MicroRNAs: pleiotropic players in congenital heart disease and regeneration
Introduction
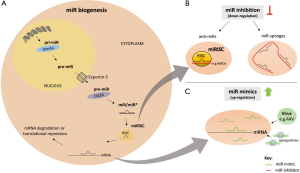
In mammals, the heart is the first functional organ that develops during embryogenesis, indicating its extraordinary importance already in early stages of life (1). Though, cardiac development is not at all an easy task to accomplish. The functionality of this process depends on highly sensitive and tightly regulated homeostatic networks of numerous proteins and other molecules (e.g., non-coding RNAs) that interact with all cardiac cell lineages to finally evolve into the three-dimensional beating heart. One important component of these networks is microRNAs (miRs), which are small non-coding RNAs with a length of ~22 bp that are able to regulate gene expression. Their regulatory function is based on either targeting mRNAs for cleavage or translational repression, leading to a down-regulation of the corresponding miR targets (2). Similar to protein-coding genes, miRs have a genomic origin, usually transcribed by RNA polymerase II as a primary transcript (so-called pri-miR) (3). After transcription and the first processing of Drosha (RNase II enzyme) in the nucleus, the precursor miR (pre-miR; ~70 bp) is exported to the cytoplasm by exportin 5, a RanGTP-dependent double-strand binding protein (4). The pre-miR is then further processed by DICER, an adenosine triphosphate-dependent RNase III, into a shorter double-stranded miR/miR* RNA complex (5). One strand of this RNA duplex further assembles with the RNase Argonaut to the RNA-induced-silencing complex (RISC) and is processed into a mature, single-stranded and regulatory miR. The remaining strand (the so-called miR*, passenger or sense strand) is released and degraded (6,7) (Figure 1A). Through this incorporation and the specific “seed regions” of miRs (2nd to 7th nucleotide at the 5’ end of the miR) the miR/RISC complex (miRISC) is able to bind to complementary mRNA (especially to regions in the 3’untranslated region) of target genes, thereby facilitating its regulatory function. In cases of perfect base pairing the target mRNA becomes degraded, whereas imperfect binding leads to translational repression (8,9). In the last decade, extensive research has revealed important functions of these small molecules that are essential for the development of various organs (10,11), including the human heart (12,13). Many cell-specific biological processes such as proliferation, differentiation and apoptosis are regulated by miRs (14). Therefore, it is not surprising that miRs are also involved in the development of cardiac disease and cardiac remodeling, respectively (15-17). Dysregulation of miRs can lead to structural and functional cardiac abnormalities (15). During early development, a loss or gain of miRNA expression may result in CHD, the most common human birth defect (18). Furthermore, impairments in the function or regulation of miRs in the adult or infant heart can lead to a spectrum of fatal acquired disorders, such as arrhythmias, cardiomyopathies, heart failure and sudden death (15,19).
miRNAs in congenital heart disease (CHD)
CHDs refer to a broad range of cardiac developmental abnormalities. CHDs are the leading cause of infant death affecting approximately 4 to 14 live births per 1,000. The underlying malformations are manifold, ranging from small ventricular septal defects (VSDs) to highly complex or severe malformations, such as tetralogy of Fallot (TOF) and hypoplastic left heart syndrome (HLHS) (18).
Initially, the requirement of miRs in normal cardiac development was revealed by a tissue-specific deletion of DICER in mice, an RNase that is essential for miR processing and biosynthesis. In vascular lineages, lethal phenotypes at embryonic day 16 (E16) to E17 have been observed associated with extensive internal hemorrhage due to thin-walled blood vessels (20). Additionally, cardiac-specific disruption of DICER led to embryonic lethality within the first four days after birth due to dilated cardiomyopathy (21). Both studies indicated the great importance of miRs during embryogenesis and cardiac development. Although DICER deletion revealed a crucial role for those small RNAs in cardiac development in vivo, no specific miR deletion has yet led to a fully penetrant lethal phenotype in mice, thus demonstrating the complex actions of miRs and compensatory mechanisms of their regulation (22). Nonetheless, several molecular tools such as antisense oligonucleotide-mediated knockdowns (anti-miRs), genetic ablation of single miRs or miR-clusters as well as miR mimics for overexpression are available to researchers to study miR function both in vivo and in vitro (Figure 1B,C). Additionally, the evaluation of tissue or blood samples from children with CHDs is attractive to determine correlating miRs and their targets which could play a role in the development of CHDs when dysregulated.
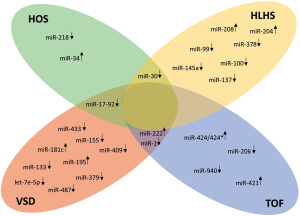
In this review, we describe the roles of selected miRs that are dysregulated in CHDs, especially focusing on those in HLHS, TOF, VSDs and Holt-Oram syndrome (HOS) (Table 1, Figure 2).
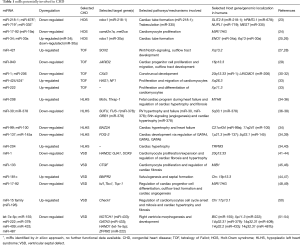
Full table
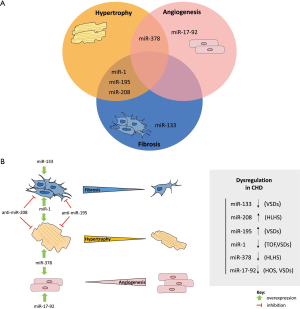
We then seek to bridge this knowledge with the possible regulatory properties of these miRs by stimulating fibrosis, angiogenesis, and hypertrophy in these patients (Figure 3A) and indicate potential miR-candidates which may be used in novel therapeutic strategies and regenerative approaches to ameliorate cardiac impairments (Figure 3B).
HOS
HOS is inherited in a penetrant dominant autosomal way. It is classified as a heart-hand syndrome, in which affected patients display both cardiac malformations and abnormalities of the upper limbs. Remarkably, approximately 70% of patients who are diagnosed with HOS exhibit a mutation in the TBX5 gene (55). To date, numerous HOS-related mutations of the TBX5 gene have been reported in humans (56). Recently, a new mutation in the DNA binding domain of TBX5 (p.Pro85Thr) that leads to a conformational change in protein structure has been described by our group (57). Interestingly, upon mutation or a dose reduction of the TBX5 gene, transgenic mice (58) and zebrafish (24) display a HOS-like phenotype, suggesting conserved function of the transcription factor TBX5 throughout vertebrate evolution. Based on mutual interactions between transcription factors and miRs (59), an in silico approach was used to identify miRs within introns of genes that are regulated by TBX5 with the help of integrated comparisons of several gene databases with miR databases. This approach identified four candidate miRs: mir-218-1, miR-678, miR-719 and miR-335 (23). Of these, miR-218-1 was highly promising due to its high cross-species conservation, its location within a TBX5-regulated gene, and the documented role of its host gene (SLIT2) in heart development. Indeed, the expression of Slit2 and miR-218-1 was clearly modulated by Tbx5 during the differentiation of mouse embryonic carcinoma cells (P19CL6) into beating cardiomyocytes (23). A functional role for miR-218-1 during heart morphogenesis is strongly supported by the fact that miR-218-1 overexpression dose-dependently induced cardiac defects in zebrafish embryos, possibly by affecting the migratory properties of cardiac precursors (23).
In another approach, several miRs were found to be dysregulated in tbx5-depleted zebrafish embryos. MiR-19a, a member of the miR-17-92 cluster, was of special interest since its injection into tbx5-depleted zebrafish embryos partially rescued the heart and fin phenotype (24). Additionally, the injection of miR-19a also rescued the overall gene expression profile of tbx5 morphants. Nearly half of the genes that were affected by tbx5 depletion appeared to be controlled by miR-19a, including camk2n1a, an inhibitor of CaMKII (calcium/calmodulin-dependent protein kinase II), and mef2ca with nine putative miR-19a binding sites (24,60). In both cases, luciferase assays confirmed a direct regulatory role for miR-19a (24). A genome-wide comparison of the miR profile in tbx5-depleted zebrafish embryos and wild-type embryos revealed various miRs that were significantly dysregulated within the first two days after fertilization (25). The most interesting candidates appeared to be miR-34 and miR-30a, which regulate the expression of genes that are involved in cardiac morphogenesis, cell proliferation, and contraction. One of the affected target genes that was identified, robo1 (roundabout guidance receptor 1), is involved in tube formation of the developing zebrafish heart (26). Robo1 is also a confirmed target gene of the aforementioned miR-218 in several organs, including the heart (26,61,62), and a putative target of miR-30a. Thus, two different miRs that are dysregulated upon tbx5-depletion in zebrafish both negatively affect the expression of robo1, a gene that is essential for proper heart development. Currently, the role of miRs in HOS is restricted to data that have been obtained in animal models. Future studies with human tissue must show whether the above-mentioned mechanisms can also be attributed to these miRs in humans.
