patients (30/110) exhibited remarkably high levels, > 100-fold. The median MIAT relative expression level in patients was 12-fold (interquartile 457.3) higher than that in control individuals (p<0.001) (Figure 1A). Regarding the MALAT1 expression profile, this was under-expressed (<1.0) in nearly half the patients, while the other half showed up-regulation. About 15% of the over-expressed samples was dramatically elevated by > 100-fold. The median (IQR) MALAT1 level in patients was 1.02 (7.47), which was nearly similar to controls (Figure 1B).
Peripheral blood expression levels of MIAT (A) and MALAT1 (B) in patients and controls. Data are represented as medians. The box defines upper and lower quartiles (25% and 75%, respectively), and the error bars indicate upper and lower adjacent limits. Relative expression levels of MIAT and MALAT1 were normalized to GAPDH expression level for each sample and calculated using the delta-delta Cq method [=2 (-DDCq)] in comparison to controls. Mann-Whitney U test was used for comparison.
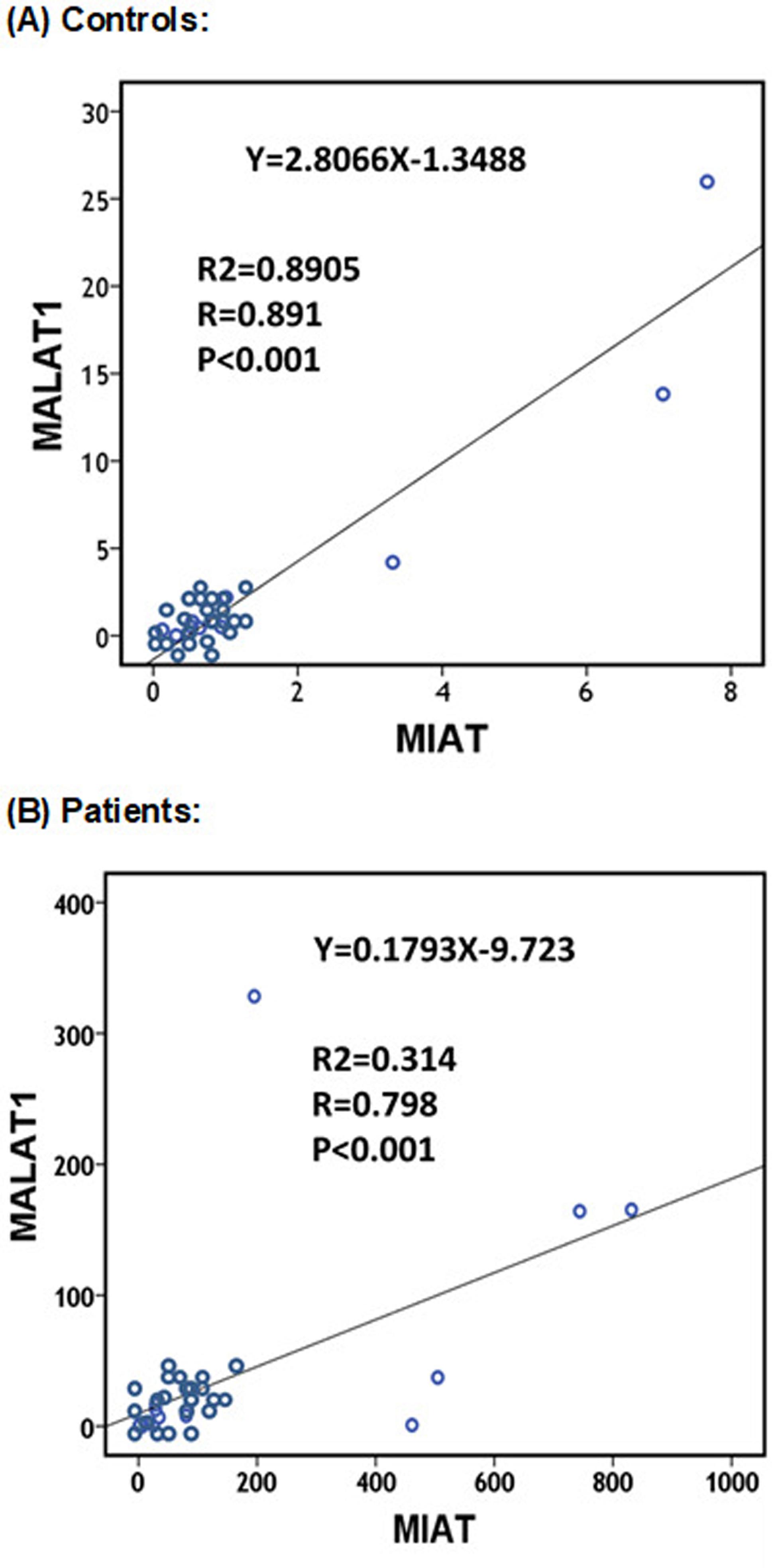
MIAT and MALAT1, being both pro-atherogenic lncRNAs (Jian et al., 2016Jian L, Jian D, Chen Q and Zhang L (2016) Long noncoding RNAs in atherosclerosis. J Atheroscler Thromb 23:376-384.), showed a positive correlation in their expression profiles in both CAD patients and controls (r=0.798 and 0.891, respectively; p<0.001) (Figure 2). However, after removing the outliers, a direct correlation between MIAT and MALAT1 expression levels still existed in the cases (r=0.620, p=0.008) but not in the control group (r=0.666, p=0.072).
