Abstract
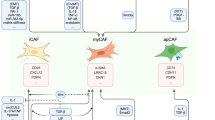
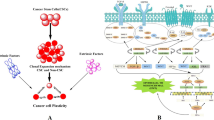
The tumor microenvironment (TME) plays an important role in supporting cancer progression. The TME is composed of tumor cells, the surrounding tumor-associated stromal cells, and the extracellular matrix (ECM). Crosstalk between the TME components contributes to tumorigenesis. Recently, one of our studies showed that pancreatic ductal adenocarcinoma (PDAC) cells can induce DNA methylation in cancer-associated fibroblasts (CAFs), thereby modifying tumor-stromal interactions in the TME, and subsequently creating a TME that supports tumor growth. Here we summarize recent studies about how DNA methylation affects tumorigenesis through regulating tumorassociated stromal components including fibroblasts and immune cells. We also discuss the potential for targeting DNA methylation for the treatment of cancers.
摘 要
肿瘤微环境主要由肿瘤细胞、肿瘤相关间质细胞 及细胞外基质组成。肿瘤细胞通过多种方式调控 肿瘤微环境中的间质细胞, 诱导间质细胞分化并 发挥促肿瘤的作用, 从而为肿瘤的生长及转移创 造一个适宜的环境。DNA 甲基化异常是肿瘤的特 点。目前关于肿瘤的甲基化调控机制已有大量报 道, 对于肿瘤细胞与微环境中间质细胞的相互作 用机制也有了一些报道。然而, 关于肿瘤细胞对 微环境间质细胞的甲基化调控机制以及这种调 控对肿瘤发生发展的影响并没有系统的论述。本 综述总结了肿瘤细胞对微环境中间质细胞甲基 化调控机制的最新研究进展, 以及间质细胞发生 的一些促肿瘤改变, 从而全面阐释了肿瘤细胞和 间质细胞间的相互作用, 同时总结了肿瘤细胞对 肿瘤微环境的表观遗传学调控, 尤其是甲基化调 控在肿瘤进展中发挥了重要的作用。干预肿瘤细 胞对微环境中间质细胞的甲基化调节过程, 可以 发挥抗肿瘤的作用。
Similar content being viewed by others
References
Ahmed, S.F., Farquharson, C., 2010. The effect of GHand IGF1 on linear growth and skeletal development and their modulation by SOCS proteins. J. Endocrinol., 206(3): 249–259. http://dx.doi.org/10.1677/JOE-10-0045
Albrengues, J., Bertero, T., Grasset, E., et al., 2015. Epigenetic switch drives the conversion of fibroblasts into proinvasive cancer-associated fibroblasts. Nat. Commun., 6:10204. http://dx.doi.org/10.1038/ncomms10204
Amodio, N., Bellizzi, D., Leotta, M., et al., 2013. miR-29b induces SOCS-1 expression by promoter demethylation and negatively regulates migration of multiple myeloma and endothelial cells. Cell Cycle, 12(23):3650–3662. http://dx.doi.org/10.4161/cc.26585
Batarseh, K.I., 2013. Antineoplastic activities, apoptotic mechanism of action and structural properties of a novel silver(I) chelate. Curr. Med. Chem., 20(18):2363–2373. http://dx.doi.org/10.2174/0929867311320180007
Berraondo, P., Minute, L., Ajona, D., et al., 2016. Innate immune mediators in cancer: between defense and resistance. Immunol. Rev., 274(1):290–306. http://dx.doi.org/10.1111/imr.12464
Bian, E.B., Huang, C., Ma, T.T., et al., 2012. DNMT1-mediated PTEN hypermethylation confers hepatic stellate cell activation and liver fibrogenesis in rats. Toxicol. Appl. Pharmacol., 264(1):13–22. http://dx.doi.org/10.1016/j.taap.2012.06.022
Bird, A., 2007. Perceptions of epigenetics. Nature, 447(7143): 396–398. http://dx.doi.org/10.1038/nature05913
Bock, C., Beerman, I., Lien, W.H., et al., 2012. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol. Cell, 47(4):633–647. http://dx.doi.org/10.1016/j.molcel.2012.06.019
Broske, A.M., Vockentanz, L., Kharazi, S., et al., 2009. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat. Genet., 41(11): 1207–1215. http://dx.doi.org/10.1038/ng.463
Cheung, P., Allis, C.D., Sassone-Corsi, P., 2000. Signaling to chromatin through histone modifications. Cell, 103(2): 263–271. http://dx.doi.org/10.1016/S0092-8674(00)00118-5
Chiappinelli, K.B., Zahnow, C.A., Ahuja, N., et al., 2016. Combining epigenetic and immunotherapy to combat cancer. Cancer Res., 76(7):1683–1689. http://dx.doi.org/10.1158/0008-5472.CAN-15-2125
Cui, H., Onyango, P., Brandenburg, S., et al., 2002. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res., 62(22):6442–6446.