TOF
TOF is characterized by a tetrad of abnormalities that occur in the infant heart, including (I) mainly large and non-restrictive VSDs, (II) pulmonary stenosis (i.e., a narrowing of the right outflow tract), (III) hypertrophy of the right ventricle and (IV) displacement of the aorta to the right side over the VSD (an overriding aorta). TOF is a classic cyanotic CHD that is caused by a subpulmonary obstruction, with shunting mainly from the right to the left ventricle via the VSD, leading to an ejection of deoxygenated blood into the aorta (63). The exact cause and pathophysiology of TOF are mainly unknown, but miRs may contribute to the development of TOF. O’Brien and colleagues investigated the expression patterns of non-coding RNAs (ncRNAs), particularly miRs and small nucleolar RNAs (snoRNAs), in the right ventricular (RV) myocardium in 16 infants with non-syndromic TOF (without deletion of 22q11.2; mean age, 276 days) and eight infants with normal cardiac development (mean age, 142 days) (64). Sixty-one miRs and 135 snoRNAs appeared to be significantly dysregulated in children with TOF when matched with cardiac tissue from normally developing infants. In addition, the same group evaluated gene expression patterns that are necessary for cardiac development. The analysis revealed a significant negative correlation between 33 miRs, the expression of which changed in the evaluated TOF samples when compared to controls, and 44 genes that are involved in the cardiac network (64). Additionally, 51% of these genes that are critical for cardiac development were found to be expressed as splice variants, mediated also by snoRNAs. This suggests that an impaired expression of ncRNAs (both miRs and snoRNAs) may contribute to the development of TOF by influencing expression, differential transcript splicing, and translation of genes and/or proteins that are important for normal heart development (64). Finally, Bittel et al. (27) analyzed the role of miR-421, which was shown to be significantly up-regulated in RV tissues in TOF infants (64). Therefore, primary cells derived from the RV of infants with TOF were transfected with siRNA to achieve a miR-421 knockdown, whereas primary cardiomyocytes from healthy infants were transfected with a miR-421 overexpression plasmid. The results showed that miR-421 overexpression in normal cardiomyocytes led to a down-regulation of SOX4 (SRF-Box 4), whereas miR-421 knockdown in primary cells that were derived from TOF patients led to an up-regulation of SOX4. This supports the hypothesis of an inverse correlation of miR-421 and SOX4, which is known as a central regulator of Notch and Wnt signaling and also has been implicated to play a role in the development of cardiac outflow tract (28,65). However, miR-421 has multiple sets of downstream targets, and the potential direct correlation between this regulated miR and the altered expression of SOX4 and its resulting impact on the Notch and Wnt pathway remains to be evaluated (27). MiR-940 appeared to be the most highly expressed miR in the normal human RV outflow tract but it is dramatically down-regulated in TOF patients (29). A functional analysis of miR-940 in isolated human Sca-1+ cardiac cells revealed that a reduction of miR-940 enhanced Sca-1+ cardiac resident cell proliferation but repressed their migration by targeting JARID2 (Jumonji and AT-rich interaction domain containing 2), a gene that may also affect the development of the cardiac outflow tract (66). However, mentionable in this context remains that a human ortholog of Sca-1 protein has not been yet identified, although it seems possible to isolate Sca-1+-like cells from the human heart using an anti-mouse Sca-1 antibody (29,67). Also alterations of the expression of CX43 (connexin 43) are known to be involved in the development of conotruncal anomalies (30). CX43 mRNA and protein were up-regulated in the myocardium of 30 TOF patients compared with controls (31). Of 10 miRNAs that were potentially associated with CX43, only miR-1 and miR-206 were significantly down-regulated in TOF patients. Both have previously been shown to impair CX43 expression (32), suggesting that both miR-1 and miR-206 may contribute to the development of TOF (31). Another microarray study that evaluated five right ventricular outflow tract (RVOT) biopsies from TOF patients and three corresponding control tissues identified 41 candidate miRs. Of these, 18 were differentially expressed in RVOT of non-syndromic TOF patients. These miRs targeted a network of genes that are involved in cardiac development and CHDs. Subsequently, in vitro data confirmed that an up-regulation of miR-424/424* enhanced proliferation and inhibited migration of embryonic mouse cardiomyocytes by suppressing HAS1 and NF1. In fact, both genes were down-regulated in RVOT samples from TOF patients (33). In contrast, miR-222 promoted cardiomyocyte proliferation and inhibited myogenic differentiation of P19 cells (33). Thus, both miR-424/424* and miR-222 may contribute to the TOF pathotype by affecting cardiomyocyte development.
HLHS
The HLHS is a rare CHD that has severe consequences in left heart structures. It is characterized by stenotic or atretic aortic and/or mitral valves, a highly hypoplastic aorta ascendens and a left ventricle that is highly hypoplastic or even completely missing (68). The severity of the disease requires surgical intervention within the first couple of days after birth. This palliative, so-called Norwood procedure includes three successive surgical interventions to generate circulation that is restricted to two chambers of the heart (63,68). In a recent study by Sucharov et al. the miR profile of the RV in HLHS patients was compared with the RV of non-failing (NF) hearts using an array that detected 754 miRs (34). A total of 93 miRs were found to be differentially regulated in HLHS hearts, which clustered separately compared to NF hearts. In a mouse model, some of these miRs have been implicated in the fetal program during heart failure (miR-208) (35) and in RV hypertrophy and RV failure (miR-30, miR-378) (36). Additionally, three miR species (miR-99a, miR-100, miR-145a) were down-regulated prior to or directly after the stage 1 operation of the Norwood procedure while the values returned to control levels after the stage 3 operation (34). This demonstrates the strong influence of altered blood flow conditions on the expression patterns of miRs in the heart. MiR-137 and miR-145 which were down-regulated in the RV of HLHS patients (34), both directly regulate the expression of FOG-2, a transcription factor that modulates the expression of GATA4, GATA5, and GATA6 (39), essential players in cardiac development. In contrast, miR-204 is up-regulated in the hypertrophic RV of HLHS patients, similar to patients who suffer from hypertrophic cardiomyopathy (40). However, despite these convincing data, it should be noted that the changes in expression of especially miR-99a, miR-100 and miR-145a after surgical interventions reflect the consequences of the disease due to altered blood flow conditions rather than shedding light on the initial molecular mechanisms that lead to HLHS which must happen much earlier during development. However, some miRs, including miR-208 and miR-378, were shown to be consistently dysregulated independent of the surgical stage compared with NF hearts. This in turn could indicate a decontrolled activation or repression of downstream targets or pathways even after the surgical intervention, potentially leading to the development of cardiac fibrosis and/or hypertrophy (see Section “Regenerative approaches in patients with CHD”).