Correlation between peripheral blood MIAT and MALAT1 in controls (A) and patients (B). Spearman’s rho binary correlation analysis was performed.
Association between the expression levels and clinical and laboratory features
MALAT1 expression was significantly higher in patients with positive FH of CAD. When patients were classified into high and low expressers for circulating MALAT1, there were no differences in the baseline characteristics of CAD patients between the subgroups. Circulating MIAT was significantly higher expressed in diabetic CAD patients. Both MIAT and MALAT1 expression levels were associated with hypertension and premature CAD (Table 2). Also, both lncRNAs showed higher relative expression in patients with a positive history of previous cardiac ischemic events. Furthermore, MALAT1 showed higher expression in patients with previous history of stroke.
Association between MIAT and MALAT1 expression levels and clinical characteristics in patients.
ROC curve analysis
The AUC (area under curve) of MIAT was significantly larger than that of MALAT1 for detecting the presence of significant CAD (Figure 3A). However, the AUC of MIAT and MALAT1 were similar in predicting the presence of multivessel disease and high Gensini score (≥ 20) in CAD patients (Figure 3B, C).
Receivers operating characteristic curves of lncRNAs MIAT and MALAT1 using relative expression levels. Peripheral blood MIAT (blue line) and MALAT1 (green line) expressions were plotted to determine their sensitivity and specificity at the cutoff fold change value for detecting the presence of significant coronary artery disease (A), multivessel disease (B), and high Gensini score > 20 (C) in CAD patients. AUC: Area under the curve; CI: Confidence interval; SE: Standard error; SEN: sensitivity; SP: specificity.
DISCUSSION
In the past decade, high-throughput sequencing analysis identified a group of lncRNAs that were found to be important regulators of gene function in many biological processes (Trapnell et al., 2010Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ and Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515.). Disease-related deregulation of their expression was evident in various clinical entities, including cardiovascular disease (Zangrando et al., 2014Zangrando J, Zhang L, Vausort M, Maskali F, Marie PY, Wagner DR and Devaux Y (2014) Identification of candidate long non-coding RNAs in response to myocardial infarction. BMC Genomics 15:460.; Zhang et al., 2016Zhang J,Yuan L,Zhang X, Hamblin MH, Zhu T, Meng F, Li Y, Chen YE and Yin KJ (2016) Altered long non-coding RNA transcriptomic profiles in brain microvascular endothelium after cerebral ischemia. Exp Neurol 277:162-170.). More specifically, CAD has been characterized by a transcriptional reprogramming, which activates a network of cardiac signals that interact and converge on cardiac transcriptional factors. Collectively, these factors activate specific temporal and spatial programs to generate cardiac pathological remodeling controlled by lncRNAs (Archer et al., 2015Archer K, Broskova Z, Bayoumi AS, Teoh JP, Davila A, Tang Y, Su H and Kim IM (2015) Long non-coding RNAs as master regulators in cardiovascular diseases. Int J Mol Sci 16:2651-23667.). Given that lncRNAs MIAT and MALAT1 are known to affect endothelial function through alteration of specific signaling pathways that affect cellular proliferation, migration, and survival (Liu et al., 2014Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ, Yan B and Jiang Q (2014) Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis 5:e1506.; Yan et al., 2015Yan B, Yao J, Liu J-Y, Li XM, Wang XQ, Li YJ, Tao ZF, Song YC, Chen Q and Jiang Q (2015) LncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res 116:1143-1156.), we speculated on their potential role in the development of atherosclerosis and subsequent CAD.