Dedeurwaerder, S., Desmedt, C., Calonne, E., et al., 2011. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol. Med., 3(12):726–741. http://dx.doi.org/10.1002/emmm.201100801 de
Wever, O., Demetter, P., Mareel, M., et al., 2008. Stromal myofibroblasts are drivers of invasive cancer growth. Int. J. Cancer, 123(10):2229–2238. http://dx.doi.org/10.1002/ijc.23925
Easwaran, H., Tsai, H.C., Baylin, S.B., 2014. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol. Cell, 54(5):716–727. http://dx.doi.org/10.1016/j.molcel.2014.05.015 El
Taghdouini, A., Sorensen, A.L., Reiner, A.H., et al., 2015. Genome-wide analysis of DNA methylation and gene expression patterns in purified, uncultured human liver cells and activated hepatic stellate cells. Oncotarget, 6(29):26729–26745. http://dx.doi.org/10.18632/oncotarget.4925
Esteller, M., 2007. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet., 8(4): 286–298. http://dx.doi.org/10.1038/nrg2005
Feig, C., Gopinathan, A., Neesse, A., et al., 2012. The pancreas cancer microenvironment. Clin. Cancer Res., 18(16): 4266–4276. http://dx.doi.org/10.1158/1078-0432.CCR-11-3114
Garzon, R., Calin, G.A., Croce, C.M., 2009. MicroRNAs in Cancer. Annu. Rev. Med., 60(1):167–179. http://dx.doi.org/10.1146/annurev.med.59.053006.104707
Gascard, P., Tlsty, T.D., 2016. Carcinoma-associated fibroblasts: orchestrating the composition of malignancy. Genes Dev., 30(9):1002–1019. http://dx.doi.org/10.1101/gad.279737.116
Gibb, E.A., Brown, C.J., Lam, W.L., 2011. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer, 10(1):38. http://dx.doi.org/10.1186/1476-4598-10-38
Gotze, S., Schumacher, E.C., Kordes, C., et al., 2015. Epigenetic changes during hepatic stellate cell activation. PLoS ONE, 10(6):e0128745. http://dx.doi.org/10.1371/journal.pone.0128745
Gronbaek, K., Hother, C., Jones, P.A., 2007. Epigenetic changes in cancer. APMIS, 115(10):1039–1059. http://dx.doi.org/10.1111/j.1600-0463.2007.apm_636.xml.x
Hanahan, D., Weinberg, R.A., 2011. Hallmarks of cancer: the next generation. Cell, 144(5):646–674. http://dx.doi.org/10.1016/j.cell.2011.02.013
Hinz, B., Phan, S.H., Thannickal, V.J., et al., 2007. The myofibroblast: one function, multiple origins. Am. J. Pathol., 170(6):1807–1816. http://dx.doi.org/10.2353/ajpath.2007.070112
Iba, K., Albrechtsen, R., Gilpin, B.J., et al., 1999. Cysteinerich domain of human ADAM 12 (meltrin a) supports tumor cell adhesion. Am. J. Pathol., 154(5):1489–1501. http://dx.doi.org/10.1016/S0002-9440(10)65403-X
Iorio, M.V., Piovan, C., Croce, C.M., 2010. Interplay between microRNAs and the epigenetic machinery: an intricate network. Biochim. Biophys. Acta, 1799(10–12):694–701. http://dx.doi.org/10.1016/j.bbagrm.2010.05.005
Ishii, G., Ochiai, A., Neri, S., 2016. Phenotypic and functional heterogeneity of cancer-associated fibroblast within the tumor microenvironment. Adv. Drug Deliv. Rev., 99(Pt B): 186–196. http://dx.doi.org/10.1016/j.addr.2015.07.007
Janson, P.C., Marits, P., Thorn, M., et al., 2008. CpG methylation of the IFNG gene as a mechanism to induce immunosuppression in tumor-infiltrating lymphocytes. J. Immunol., 181(4):2878–2886. http://dx.doi.org/10.4049/jimmunol.181.4.2878
Jiang, L., Gonda, T.A., Gamble, M.V., et al., 2008. Global hypomethylation of genomic DNA in cancer-associated myofibroblasts. Cancer Res., 68(23):9900–9908. http://dx.doi.org/10.1158/0008-5472.CAN-08-1319
Karagiannis, G.S., Poutahidis, T., Erdman, S.E., et al., 2012. Cancer-associated fibroblasts drive the progression of metastasis through both paracrine and mechanical pressure on cancer tissue. Mol. Cancer Res., 10(11):1403–1418. http://dx.doi.org/10.1158/1541-7786.MCR-12-0307