VSDs
VSDs are defined by discontinuation in the septal wall that divides the left and right ventricles of the heart. VSDs are by far the most common form of CHDs (18). However, small VSDs can be asymptomatic and close spontaneously within the first year after birth. In contrast, large defects in the cardiac septum can lead to severe heart failure in infants due to volume overload that results in pulmonary hypertension (69). Several miRs have been implicated to participate in the development of VSDs, mainly derived from specific miR knockdown or overexpression studies in animal models. MiR-1 was the first miR that was shown to regulate fundamental steps of heart development (41). This muscle-specific small RNA is highly abundant in the mammalian heart and accounts for almost 40% of all miRs in the myocardium (70). Mice with increased miR-1 expression in the developing heart showed a decreased set of proliferating ventricular cardiomyocytes, proposed to be mediated by the translational inhibition of Hand2, a bHLH transcription factor that is required for ventricular cardiomyocyte expansion (41,42). Consistent with this, injection of miR-1 into Xenopus embryos also blocked cardiac development (71). In contrast, depletion of miR-1-2 in mice resulted in 50% embryonic lethality, largely due to VSDs (43). Additionally, miR-1-1 was shown to be significantly dysregulated in cardiac tissue samples from patients with VSDs (44). It was demonstrated that this down-regulation of miR-1-1 in such patients directly correlated with an increased level of its predicted target genes GJA1 (Gap junction alpha-1) and SOX9 (SRY-Box 9). Likewise, this microarray analysis of VSD samples also identified an up-regulation of miR-181c, which regulates the expression of BMPR2 (Bone morphogenetic protein receptor 2), a protein which is involved in valvulogenesis and septal formation (44,47). Transcribed from the same locus in a bicistronic manner miR-133a-1/miR-1-2 and miR-133a-2/miR-1-1 are expressed throughout the ventricular myocardium and interventricular septum from E8.5 until adulthood. In 2008, Liu et al. (45) illustrated the role of miR-133 in genetically engineered mice that were deficient for either miR-133a-1 or miR-133a-2, or both, as well as mice overexpressing miR-133a. Surprisingly, mice with deletion of either miR-133a-1 or miR-133a-2 were phenotypically normal, demonstrating that miR-133a-1 and miR-133a-2 may play overlapping, and potentially compensatory roles during cardiac development. In contrast, miR-133a double-mutant mice exhibited lethal VSDs in approximately 50% of genetically modified embryos or neonates. The residual mice that survived until adulthood appeared to manifest dilated cardiomyopathy, severe fibrosis and heart failure (45). Neonatal lethality due to VSDs and lung hypoplasia is also observed in mice with targeted deletion of the miR-17-92 cluster (48). This cluster and its paralogue clusters, miR-106a-363 and 106b-25, belong to a family of highly conserved miR clusters that consist of six members: miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1. Knockout mice that lacked the paralogue clusters miR-106a-363 and 106b-25 did not display an obvious phenotype, whereas mice with a loss of both miR-17-92 and miR-106b-25 developed exacerbated cardiac failings, including ventricular wall thinning and VSDs (48). Furthermore, the miR-17-92 cluster was shown to regulate myocardial differentiation of cardiac progenitors by affecting signaling cascades of the second heart field which is required for appropriate outflow tract formation (49). It has been shown that BMP (Bone morphogenetic protein) signaling enhances the transcription of several members of the miR-17-92 cluster through conserved Smad-binding sites. In turn elevated miR-17-92 levels inhibit the expression of important cardiac progenitor genes, namely Isl1 (ISL LIM homeobox 1) and Tbx1 (T-box 1), thereby facilitating myocardial differentiation.
Another miR that may be involved in the formation of VSDs is miR-195, a member of the miR-15 family. Early cardiac muscle-specific overexpression of miR-195 under the control of the β-myosin heavy chain promoter during embryogenesis was associated with ventricular hypoplasia and VSDs in hearts of these transgenic mice on postnatal day 1 (P1) to P3. Transgenic mice were generated by subcloning a mouse genomic fragment flanking miR-195 into a cardiac-specific expression plasmid containing 7kb of upstream regulatory sequences from β-myosin heavy chain promoter (72). Additionally, miR-195 overexpressing hearts showed a marked decrease in cells that underwent mitosis and an increase in cardiomyocyte multinucleosation, indicating that miR-195 could play a role in cell cycle arrest and the mitotic regulation of cardiomyocytes (50). Li and colleagues detected dysregulated expression of 36 circulating miRs in plasma from three VSD patients compared with controls (51). The expression of eight of these miRs was then further validated by qRT-PCR in plasma from 20 unrelated VSD patients. Seven of these miRs, including hsa-let-7e, miR-155, miR-222, miR-379, miR-409, miR-433 and miR-487, were shown to be down-regulated except one (miR-498), which was up-regulated when compared with controls. Gene Ontology (GO) analysis revealed that target genes of these miRs include NOTCH1, HAND1, ZFPM2 (also known as transcription factor GATA4/FOG-2) and GATA3, genes known to be involved in cardiac right ventricle development and right ventricle morphogenesis (52-54). Dysregulation of these genes by miRs may therefore contribute to an altered cardiac morphogenesis that result in the development of VSDs (51). An overview of selected target genes and/or their miRs that are potentially involved in CHDs is provided in Table 1 and Figure 2.
miRNA modulation for therapeutic intervention
Given the increasing number of studies that focused on the role of specific miRs in the development or progression of CHDs, it appears that their therapeutic regulation could be a promising approach to improve clinical outcomes in patients after surgical interventions. Currently, two major miR-modulation strategies are employed: (I) inhibition of miR activity by chemically modified anti-miR oligonucleotides (AMOs) or miR-sponges (Figure 1B) or (II) restoration of the function of a miR using viral vector-based overexpression or delivery of synthetic double-stranded miRs (Figure 1C). This section focuses on the currently available portfolio of miR therapeutic technologies.
Anti-miRs and miR-sponges
The most common transient miR-inhibitors are AMOs (Figure 1B). Mostly designed as single-stranded RNAs that generally exhibit a sequence that is fully complementary to the mature miR sequence, several chemically modified AMOs are available to improve their stability, binding affinity, and resistance to endonucleases. However, some AMOs, including those with modifications like methylation of nucleoside ribose 2’-hydroxyl groups (“OMe’s”), inclusion of phosphorothiolate linkages (PS-modified OMe oligos), addition of N,N-dietyl-4-(4-nitronaphtalen-1-ylazo)-phenylamine (“ZEN”-modified OMe oligos), and 3-end substitution to cholesterol (“AntagomiRs”)“, have both advantages and disadvantages when used for miR inhibition (recently reviewed by Beavers et al.) (73).
Nevertheless, the most effective classes of AMOs are locked nucleotide acids (“LNAs”), phosphorodiamidate morpholino oligonucleotides (“PMOs”) and peptide nucleic acids (“PNAs”). Their modifications ensure high binding affinity to the targeted miRs and equip them with improved nuclease resistance (73). Next to the transient inhibition of miRs by AMOs, another class of stable miR inhibitors is raising interest: miR-sponges. They are intracellularly expressed from transgenes (e.g., viral vectors) to generate high levels of competitive miR inhibitor transcripts (Figure 1B) (74). Modifications such as “bulged” sequences can make these sponges more effective in targeting a specific miR by having imperfect complementarity to the miR sequence. Additionally, this imparts them with higher stability against RISC-mediated degradation (74,75). Compared with individual anti-miRs which exclusively inhibit one miR by sharing the particular complementary seed region, miR sponges are able to inhibit an entire miR-family. Several sponge-related miR targeting systems have been successfully developed and tested, including tough decoys (“TuDs”) (76), and “miRNA erasers” (77).
miR mimics
Restoring the activity and function of a miR that is potentially down-regulated or even absent in pathophysiological settings has become increasingly important. The delivery of synthetic RNA duplexes, which mimics the affected miR is therefore one therapeutic option (Figure 1C). When applied, one strand (guide strand, or antisense) of the duplex is ideally recognized from miRISC as the miR of interest (by representing the same sequence), whereas the other complementary strand (passenger strand, or sense) will be degraded. Hence, the possibilities to chemically modify these RNA duplexes, especially the guide strand, are restricted due to the miRISC recognition process (78). Nevertheless, some modifications have been developed for the passenger strand to ensure optimal cellular uptake (e.g., by linking cholesterol or 5’-O-methylation to enhance guide strand selectivity) (79). Further modifications, including 2’-fluoro (2’-F) modifications, have been shown to impact exonuclease resistance and miR-mimic stability, respectively (80). Two major approaches have been tested to deliver miR-mimics: (I) miR-mimic viral based overexpression (Figure 1C) and (II) injection (intravenously or directly into the affected tissue) of miR-mimics that are linked to liposomal nanoparticles, atelocollagen or polyethyleneimine (81-83) (Figure 1C). Systemically viral based overexpression of a miR-mimic can be performed by using adeno-associated viruses (AAVs) (84,85) (Figure 1C). Due to the natural tropism of AAVs to cell types, receptors or organs, a cardiac-specific expression of the miR-mimicking constructs could also be realized (86). In addition to serotype that are used to enhance cell type specificity, the choice of a strong or tissue-specific promoter within the viral vector system could furthermore increase the efficiency of miR-mimic expression (e.g., CMV or TnnT promoter) (87,88). Several AAV approaches expressing miR-mimics have already undergone clinical trials. Their feasibility and safety profiles are promising and miR-mimics overexpression may be a promising approach to ameliorate poor clinical outcomes and complications in CHD patients and enhance their life expectancy (89).