In the present study, MIAT relative expression levels were elevated in peripheral blood of CAD patients, with a 12-fold higher relative expression level compared to healthy controls. In contrast, Vausort et al. (2014)Vausort M, Wagner DR and Devaux Y (2014) Long noncoding RNAs in patients with acute myocardial infarction. Circ Res 115:668-677. found that MIAT was equally expressed in peripheral blood of both MI patients and healthy individuals, although they found a significant difference in its relative expression between ST elevation MI (STEMI) and Non-ST segment MI (NSTEMI) subgroups. While these results might seem contradictory to ours, we propose that this could be attributed to the time of MIAT relative expression quantification. The previous authors quantified MIAT expression levels within hours from the onset of the acute attack, while our assessments were made in already diagnosed stable CAD patients presenting for angiography, and not necessarily during the acute attack. This may suggest that MIAT levels are not initially raised during the acute attack, but rise later and remain elevated for an extended time. The significance of this, however, is yet unclear, but may point to a possible role of MIAT as an adaptive mechanism similar to the activation of the renin-angiotensin-aldosterone system that occurs in post MI patients (Sutton and Sharpe, 2000Sutton M and Sharpe N (2000) Left ventricular remodeling after myocardial infarction: Pathophysiology and therapy. Circulation 101:2981-2988.). Another finding in the current study that could further support this hypothesis is that patients with previous cardiac ischemic events have significantly higher levels of MIAT than those without.
Interestingly, it has been reported recently that MIAT functions as a miR-150-5p sponge that affects the repression of VEGF (vascular endothelial growth factor), which promotes inflammatory cellular migration into the intima, as well as inducing pathological angiogenesis (Zhu et al., 2018Zhu M, Li N, Luo P, Jing W, Wen X, Liang C and Tu J (2018) Peripheral blood leukocyte expression of lncRNA MIAT and its diagnostic and prognostic value in ischemic stroke. J Stroke Cerebrovasc Dis 27:326-337.). During the process of angiogenesis, which is a natural phenomenon after a CAD event, MIAT could significantly be up-regulated, subsequently sponging miR-150-5p, thereby up-regulating the level of VEGF and other factors contributing to atherosclerosis. Moreover, MIAT was recently found to be up-regulated in patients with ischemic stroke, a disease that shares a similar pathogenesis with CAD (Sun and Wong, 2016Sun X and Wong D (2016) Long non-coding RNA-mediated regulation of glucose homeostasis and diabetes. Am J Cardiovasc Dis 6:17-25.).
On the other hand, in the current study, circulatory MALAT1 relative expression levels were up-regulated in half the patients undergoing coronary angiography. These relative expression levels were markedly elevated in patients with a history of previous ischemic events. This is consistent with the hypoxia-induced up-regulation of lncRNA MALAT1 seen in ischemic limbs (Watts et al., 2013Watts R, Johnsen VL, Shearer J and Hittel DS (2013) Myostatin-induced inhibition of the long noncoding RNA MALAT1 is associated with decreased myogenesis. Am J Physiol Cell Physiol 304:C995-1001.). Recently, Sallé-Lefort et al. (2016)Sallé-Lefort S, Miard S, Nolin MA, Boivin L, Paré MÈ, Debigaré R and Picard F (2016) Hypoxia upregulates MALAT1 expression through a CaMKK/AMPK/HIF-1α axis. Int J Oncol 49:1731-1736. indicated that the enhanced MALAT1 gene transcription upon low oxygen conditions was under the control of hypoxia-inducible factor 1α (HIF-1α), regulated by the activation of the CaMKK/AMPK (calcium/calmodulin-dependent protein kinase kinase/AMP-activated protein kinase) complex. At the cellular level, MALAT1 was found to act as a decoy that binds to and interferes with the function of other RNAs or proteins (Tripathi et al., 2010Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA et al. (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39:925–938.). In addition, it has been found to regulate genes that induce proliferation in endothelial cells, through enhancing cell cycle regulatory genes, particularly CCNA2 (Cycline A2) gene, CCNB1, and CCNB2, and repressing the cell cycle inhibitor genes p21 and p27Kip1 (Michalik et al., 2014Michalik KM, You X, Manavski Y, Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S et al. (2014) Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 114:1389-1397.).