Ke, X., Zhang, S., Xu, J., et al., 2016. Non-small-cell lung cancer-induced immunosuppression by increased human regulatory T cells via Foxp3 promoter demethylation. Cancer Immunol. Immunother., 65(5):587–599. http://dx.doi.org/10.1007/s00262-016-1825-6
Kouzarides, T., 2007. Chromatin modifications and their function. Cell, 128(4):693–705. http://dx.doi.org/10.1016/j.cell.2007.02.005
Ling, H., Spizzo, R., Atlasi, Y., et al., 2013. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res., 23(9):1446–1461. http://dx.doi.org/10.1101/gr.152942.112
Liu, H.X., Li, X.L., Dong, C.F., 2015. Epigenetic and metabolic regulation of breast cancer stem cells. J. Zhejiang Univ.-Sci. B (Biomed. & Biotechnol.), 16(1):10–17. http://dx.doi.org/10.1631/jzus.B1400172
Luperchio, T.R., Wong, X., Reddy, K.L., 2014. Genome regulation at the peripheral zone: lamina associated domains in development and disease. Curr. Opin. Genet. Dev., 25: 50–61. http://dx.doi.org/10.1016/j.gde.2013.11.021
Mann, J., Chu, D.C., Maxwell, A., et al., 2010. MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology, 138(2): 705–714. http://dx.doi.org/10.1053/j.gastro.2009.10.002
Mueller, M.M., Fusenig, N.E., 2004. Friends or foes—bipolar effects of the tumour stroma in cancer. Nat. Rev. Cancer, 4(11):839–849. http://dx.doi.org/10.1038/nrc1477
Neesse, A., Algul, H., Tuveson, D.A., et al., 2015. Stromal biology and therapy in pancreatic cancer: a changing paradigm. Gut, 64(9):1476–1484. http://dx.doi.org/10.1136/gutjnl-2015-309304
Page, A., Paoli, P., Moran, S.E., et al., 2016. Hepatic stellate cell transdifferentiation involves genome-wide remodeling of the DNA methylation landscape. J. Hepatol., 64(3): 661–673. http://dx.doi.org/10.1016/j.jhep.2015.11.024
Peric-Hupkes, D., Meuleman, W., Pagie, L., et al., 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell, 38(4):603–613. http://dx.doi.org/10.1016/j.molcel.2010.03.016
Pinzani, M., Rombouts, K., Colagrande, S., 2005. Fibrosis in chronic liver diseases: diagnosis and management. J. Hepatol., 42(Suppl. 1):S22–S36. http://dx.doi.org/10.1016/j.jhep.2004.12.008
Pleyer, L., Greil, R., 2015. Digging deep into “dirty” drugs— modulation of the methylation machinery. Drug Metab. Rev., 47(2):252–279. http://dx.doi.org/10.3109/03602532.2014.995379
Rabinovich, E.I., Kapetanaki, M.G., Steinfeld, I., et al., 2012. Global methylation patterns in idiopathic pulmonary fibrosis. PLoS ONE, 7(4):e33770. http://dx.doi.org/10.1371/journal.pone.0033770
Rucki, A.A., Zheng, L., 2014. Pancreatic cancer stroma: understanding biology leads to new therapeutic strategies. World J. Gastroenterol., 20(9):2237–2246. http://dx.doi.org/10.3748/wjg.v20.i9.2237
Schuyler, R.P., Merkel, A., Raineri, E., et al., 2016. Distinct trends of DNA methylation patterning in the innate and adaptive immune systems. Cell Rep., 17(8):2101–2111. http://dx.doi.org/10.1016/j.celrep.2016.10.054
Sharma, S., Kelly, T.K., Jones, P.A., 2010. Epigenetics in cancer. Carcinogenesis, 31(1):27–36. http://dx.doi.org/10.1093/carcin/bgp220
Sido, J.M., Yang, X., Nagarkatti, P.S., et al., 2015. ?9-Tetrahydrocannabinol-mediated epigenetic modifications elicit myeloid-derived suppressor cell activation via STAT3/S100A8. J. Leukoc. Biol., 97(4):677–688. http://dx.doi.org/10.1189/jlb.1A1014-479R
Smith, Z.D., Meissner, A., 2013. DNA methylation: roles in mammalian development. Nat. Rev. Genet., 14:204–220. http://dx.doi.org/10.1038/nrg3354
Sorensen, A.L., Timoskainen, S., West, F.D., et al., 2010. Lineage-specific promoter DNA methylation patterns segregate adult progenitor cell types. Stem Cells Dev., 19(8):1257–1266. http://dx.doi.org/10.1089/scd.2009.0309