Regenerative approaches in patients with CHD
Through advancements in surgical techniques and clinical care, the number of patients with grown-up congenital heart disease (GUCH) is increasing rapidly. As recently reviewed by Webb et al. (90), the number of adults with CHD (ACHD) is estimated to reach 2.3 million compared to 1.9 million cases in children out of a total population of 800 million across Europe. In fact, in most Western countries, the number of GUCH cases already outpaces the number of children born with CHDs and the gap continues to widen (90). This presents new challenges for pediatric cardiologists and heart surgeons, because these patients require complex and life-long intensive care to prevent late morbidity and mortality (91). However, a high number of GUCH patients develop cardiac hypertrophy (CH) (92) and fibrosis (93) in response to altered blood perfusion, abnormal pulmonary pressure or other malformations associated with severe CHDs. Additionally, underdeveloped or abnormal vascularization can trigger severe long-term cardiac complications, including heart failure (94,95). Thus, an adjuvant, miR-based therapeutic option to treat patients facing those problems could be a promising approach to support existing palliative care practices. In this review, we provide an overview of potential miR candidates that may be used for miR-based therapies in CHD patients, including GUCH patients. Although, the selected miRs were shown to be dysregulated in children with CHDs, the expression pattern in ACHD patients has yet to be investigated.
Regulation of fibrosis
Myocardial fibrosis, which can be observed not only after myocardial infarction (MI) (96), but also in patients with CHD (97), could contribute, at least partially, to the observed poor clinical outcomes by altering the functional status of the heart or regulating arrhythmias (93). New promising cardiac imaging technologies, recently reviewed by Rathod et al., have allowed the identification of different types of myocardial fibrosis, including reactive interstitial fibrosis and replacement fibrosis (97). In particular, reactive interstitial fibrosis, which is characterized by a progressive chronic course of diffuse collagen deposition, may be found in patients with CHD. It occurs mainly after hypertension, aortic stenosis (AS), chronic heart insufficiency or shunts—cardiac pathologies that are also found in CHD patients (97). Therefore, the development of anti-fibrotic drugs may have tremendous therapeutic potential and ameliorate the clinical outcome of many CHD patients. Several miRs are known to play an important role in the development and/or progression of fibrosis. Given the growing numbers of miRs that have been shown to influence cardiac fibrosis, we focus on a few miRs that are also found to be dysregulated in CHDs. The first pro-fibrotic miR found to play a key role in fibrogenesis after tissue injury was the cardiac-specific miR-208. This miR is encoded by intron 27 of the α-MHC (alpha Myosin heavy chain) gene in humans and mice. It is highly conserved in all mammals and specifically expressed in the heart (98,99). Van Rooij et al. (99) investigated cardiac responses of wild-type and miR-208-/- mice after induction of cardiac stress by thoracic aortic banding (TAB). They could show that wild-type mice displayed stress-dependent ventricular cardiac fibrosis after TAB, whereas mice with deletion of miR-208 were almost devoid of fibrosis (99). In addition, therapeutic inhibition of miR-208 by antisense oligonucleotide (anti-miR) delivery during hypertension-induced heart failure in Dahl hypertensive rats was shown to prevent cardiac remodeling, including cardiac fibrosis (100). Interestingly, miR-208 is known to be up-regulated in patients with HLHS, as mentioned above (34). Therefore, this miR could be an interesting target for anti-fibrotic drugs, and possibly also in patients with CHD (Figure 3B).
Another miR cluster with pro-fibrotic potential in the heart is the miR-15 family. Demonstrated to have an influence on the development of VSDs in hearts of transgenic mice while overexpressed (50), this miR-family, including miR-195, were also shown to be consistently up-regulated in different settings of heart disease [reviewed by Small et al. (101)]. Anti-miR studies have provided new insights into the regulatory function of this miR family in cardiac fibrosis (101). Hullinger et al. (102) demonstrated that miR-15 family members are up-regulated in infarcted regions of the porcine and murine heart in response to ischemia-reperfusion injury. Therapeutic blocking of miR-15 family members using LNA-modified anti-miRs after MI led to a reduction of infarct size, cardiac remodeling and enhanced cardiac function which corresponded with a decrease in cardiac fibrosis (102). Since many CHD patients present specific hemodynamic abnormalities, including left or RV volume overload and pulmonary hypertension as a conceivable consequence for instance of large VSDs, a resulting activation of fibroblasts may occur which in turn could facilitate cardiac fibrosis (103).
A second strategy to regulate fibrosis could be realized by an overexpression of anti-fibrotic miRs. Such miRs, including miR-1 and miR-133, are normally shown to be down-regulated in pathophysiological settings in the heart. Increasing their expression, through miR-mimics, may therefore positively influence the development of fibrosis (Figure 3B).
As mentioned above, adult miR-133a double knockout mice displayed severe fibrosis and heart failure (45). This fibrotic response could be explained by the interaction of miR-133 with its potential target gene CTGF (connective tissue growth factor). In a disease setting in which miR-133 is down-regulated, this may lead to enhanced expression of its target gene in cardiomyocytes, which in turn could stimulate extracellular matrix synthesis in fibroblasts and therefore support the development of fibrosis (46). Another study demonstrated that miR-1 replacement therapy can attenuate cardiac remodeling, including fibrosis (104). Therefore, Sprague-Dawley rats with pressure overload induced CH received either miR-1 overexpressing AAV9 or AAV9-GFP control virus via single bolus tail vein injection two weeks after surgical intervention. Animals with AAV9-miR-1 delivery showed a marked reduction of myocardial fibrosis after 7 weeks of treatment when compared to control animals (104). Interestingly, miR-1 is known to be significantly down-regulated in cardiac pathological settings, including TOF and VSDs (31,44) (Table 1, Figure 3B). Therefore, it is tempting to speculate that therapeutic modulation of fibro-miRs, such as miR-208, miR-1, miR-133 or the miR-15 family may open new options for the treatment of fibrosis in CHD in the near future.