In the current work, the MALAT1 gene expression profile correlated strongly with MIAT expression levels in CAD patients, both in the control and CAD patient groups, supporting the shared significant roles these lncRNAs could play in vascular physiological or pathological processes (Jian et al., 2016Jian L, Jian D, Chen Q and Zhang L (2016) Long noncoding RNAs in atherosclerosis. J Atheroscler Thromb 23:376-384.). Altered MALAT1 expression was reported in previous studies and was closely related to various diseases, ranging from diabetic complications (Wu et al., 2015Wu Y, Huang C, Meng X and Li J (2015) Long noncoding RNA MALAT1: Insights into its biogenesis and implications in human disease. Curr Pharm Des 21:5017-5028.), to a wide array of cancers (Ren et al., 2016Ren D, Li H, Li R, Sun J, Guo P, Han H, Yang Y and Li J (2016) Novel insight into MALAT-1 in cancer: Therapeutic targets and clinical applications. Oncol Lett 11:1621-1630.; Toraih et al., 2018Toraih EA, Ellawindy A, Fala SY, Al Ageeli E, Gouda NS, Fawzy MS and Hosny S (2018) Oncogenic long noncoding RNA MALAT1 and HCV-related hepatocellular carcinoma. Biomed Pharmacother 102:653-669.). Furthermore, MALAT1 was one of the most up-regulated oxygen deprivation-responsive endothelial lncRNAs in ischemic stroke, implying a critical role in protecting the cerebral microvasculature from cerebral ischemic insults in mice (Zhang et al., 2016Jian L, Jian D, Chen Q and Zhang L (2016) Long noncoding RNAs in atherosclerosis. J Atheroscler Thromb 23:376-384.).
Although correlation of MIAT and MALAT1 levels with age was not statistically significant, it was found that their levels were significantly higher in premature CAD cases, being highly significant in case of MIAT and borderline in case of MALAT1. Circulating levels of MIAT and MALAT1, in addition, correlate with the severity of CAD using the gold standard, coronary angiography expressed in terms of the Gensini score, with MIAT being more sensitive in the detection of the presence of significant CAD. Another finding in the current study, was the link between positive family history for CAD and MALAT1 levels, highlighting the presence of a genetic heritable factor and suggesting MALAT1 as a putative risk factor for CAD. Moreover, the higher relative expression observed in CAD diabetic and hypertensive patient subgroups suggest a possible role of these types of lncRNA in the mechanisms by which diabetes and hypertension produce endothelial dysfunction and accelerate the rate of atherosclerosis (Dharmashankar and Widlansky, 2010Dharmashankar K and Widlansky ME (2010) Vascular endothelial function and hypertension: Insights and directions. Curr Hypertens Rep 12:448-455.; Kolluru et al., 2012Kolluru GK, Bir SC and Kevil CG (2012) Endothelial dysfunction and diabetes: Effects on angiogenesis, vascular remodeling, and wound healing. Int J Vasc Med 2012:918267.). Two recent studies reported the possible contribution of MIAT and MALAT1 to endothelial dysfunction in diabetic patients (Puthanveetil et al., 2015Puthanveetil P, Chen S, Feng B, Gautam A and Chakrabarti S (2015) Long non-coding RNA MALAT1 regulates hyperglycemia induced inflammatory process in the endothelial cells. J Cell Mol Med 19:1418-1425.; Yan et al., 2015Yan B, Yao J, Liu J-Y, Li XM, Wang XQ, Li YJ, Tao ZF, Song YC, Chen Q and Jiang Q (2015) LncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res 116:1143-1156.). The first one showed that MIAT expression can be induced by hyperglycemia and is associated with the microvascular dysfunction occurring in diabetes. This could partially explain the high MIAT relative expression in our CAD diabetic patients versus the non-diabetic subgroup. The second study found an increase in endothelial MALAT1 levels in response to hyperglycemia resulting in inflammation and subsequent endothelial cell dysfunction (Puthanveetil et al., 2015Puthanveetil P, Chen S, Feng B, Gautam A and Chakrabarti S (2015) Long non-coding RNA MALAT1 regulates hyperglycemia induced inflammatory process in the endothelial cells. J Cell Mol Med 19:1418-1425.).