Sukari, A., Nagasaka, M., Al-Hadidi, A., et al., 2016. Cancer immunology and immunotherapy. Anticancer Res., 36(11): 5593–5606. http://dx.doi.org/10.21873/anticanres.11144
Tampe, B., Tampe, D., Muller, C.A., et al., 2014. Tet3-mediated hydroxymethylation of epigenetically silenced genes contributes to bone morphogenic protein 7-induced reversal of kidney fibrosis. J. Am. Soc. Nephrol., 25(5): 905–912. http://dx.doi.org/10.1681/ASN.2013070723
Trikha, P., Carson, W.R., 2014. Signaling pathways involved in MDSC regulation. Biochim. Biophys. Acta, 1846(1): 55–65. http://dx.doi.org/10.1016/j.bbcan.2014.04.003
Trimboli, A.J., Cantemir-Stone, C.Z., Li, F., et al., 2009. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature, 461(7267):1084–1091. http://dx.doi.org/10.1038/nature08486 van
Kampen, J.G.M., Marijnissen-van Zanten, M.A.J., Simmer, F., et al., 2014. Epigenetic targeting in pancreatic cancer. Cancer Treat. Rev., 40(5):656–664. http://dx.doi.org/10.1016/j.ctrv.2013.12.002
Vincent, A., Omura, N., Hong, S.M., et al., 2011. Genomewide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clin. Cancer Res., 17(13):4341–4354. http://dx.doi.org/10.1158/1078-0432.CCR-10-3431
Vizoso, M., Puig, M., Carmona, F.J., et al., 2015. Aberrant DNA methylation in non-small cell lung cancerassociated fibroblasts. Carcinogenesis, 36(12):1453–1463. http://dx.doi.org/10.1093/carcin/bgv146
Wang, Z., Gao, Z., Shi, Y., et al., 2007. Inhibition of Smad3 expression decreases collagen synthesis in keloid disease fibroblasts. J. Plast. Reconstr. Aesthet. Surg., 60(11): 1193–1199. http://dx.doi.org/10.1016/j.bjps.2006.05.007
Wieczorek, G., Asemissen, A., Model, F., et al., 2009. Quantitative DNA methylation analysis of FOXP3 as a new method for counting regulatory T cells in peripheral blood and solid tissue. Cancer Res., 69(2):599–608. http://dx.doi.org/10.1158/0008-5472.CAN-08-2361
Xiang, J.F., Yin, Q.F., Chen, T., et al., 2014. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res., 24(5): 513–531. http://dx.doi.org/10.1038/cr.2014.35
Xiao, Q., Zhou, D., Rucki, A.A., et al., 2016. Cancerassociated fibroblasts in pancreatic cancer are reprogrammed by tumor-induced alterations in genomic DNA methylation. Cancer Res., 76(18):5395–5404. http://dx.doi.org/10.1158/0008-5472.CAN-15-3264
Xing, Y., Zhao, S., Zhou, B.P., et al., 2015. Metabolic reprogramming of the tumour microenvironment. Febs. J., 282(20):3892–3898. http://dx.doi.org/10.1111/febs.13402
Yu, J., Walter, K., Omura, N., et al., 2012. Unlike pancreatic cancer cells pancreatic cancer associated fibroblasts display minimal gene induction after 5-Aza-2'-deoxycytidine. PLoS ONE, 7(9):e43456. http://dx.doi.org/10.1371/journal.pone.0043456
Zhi, K., Shen, X., Zhang, H., et al., 2010. Cancer-associated fibroblasts are positively correlated with metastatic potential of human gastric cancers. J. Exp. Clin. Cancer Res., 29(1):66. http://dx.doi.org/10.1186/1756-9966-29-66
Author information
Authors and Affiliations
Corresponding authors
Additional information
Project supported by the National Cancer Institute (Nos. R01CA169702, P50CA062924, and R01CA197296), the United States
ORCID: Meng-wen ZHANG, http://orcid.org/0000-0003-1589-9314
Rights and permissions
About this article
Cite this article
Zhang, Mw., Fujiwara, K., Che, X. et al. DNA methylation in the tumor microenvironment. J. Zhejiang Univ. Sci. B 18, 365–372 (2017). https://doi.org/10.1631/jzus.B1600579
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1631/jzus.B1600579
Key words
- Tumor microenvironment (TME)
- DNA methylation
- Cancer-associated fibroblasts
- Cancer-associated immune cells
- Epigenetic therapy