Regulation of angiogenesis
Angiogenesis is a complex process that depends on a well-balanced equilibrium of both pro- and anti-angiogenic modulators that influence proliferation or quiescence of endothelial cells (ECs) within the myocardium. An imbalance of these factors can lead to impaired angiogenesis, also found in patients with CHD (94,95). Therapeutic treatment options in a pro- or anti-angiogenic manner could therefore help to avoid abnormal blood vessel proliferation or enhance normal angiogenic growth within the myocardium of affected patients. Several in vitro and in vivo studies have indicated a fundamental role of miRs in angiogenic processes nearly a decade ago (105-108). For instance, in vitro EC-specific silencing of DICER, which leads to a general reduction of miRs, demonstrated that EC function is strongly influenced by an absence of these tiny regulators (106,107). Mice with a global Dicer knockout revealed angiogenic impairment, e.g., defective blood vessels, resulting in late embryonic lethality (105,108). However, EC-specific Dicer hypomorphic mice survived but showed a reduced postnatal angiogenic response to a variety of stimuli like VEGF (vascular endothelial growth factor) or ischemia, indicating that EC-specific miRs are rather required for postnatal angiogenic processes than for developmental angiogenesis (109). In addition, this EC-specific deletion of Dicer led to an up-regulation (mRNA and/or protein) of various regulators of angiogenesis including TEK/Tie2 (tyrosine kinase with immunoglobulin like and epidermal growth factor homology domains-2), KDR/VEGFR2 (VEGF receptor-2), Tie-1 (tyrosine kinase with immunoglobulin like and epidermal growth factor homology domains-1), eNOS (endothelial nitric oxide synthase) and Tsp-1 (thrombospondin-1) and a down-regulation of IL-8 (interleukin-8) (106,107,109). Interestingly, Tsp-1 is a predicted target of the miR-17-92 cluster (Table 1), which is down-regulated in HOS and VSDs possibly indicating an up-regulation of Tsp-1 in these patients, though this still has to be validated. It was shown that an inhibition of the miR-17-92 cluster reduces tube formation and EC sprouting in matrigel in vitro (109). In contrast, an induced over-expression of members of the miR-17-92 family, especially miR-18a, led to a reversal of impaired EC proliferation and morphogenesis and was able to rescue the initiated overexpression of Tsp-1 caused by Dicer deletion. Another potent stimulator of angiogenesis, EC apoptosis and cell arrest is hypoxia. Quite commonly found in various CHDs (110,111), hypoxia may also have an influence on the angiogenic processes in affected children. Indeed, hypoxia has been shown to regulate the expression of different miRs, which have been identified in several cancer cell lines or ECs (112-114). Regulation of the expression of several miRs was found to be mediated by VEGF, including the modulation of miR-155 and members of the miR-17-92 family such as miR-18a and miR-20a (109). Recently, Yin and colleagues published the correlation of several gene expression patterns of angiogenic genes in CHD, including cases which concomitantly showed hypoxia (95). It was shown, that under hypoxic conditions, HIF-1α (hypoxia-inducible factor-1α) is accumulating rapidly combined with HIF-1β, thus activating downstream genes like VEGF and ET-1 (endothelin-1) which induced proliferation, sprouting and migration of ECs. Especially in patients with cyanotic CHD, consistently elevated HIF-1α levels may induce the development of systemic to pulmonary collateral arteries (95,115,116). However, miR-155 was shown to be down-regulated in patients with VSDs, indicating that VEGF levels were not increased in the cohort and thus did not activate their downstream miRs. This could be explained by the fact that the VSD cohort included in the study appeared to have relatively small shunt diameters (5–8 mm) (51,117), which in turn do not necessarily induce hypoxia. Therefore, miR-155 likely has a predominant impact on cardiac right ventricle development and right ventricle morphogenesis in patients with small VSDs rather than being involved in angiogenesis. Nevertheless, larger VSDs without surgical correction do have a greater chance of developing cyanotic pathologies and hypoxia due to increased pulmonary pressure, the right to left shunt, and the resulting occurrence of unoxygenated blood in the left ventricle. This may have an influence on the activation of VEGF and its downstream targets. Additionally, VSDs were observed in mouse heart with targeted deletion of the miR-17-92 cluster (48). However, whether there might be a direct link between these miRs, VEGF, and angiogenic processes in patients with CHD remains unknown and little is known about the precise function of these miRs that regulate angiogenesis in other CHDs like TOF and HLHS.
In conclusion, it is tempting to speculate that these miRs might be potential candidates for stimulation of angiogenesis in patients with CHDs. However, therapeutic approaches that especially target the miR-17-92 cluster (Figure 3B) should be considered carefully. The miR-17-92 cluster harbors several oncomirs that are associated with tumor progression and it is considered as a bona fide oncogene in lymphomagenesis (118,119). Therefore, such a strategy must be considered judiciously prior to potential application. Further research is needed to discern the distinct roles of this miR-cluster in angiogenesis (120).
HLHS is one of the most challenging CHDs, resulting in problems including RV failure and insufficient growth of pulmonary vasculature (121). MiRs such as miR-378 [down-regulated in HLHS (34)] (Table 1, Figure 2) have been implicated to regulate the pathophysiological development of the RV (36). Accumulating evidence indicates a role for miR-378 in the regulation of angiogenesis which may in turn influence the impairment of vasculature in these patients (37). When overexpressed in cancer cell lines, miR-378 was able to enhance cell survival and reduce cell death. In contrast, an inhibition of miR-378 showed a significant reduction of cell survival in these cells (37). Interestingly, miR-378 was shown to target the tumor suppressors SUFU (Suppressor of fused) and FUS-1 (Fused in sarcoma). Especially SUFU is known as a negative regulator of the Shh (Sonic hedgehog) signaling pathway, which implicates that this pathway is not only important for normal embryonic development but also for postnatal regulatory actions of angiogenesis (37). Furthermore, it is known that Shh regulates the expression of angiogenic cytokines such as VEGF, Ang-1 and Ang-2 (angiopoietin-1 and -2) which may contribute to the induction of robust angiogenesis, observed after Shh signaling activation, characterized by distinct large-diameter vessels (122).
In conclusion, miR-378 is able to regulate angiogenesis by indirect up-regulation of angiogenic factors and promotes cell survival by regulating the tumor suppressors SuFu and Fus-1. Apart from miR-378, many other miRs have been shown to participate in the process of angiogenesis (123). New therapeutic strategies that would improve blood perfusion in patients with CHDs may enhance long term clinical outcomes and reduce complications due to reduced perfusion of the most important organ of the body.
Regulation of hypertrophy
Although CH can emerge in a normal physiological context (e.g., as a reaction to elevated workload, exercise, or pregnancy), it often correlates with cardiac diseases such as hypertension, AS, MI or valvular dysfunction. Known as a compensatory mechanism of action in early stages of development (due to the above-mentioned stimuli), CH in a chronic and manifested manner with eccentric growth of cardiomyocytes can lead to heart failure. Especially in CHD pathologies like TOF, RV hypertrophy reduces clinical outcomes by triggering RV failure. But also other infants with CHDs who underwent surgical interventions have a higher risk of mortality in adolescence, often due to RV failure caused by chronic hypertrophy (92,121,124). Many miRs are known to play important roles in the development or progression of CH (125,126) and could therefore be promising therapeutic targets. The strategy of therapeutic intervention depends on the specific regulatory properties of these miRs (i.e., do they exhibit pro- or anti-hypertrophic potential). MiR-208, also involved in the regulation of fibrosis, displays pro-hypertrophic properties. In vivo overexpression of miR-208 stimulated fibrosis (99) and enhanced CH. Additionally, this miR is implicated to regulate heart failure in mice (36).
Overexpression of miR-208 resulted in a remarkable down-regulation of its target genes Mstn (myostatin) and Thrap-1 (thyroid hormone associated protein 1), which both function as repressors of hypertrophy and muscle growth (35,99). In addition, it was shown that a potent therapeutic inhibition of miR-208a by antisense oligonucleotides in the heart of Dahl hypertensive rats prevents pathological myosin switching and cardiac remodeling, including CH (100). Thus, regulating miR-208 with an antisense strategy might improve the clinical outcome in CHD patients (e.g., HLHS) at risk of developing heart failure due to progressive CH (Figure 3B). Another miR with pro-hypertrophic potential is miR-195. Cardiac-specific overexpression of this miR under the control of the α-MHC promoter provided insights in its regulatory action during CH in vivo. When overexpressed in mice, initial induction of cardiac growth was observed. This was accompanied by a disorganization of cardiomyocytes and subsequently a dilated phenotype of the whole heart at 6 weeks of age. MiR-195 overexpressing transgenic mice displayed a remarkable increase of cardiomyocyte cell size when compared to wild-type mice. This study provides evidence that a cardiac-specific delivery of miR-195 is able to sufficiently stimulate cardiac growth and drive CH, respectively (127). Interestingly, miR-195 overexpression during embryogenesis was associated with ventricular hypoplasia and VSDs in hearts of transgenic mice (50). Although proofed to regulate the development of VSDs and hypoplasia during early stages of life and embryogenesis, this miR could also play a role in the development of CH e.g., in patients with prolonged non-treated larger septal defects, respectively. Since larger VSDs or AVSDs (atrioventricular septal defects) can lead to elevated pulmonary pressure, a commonly known cause of CH (128), this miR might be differentially expressed or could be reactivated later in life, thus regulating CH. However, the distinct role of miR-195, especially in patients with VSD and pulmonary hypertension still has to be evaluated as well as the possibility of regulating this miR therapeutically by anti-sense strategies.