Conclusions
Collectively, whether lncRNAs MIAT and MALAT1 have a causative role in CAD is still unknown, but our study confirms the possible association of their circulatory relative expression levels with coronary artery disease in our population. This association was more prominent in CAD patients with previous cardiac ischemic events. The current study was limited by the relatively small sample size and the fact that the samples were drawn only from outpatients presenting for coronary angiography, and not during the acute attack. We recommend further studies of a larger scale and serial measurements during the acute attack and at regular intervals afterwards. In addition, the molecular basis of the possible role of these lncRNAs as an adaptive mechanism should be further explored and established, as this might be an interesting target for future therapeutic interventions in post myocardial infarction patients.
Acknowledgments
The authors would like to thank all study participants and the Oncology Diagnostic Unit, and Center of Excellence in Molecular and Cellular Medicine; Suez Canal University, Egypt, for providing the facilities for performing the current work.
Conflict of Interest
The authors declare no conflict of interest.
Author contributions
EAT, MSF, AE-W and ME-W. conceived and designed the experiments; ME-W supplied the study samples. EAT and MSF. performed the experiments; EAT performed the statistical analysis. All authors contributed reagents/materials/analysis tools, and all authors contributed in writing, reading and approval of the final manuscript.
References
- Antman EM, Cohen M, Bernink PJ, McCabe CH, Horacek T, Papuchis G, Mautner B, Corbalan R, Radley D and Braunwald E (2000) The TIMI risk score for unstable angina/non-ST elevation MI: A method for prognostication and therapeutic decision making. JAMA 284:835-842.
- Archer K, Broskova Z, Bayoumi AS, Teoh JP, Davila A, Tang Y, Su H and Kim IM (2015) Long non-coding RNAs as master regulators in cardiovascular diseases. Int J Mol Sci 16:2651-23667.
- Boon RA, Jaé N, Holdt L and Dimmeler S (2016) Long noncoding RNAs: From clinical genetics to therapeutic targets? J Am Coll Cardiol 67:1214-1226.
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL et al. (2009) The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622.
- Dharmashankar K and Widlansky ME (2010) Vascular endothelial function and hypertension: Insights and directions. Curr Hypertens Rep 12:448-455.
- Fawzy MS, Toraih EA, Aly NM, Fakhr-Eldeen A, Badran DI and Hussein MH (2017a) Atherosclerotic and thrombotic genetic and environmental determinants in Egyptian coronary artery disease patients: A pilot study. BMC Cardiovasc Disord 17:26.
- Fawzy MS, Toraih EA and Abdallah HY (2017b) Long noncoding RNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1): A molecular predictor of poor survival in glioblastoma multiforme in Egyptian patients. Egypt J Med Hum Genet 18:231-239.
- Fawzy MS, Toraih EA, Hamed EO, Hussein MH and Ismail HM (2018) Association of MIR-499a expression and seed region variant (rs3746444) with cardiovascular disease in Egyptian patients. Acta Cardiol 73:131-140.
- Ford ES, Giles WH and Mokdad AH (2004) The distribution of 10-Year risk for coronary heart disease among US adults: Findings from the National Health and Nutrition Examination Survey III. J Am Coll Cardiol 43:1791-1796.
- Friedewald WT, Levy RI and Fredrickson DS (1972) Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 18:499-502.
- Huang B and Zhang R (2014) Regulatory non-coding RNAs: Revolutionizing the RNA world. Mol Biol Rep 41:3915-3923.
- Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M et al. (2006) Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 51:1087-1099.
- Jellinger PS, Handelsman Y, Rosenblit PD, Bloomgarden ZT, Fonseca VA, Garber AJ, Grunberger G, Guerin CK, Bell DSH, Mechanick JI et al. (2017) American Association of Clinical Endocrinologists and American College of Endocrinology guidelines for management of dyslipidemia and prevention of atherosclerosis. Endocr Pract 23:479-497.
- Jian L, Jian D, Chen Q and Zhang L (2016) Long noncoding RNAs in atherosclerosis. J Atheroscler Thromb 23:376-384.
- Kolluru GK, Bir SC and Kevil CG (2012) Endothelial dysfunction and diabetes: Effects on angiogenesis, vascular remodeling, and wound healing. Int J Vasc Med 2012:918267.
- Li X, Wu Z, Fu X and Han W (2013) Long noncoding RNAs: Insights from biological features and functions to diseases. Med Res Rev 33:517-553.