Several other studies identified anti-hypertrophic miRs such as miR-1 and miR-378. MiR-1 was shown to be significantly down-regulated after pressure-overload-induced CH in rat and mouse models (129,130). Interestingly, this down-regulation of miR-1 was observed as one of the earliest events in the heart after aortic constriction-induced hypertrophy—even before the increase of cardiomyocyte cell mass emerges (131). Similarly, miR-378 appeared to be down-regulated during CH and heart failure (38) (Table 1). Overexpression studies in primary cardiomyocyte cultures suggest that miR-378 targets two major growth-promoting signaling pathways: PI3K-AKT and RAF1-MEK1-ERK1/2. Both act downstream of Ras signaling which was blocked by down-regulation of Grb2, a direct target gene of miR-378, that is accompanied by a repression of fetal gene expression. Taken together, these data suggest that miR-378 deficiency favors the development of CH (38). Particularly noteworthy is that both miR-1 and miR-378 appear to be down-regulated in CHD patients, including those diagnosed with TOF (31,36) (Table 1). Patients with this phenotype struggle with poor clinical outcomes, partially due to the presence of CH (92). Especially in HLHS patients, the single RV is exposed to chronic pressure overload, facilitating CH and leading to an emerging number of RV failure in HLHS patients (132). Therefore, therapeutic intervention which delivers miR-1 or miR-378 or mimicking molecules thereof may be a promising therapeutic option for the treatment of CH in the future (Figure 3B).
Conclusions and perspectives
Several studies have underlined the role of miRNAs in pathological conditions in infants, including CHDs such as TOF, HLHS, VSDs and HOS. Many of these miRs appear to be dysregulated in CHD patients; however, most of the studies elucidated changes in miR expression as a result of the disease. Whether this dysregulation of miRs causally contributes to the development of CHD remains unclear. A number of studies in this review have demonstrated the promising feasibility of therapeutic modulation of miR expression during cardiac malformations by either inhibition by anti-miRs or overexpression by miR-mimics in animal models and were able to prevent or reverse cardiac fibrosis, inhibit CH and initiate angiogenesis. Thus, therapeutic intervention of miR and/or their target gene expression could be a promising approach in future to enhance clinical outcomes in CHD patients after surgical intervention.
Acknowledgements
Funding: R Lange is supported by the Bayerische Forschungsstiftung (AZ-1012-12). M Krane is supported by the Deutsche Stiftung für Herzforschung (F/37/11), Deutsches Zentrum für Herz Kreislauf Forschung (DZHK B 15-005, DZHK B 15-039SE) and Deutsche Forschungsgemeinschaft – Sachmittelantrag (KR3770/7-1, KR3770/9-1).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Zaffran S, Frasch M. Early signals in cardiac development. Circ Res 2002;91:457-69. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004;23:4051-60. [Crossref] [PubMed]
- Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004;10:185-91. [Crossref] [PubMed]
- Lee Y, Jeon K, Lee JT, et al. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 2002;21:4663-70. [Crossref] [PubMed]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003;115:209-16. [Crossref] [PubMed]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009;10:126-39. [Crossref] [PubMed]
- Ambros V. microRNAs: tiny regulators with great potential. Cell 2001;107:823-6. [Crossref] [PubMed]
- Winter J, Jung S, Keller S, et al. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 2009;11:228-34. [Crossref] [PubMed]
- Chen XM. MicroRNA signatures in liver diseases. World J Gastroenterol 2009;15:1665-72. [Crossref] [PubMed]
- Krichevsky AM, King KS, Donahue CP, et al. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 2003;9:1274-81. [Crossref] [PubMed]
- Thum T, Galuppo P, Wolf C, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation 2007;116:258-67. [Crossref] [PubMed]
- Wong LL, Wang J, Liew OW, et al. MicroRNA and Heart Failure. Int J Mol Sci 2016;17:502. [Crossref] [PubMed]
- Gurha P. MicroRNAs in cardiovascular disease. Curr Opin Cardiol 2016;31:249-54. [Crossref] [PubMed]
- Bruneau BG. The developmental genetics of congenital heart disease. Nature 2008;451:943-8. [Crossref] [PubMed]
- Cordes KR, Srivastava D. MicroRNA regulation of cardiovascular development. Circ Res 2009;104:724-32. [Crossref] [PubMed]
- Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell 2010;18:510-25. [Crossref] [PubMed]
- Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol 2002;39:1890-900. [Crossref] [PubMed]
- Olson EN, Schneider MD. Sizing up the heart: development redux in disease. Genes Dev 2003;17:1937-56. [Crossref] [PubMed]
- Albinsson S, Suarez Y, Skoura A, et al. MicroRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler Thromb Vasc Biol 2010;30:1118-26. [Crossref] [PubMed]
- Chen JF, Murchison EP, Tang R, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A 2008;105:2111-6. [Crossref] [PubMed]
- Boettger T, Braun T. A new level of complexity: the role of microRNAs in cardiovascular development. Circ Res 2012;110:1000-13. [Crossref] [PubMed]
- Chiavacci E, Dolfi L, Verduci L, et al. MicroRNA 218 mediates the effects of Tbx5a over-expression on zebrafish heart development. PLoS One 2012;7:e50536. [Crossref] [PubMed]
- Chiavacci E, D'Aurizio R, Guzzolino E, et al. MicroRNA 19a replacement partially rescues fin and cardiac defects in zebrafish model of Holt Oram syndrome. Sci Rep 2015;5:18240. [Crossref] [PubMed]
- D'Aurizio R, Russo F, Chiavacci E, et al. Discovering miRNA Regulatory Networks in Holt-Oram Syndrome Using a Zebrafish Model. Front Bioeng Biotechnol 2016;4:60. [PubMed]
- Fish JE, Wythe JD, Xiao T, et al. A Slit/miR-218/Robo regulatory loop is required during heart tube formation in zebrafish. Development 2011;138:1409-19. [Crossref] [PubMed]
- Bittel DC, Kibiryeva N, Marshall JA, et al. MicroRNA-421 Dysregulation is Associated with Tetralogy of Fallot. Cells 2014;3:713-23. [Crossref] [PubMed]
- Paul MH, Harvey RP, Wegner M, et al. Cardiac outflow tract development relies on the complex function of Sox4 and Sox11 in multiple cell types. Cell Mol Life Sci 2014;71:2931-45. [Crossref] [PubMed]
- Liang D, Xu X, Deng F, et al. miRNA-940 reduction contributes to human Tetralogy of Fallot development. J Cell Mol Med 2014;18:1830-9. [Crossref] [PubMed]
- Salameh A, Blanke K, Daehnert I. Role of connexins in human congenital heart disease: the chicken and egg problem. Front Pharmacol 2013;4:70. [Crossref] [PubMed]
- Wu Y, Ma XJ, Wang HJ, et al. Expression of Cx43-related microRNAs in patients with tetralogy of Fallot. World J Pediatr 2014;10:138-44. [Crossref] [PubMed]
- Anderson C, Catoe H, Werner R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res 2006;34:5863-71. [Crossref] [PubMed]
- Zhang J, Chang JJ, Xu F, et al. MicroRNA deregulation in right ventricular outflow tract myocardium in nonsyndromic tetralogy of fallot. Can J Cardiol 2013;29:1695-703. [Crossref] [PubMed]
- Sucharov CC, Sucharov J, Karimpour-Fard A, et al. Micro-RNA expression in hypoplastic left heart syndrome. J Card Fail 2015;21:83-8. [Crossref] [PubMed]
- Callis TE, Pandya K, Seok HY, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest 2009;119:2772-86. [Crossref] [PubMed]
- Reddy S, Zhao M, Hu DQ, et al. Dynamic microRNA expression during the transition from right ventricular hypertrophy to failure. Physiol Genomics 2012;44:562-75. [Crossref] [PubMed]
- Lee DY, Deng Z, Wang CH, et al. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A 2007;104:20350-5. [Crossref] [PubMed]
- Nagalingam RS, Sundaresan NR, Gupta MP, et al. A cardiac-enriched microRNA, miR-378, blocks cardiac hypertrophy by targeting Ras signaling. J Biol Chem 2013;288:11216-32. [Crossref] [PubMed]
- Holder AM, Klaassens M, Tibboel D, et al. Genetic factors in congenital diaphragmatic hernia. Am J Hum Genet 2007;80:825-45. [Crossref] [PubMed]
- Kuster DW, Mulders J, Ten Cate FJ, et al. MicroRNA transcriptome profiling in cardiac tissue of hypertrophic cardiomyopathy patients with MYBPC3 mutations. J Mol Cell Cardiol 2013;65:59-66. [Crossref] [PubMed]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 2005;436:214-20. [Crossref] [PubMed]
- Srivastava D, Cserjesi P, Olson EN. A subclass of bHLH proteins required for cardiac morphogenesis. Science 1995;270:1995-9. [Crossref] [PubMed]
- Zhao Y, Ransom JF, Li A, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 2007;129:303-17. [Crossref] [PubMed]
- Li J, Cao Y, Ma XJ, et al. Roles of miR-1-1 and miR-181c in ventricular septal defects. Int J Cardiol 2013;168:1441-6. [Crossref] [PubMed]
- Liu N, Bezprozvannaya S, Williams AH, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev 2008;22:3242-54. [Crossref] [PubMed]
- Duisters RF, Tijsen AJ, Schroen B, et al. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res 2009;104:170-8, 6p following 178.
- Beppu H, Malhotra R, Beppu Y, et al. BMP type II receptor regulates positioning of outflow tract and remodeling of atrioventricular cushion during cardiogenesis. Dev Biol 2009;331:167-75. [Crossref] [PubMed]
- Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 2008;132:875-86. [Crossref] [PubMed]
- Wang J, Greene SB, Bonilla-Claudio M, et al. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a MicroRNA-mediated mechanism. Dev Cell 2010;19:903-12. [Crossref] [PubMed]
- Porrello ER, Johnson BA, Aurora AB, et al. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res 2011;109:670-9. [Crossref] [PubMed]
- Li D, Ji L, Liu L, et al. Characterization of circulating microRNA expression in patients with a ventricular septal defect. PLoS One 2014;9:e106318. [Crossref] [PubMed]
- Kratsios P, Catela C, Salimova E, et al. Distinct roles for cell-autonomous Notch signaling in cardiomyocytes of the embryonic and adult heart. Circ Res 2010;106:559-72. [Crossref] [PubMed]
- Lu JR, McKinsey TA, Xu H, et al. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol Cell Biol 1999;19:4495-502. [Crossref] [PubMed]
- Vincentz JW, Barnes RM, Firulli AB. Hand factors as regulators of cardiac morphogenesis and implications for congenital heart defects. Birth Defects Res A Clin Mol Teratol 2011;91:485-94. [Crossref] [PubMed]
- Lichiardopol C, Militaru C, Popescu B, et al. Holt-Oram syndrome. Rom J Morphol Embryol 2007;48:67-70. [PubMed]
- Al-Qattan MM, Abou Al-Shaar H. Molecular basis of the clinical features of Holt-Oram syndrome resulting from missense and extended protein mutations of the TBX5 gene as well as TBX5 intragenic duplications. Gene 2015;560:129-36. [Crossref] [PubMed]
- Dreßen M, Lahm H, Lahm A, et al. A novel de novo TBX5 mutation in a patient with Holt-Oram syndrome leading to a dramatically reduced biological function. Mol Genet Genomic Med 2016;4:557-67. [Crossref] [PubMed]
- Bruneau BG, Nemer G, Schmitt JP, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 2001;106:709-21. [Crossref] [PubMed]
- Yu X, Lin J, Zack DJ, et al. Analysis of regulatory network topology reveals functionally distinct classes of microRNAs. Nucleic Acids Res 2008;36:6494-503. [Crossref] [PubMed]
- Rothschild SC, Easley CA 4th, Francescatto L, et al. Tbx5-mediated expression of Ca(2+)/calmodulin-dependent protein kinase II is necessary for zebrafish cardiac and pectoral fin morphogenesis. Dev Biol 2009;330:175-84. [Crossref] [PubMed]
- Alajez NM, Lenarduzzi M, Ito E, et al. MiR-218 suppresses nasopharyngeal cancer progression through downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res 2011;71:2381-91. [Crossref] [PubMed]
- Small EM, Sutherland LB, Rajagopalan KN, et al. MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circ Res 2010;107:1336-44. [Crossref] [PubMed]
- Lahm H, Schon P, Doppler S, et al. Tetralogy of Fallot and Hypoplastic Left Heart Syndrome - Complex Clinical Phenotypes Meet Complex Genetic Networks. Curr Genomics 2015;16:141-58. [Crossref] [PubMed]
- O'Brien JE Jr, Kibiryeva N, Zhou XG, et al. Noncoding RNA expression in myocardium from infants with tetralogy of Fallot. Circ Cardiovasc Genet 2012;5:279-86. [Crossref] [PubMed]
- Schilham MW, Oosterwegel MA, Moerer P, et al. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature 1996;380:711-4. [Crossref] [PubMed]
- Barth JL, Clark CD, Fresco VM, et al. Jarid2 is among a set of genes differentially regulated by Nkx2.5 during outflow tract morphogenesis. Dev Dyn 2010;239:2024-33. [Crossref] [PubMed]
- Valente M, Nascimento DS, Cumano A, et al. Sca-1+ cardiac progenitor cells and heart-making: a critical synopsis. Stem Cells Dev 2014;23:2263-73. [Crossref] [PubMed]
- Barron DJ, Kilby MD, Davies B, et al. Hypoplastic left heart syndrome. Lancet 2009;374:551-64. [Crossref] [PubMed]
- Smith T, Rajakaruna C, Caputo M, et al. MicroRNAs in congenital heart disease. Ann Transl Med 2015;3:333. [PubMed]
- Rao PK, Toyama Y, Chiang HR, et al. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res 2009;105:585-94. [Crossref] [PubMed]
- Chen JF, Mandel EM, Thomson JM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 2006;38:228-33. [Crossref] [PubMed]
- Rindt H, Gulick J, Knotts S, et al. In vivo analysis of the murine beta-myosin heavy chain gene promoter. J Biol Chem 1993;268:5332-8. [PubMed]
- Beavers KR, Nelson CE, Duvall CL. MiRNA inhibition in tissue engineering and regenerative medicine. Adv Drug Deliv Rev 2015;88:123-37. [Crossref] [PubMed]
- Tay FC, Lim JK, Zhu H, et al. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv Drug Deliv Rev 2015;81:117-27. [Crossref] [PubMed]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 2007;4:721-6. [Crossref] [PubMed]
- Haraguchi T, Ozaki Y, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res 2009;37:e43. [Crossref] [PubMed]
- Sayed D, Rane S, Lypowy J, et al. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol Biol Cell 2008;19:3272-82. [Crossref] [PubMed]
- van Rooij E, Kauppinen S. Development of microRNA therapeutics is coming of age. EMBO Mol Med 2014;6:851-64. [Crossref] [PubMed]
- Chen PY, Weinmann L, Gaidatzis D, et al. Strand-specific 5'-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA 2008;14:263-74. [Crossref] [PubMed]
- Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA 2003;9:1034-48. [Crossref] [PubMed]
- Ibrahim AF, Weirauch U, Thomas M, et al. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res 2011;71:5214-24. [Crossref] [PubMed]
- Pramanik D, Campbell NR, Karikari C, et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther 2011;10:1470-80. [Crossref] [PubMed]
- Takeshita F, Patrawala L, Osaki M, et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol Ther 2010;18:181-7. [Crossref] [PubMed]
- Kota J, Chivukula RR, O'Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009;137:1005-17. [Crossref] [PubMed]
- Miyazaki Y, Adachi H, Katsuno M, et al. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nat Med 2012;18:1136-41. [Crossref] [PubMed]
- Srivastava A. In vivo tissue-tropism of adeno-associated viral vectors. Curr Opin Virol 2016;21:75-80. [Crossref] [PubMed]
- Chen BD, He CH, Chen XC, et al. Targeting transgene to the heart and liver with AAV9 by different promoters. Clin Exp Pharmacol Physiol 2015;42:1108-17. [Crossref] [PubMed]
- Prasad KM, Smith RS, Xu Y, et al. A single direct injection into the left ventricular wall of an adeno-associated virus 9 (AAV9) vector expressing extracellular superoxide dismutase from the cardiac troponin-T promoter protects mice against myocardial infarction. J Gene Med 2011;13:333-41. [Crossref] [PubMed]
- Aalbers CJ, Tak PP, Vervoordeldonk MJ. Advancements in adeno-associated viral gene therapy approaches: exploring a new horizon. F1000 Med Rep 2011;3:17. [Crossref] [PubMed]