- Liao J, He Q, Li M, Chen Y, Liu Y and Wang J (2016) LncRNA MIAT: Myocardial infarction associated and more. Gene 578:158-161.
- Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ, Yan B and Jiang Q (2014) Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis 5:e1506.
- Livak KJ and Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) method. Methods 25:402–408.
- Michalik KM, You X, Manavski Y, Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S et al. (2014) Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 114:1389-1397.
- Puthanveetil P, Chen S, Feng B, Gautam A and Chakrabarti S (2015) Long non-coding RNA MALAT1 regulates hyperglycemia induced inflammatory process in the endothelial cells. J Cell Mol Med 19:1418-1425.
- Ren D, Li H, Li R, Sun J, Guo P, Han H, Yang Y and Li J (2016) Novel insight into MALAT-1 in cancer: Therapeutic targets and clinical applications. Oncol Lett 11:1621-1630.
- Rinn JL and Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81:145-166
- Saha T, Khalequzzaman M, Akanda MAK, Saha S, Tushar AZ, Ahmed R, Saha GK and Ullah M (2015) Association of GRACE risk score with angiographic severity of coronary artery disease in patients with ST elevation myocardial infarction. Cardiovasc J 8:30-34.
- Sallé-Lefort S, Miard S, Nolin MA, Boivin L, Paré MÈ, Debigaré R and Picard F (2016) Hypoxia upregulates MALAT1 expression through a CaMKK/AMPK/HIF-1α axis. Int J Oncol 49:1731-1736.
- Schussler JM (2011) Effectiveness and safety of transradial artery access for cardiac catheterization.Proc (Bayl Univ Med Cent) 24:205-209.
- Sun C, Huang L, Li Z, Leng K, Xu Y, Jiang X and Cui Y (2018) Long non-coding RNA MIAT in development and disease: A new player in an old game. J Biomed Sci 25:23.
- Sun X and Wong D (2016) Long non-coding RNA-mediated regulation of glucose homeostasis and diabetes. Am J Cardiovasc Dis 6:17-25.
- Sutton M and Sharpe N (2000) Left ventricular remodeling after myocardial infarction: Pathophysiology and therapy. Circulation 101:2981-2988.
- Toraih EA, Ellawindy A, Fala SY, Al Ageeli E, Gouda NS, Fawzy MS and Hosny S (2018) Oncogenic long noncoding RNA MALAT1 and HCV-related hepatocellular carcinoma. Biomed Pharmacother 102:653-669.
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ and Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515.
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA et al. (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39:925–938.
- Uchida S and Dimmeler S (2015) Long noncoding RNAs in cardiovascular diseases. Circ Res 116:737-750.
- Vausort M, Wagner DR and Devaux Y (2014) Long noncoding RNAs in patients with acute myocardial infarction. Circ Res 115:668-677.
- Watts R, Johnsen VL, Shearer J and Hittel DS (2013) Myostatin-induced inhibition of the long noncoding RNA MALAT1 is associated with decreased myogenesis. Am J Physiol Cell Physiol 304:C995-1001.
- Wu Y, Huang C, Meng X and Li J (2015) Long noncoding RNA MALAT1: Insights into its biogenesis and implications in human disease. Curr Pharm Des 21:5017-5028.
- Yan B, Yao J, Liu J-Y, Li XM, Wang XQ, Li YJ, Tao ZF, Song YC, Chen Q and Jiang Q (2015) LncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res 116:1143-1156.
- Zangrando J, Zhang L, Vausort M, Maskali F, Marie PY, Wagner DR and Devaux Y (2014) Identification of candidate long non-coding RNAs in response to myocardial infarction. BMC Genomics 15:460.
- Zhang J,Yuan L,Zhang X, Hamblin MH, Zhu T, Meng F, Li Y, Chen YE and Yin KJ (2016) Altered long non-coding RNA transcriptomic profiles in brain microvascular endothelium after cerebral ischemia. Exp Neurol 277:162-170.
- Zhu M, Li N, Luo P, Jing W, Wen X, Liang C and Tu J (2018) Peripheral blood leukocyte expression of lncRNA MIAT and its diagnostic and prognostic value in ischemic stroke. J Stroke Cerebrovasc Dis 27:326-337.
-
Associate Editor: Emmanuel Dias Neto
Publication Dates
-
Publication in this collection
14 Nov 2019 -
Date of issue
Jul-Sep 2019
History
-
Received
20 June 2018 -
Accepted
25 Nov 2018