- Webb G, Mulder BJ, Aboulhosn J, et al. The care of adults with congenital heart disease across the globe: Current assessment and future perspective: A position statement from the International Society for Adult Congenital Heart Disease (ISACHD). Int J Cardiol 2015;195:326-33. [Crossref] [PubMed]
- Gatzoulis MA, Balaji S, Webber SA, et al. Risk factors for arrhythmia and sudden cardiac death late after repair of tetralogy of Fallot: a multicentre study. Lancet 2000;356:975-81. [Crossref] [PubMed]
- Iacobazzi D, Suleiman MS, Ghorbel M, et al. Cellular and molecular basis of RV hypertrophy in congenital heart disease. Heart 2016;102:12-7. [Crossref] [PubMed]
- Babu-Narayan SV, Kilner PJ, Li W, et al. Ventricular fibrosis suggested by cardiovascular magnetic resonance in adults with repaired tetralogy of fallot and its relationship to adverse markers of clinical outcome. Circulation 2006;113:405-13. [Crossref] [PubMed]
- Hu J, Sun P, Ruan X, et al. Mechanism of myocardial microvessel formation in cyanotic congenital heart disease. Circ J 2005;69:1089-93. [Crossref] [PubMed]
- Yin HL, Luo CW, Dai ZK, et al. Hypoxia-inducible factor-1alpha, vascular endothelial growth factor, inducible nitric oxide synthase, and endothelin-1 expression correlates with angiogenesis in congenital heart disease. Kaohsiung J Med Sci 2016;32:348-55. [Crossref] [PubMed]
- Chistiakov DA, Orekhov AN, Bobryshev YV. The role of cardiac fibroblasts in post-myocardial heart tissue repair. Exp Mol Pathol 2016;101:231-40. [Crossref] [PubMed]
- Rathod RH, Powell AJ, Geva T. Myocardial Fibrosis in Congenital Heart Disease. Circ J 2016;80:1300-7. [Crossref] [PubMed]
- Morkin E. Control of cardiac myosin heavy chain gene expression. Microsc Res Tech 2000;50:522-31. [Crossref] [PubMed]
- van Rooij E, Sutherland LB, Qi X, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 2007;316:575-9. [Crossref] [PubMed]
- Montgomery RL, Hullinger TG, Semus HM, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation 2011;124:1537-47. [Crossref] [PubMed]
- Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation 2010;121:1022-32. [Crossref] [PubMed]
- Hullinger TG, Montgomery RL, Seto AG, et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res 2012;110:71-81. [Crossref] [PubMed]
- Sugimoto M, Kuwata S, Kurishima C, et al. Cardiac biomarkers in children with congenital heart disease. World J Pediatr 2015;11:309-15. [Crossref] [PubMed]
- Karakikes I, Chaanine AH, Kang S, et al. Therapeutic cardiac-targeted delivery of miR-1 reverses pressure overload-induced cardiac hypertrophy and attenuates pathological remodeling. J Am Heart Assoc 2013;2:e000078. [Crossref] [PubMed]
- Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat Genet 2003;35:215-7. [Crossref] [PubMed]
- Kuehbacher A, Urbich C, Zeiher AM, et al. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res 2007;101:59-68. [Crossref] [PubMed]
- Suárez Y, Fernández-Hernando C, Pober JS, et al. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res 2007;100:1164-73. [Crossref] [PubMed]
- Yang WJ, Yang DD, Na S, et al. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem 2005;280:9330-5. [Crossref] [PubMed]
- Suárez Y, Fernández-Hernando C, Yu J, et al. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A 2008;105:14082-7. [Crossref] [PubMed]
- Corno AF, Milano G, Samaja M, et al. Chronic hypoxia: a model for cyanotic congenital heart defects. J Thorac Cardiovasc Surg 2002;124:105-12. [Crossref] [PubMed]
- Lemus-Varela ML, Flores-Soto ME, Cervantes-Munguia R, et al. Expression of HIF-1 alpha, VEGF and EPO in peripheral blood from patients with two cardiac abnormalities associated with hypoxia. Clin Biochem 2010;43:234-9. [Crossref] [PubMed]
- Fasanaro P, D'Alessandra Y, Di Stefano V, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem 2008;283:15878-83. [Crossref] [PubMed]
- Hua Z, Lv Q, Ye W, et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One 2006;1:e116. [Crossref] [PubMed]
- Kulshreshtha R, Ferracin M, Wojcik SE, et al. A microRNA signature of hypoxia. Mol Cell Biol 2007;27:1859-67. [Crossref] [PubMed]
- Himeno W. Angiogenic growth factors in patients with cyanotic congenital heart disease and in normal children. Kurume Med J 2001;48:111-6. [Crossref] [PubMed]
- Ylä-Herttuala S, Alitalo K. Gene transfer as a tool to induce therapeutic vascular growth. Nat Med 2003;9:694-701. [Crossref] [PubMed]
- Zhang J, Ko JM, Guileyardo JM, et al. A review of spontaneous closure of ventricular septal defect. Proc (Bayl Univ Med Cent) 2015;28:516-20. [PubMed]
- Fuziwara CS, Kimura ET. Insights into Regulation of the miR-17-92 Cluster of miRNAs in Cancer. Front Med (Lausanne) 2015;2:64. [Crossref] [PubMed]
- Jin HY, Oda H, Lai M, et al. MicroRNA-17~92 plays a causative role in lymphomagenesis by coordinating multiple oncogenic pathways. EMBO J 2013;32:2377-91. [Crossref] [PubMed]
- Dews M, Homayouni A, Yu D, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet 2006;38:1060-5. [Crossref] [PubMed]
- Arnold RR, Loukanov T, Gorenflo M. Hypoplastic left heart syndrome - unresolved issues. Front Pediatr 2014;2:125. [Crossref] [PubMed]
- Pola R, Ling LE, Silver M, et al. The morphogen Sonic hedgehog is an indirect angiogenic agent upregulating two families of angiogenic growth factors. Nat Med 2001;7:706-11. [Crossref] [PubMed]
- Pourrajab F, Vakili Zarch A, Hekmatimoghaddam S, et al. MicroRNAs; easy and potent targets in optimizing therapeutic methods in reparative angiogenesis. J Cell Mol Med 2015;19:2702-14. [Crossref] [PubMed]
- Knowles RL, Bull C, Wren C, et al. Mortality with congenital heart defects in England and Wales, 1959-2009: exploring technological change through period and birth cohort analysis. Arch Dis Child 2012;97:861-5. [Crossref] [PubMed]
- Da Costa Martins PA, De Windt LJ. MicroRNAs in control of cardiac hypertrophy. Cardiovasc Res 2012;93:563-72. [Crossref] [PubMed]
- Wang J, Liew OW, Richards AM, et al. Overview of MicroRNAs in Cardiac Hypertrophy, Fibrosis, and Apoptosis. Int J Mol Sci 2016;17. [Crossref] [PubMed]
- van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A 2006;103:18255-60. [Crossref] [PubMed]
- Naeije R, Manes A. The right ventricle in pulmonary arterial hypertension. Eur Respir Rev 2014;23:476-87. [Crossref] [PubMed]
- Hua Y, Zhang Y, Ren J. IGF-1 deficiency resists cardiac hypertrophy and myocardial contractile dysfunction: role of microRNA-1 and microRNA-133a. J Cell Mol Med 2012;16:83-95. [Crossref] [PubMed]
- Li Q, Song XW, Zou J, et al. Attenuation of microRNA-1 derepresses the cytoskeleton regulatory protein twinfilin-1 to provoke cardiac hypertrophy. J Cell Sci 2010;123:2444-52. [Crossref] [PubMed]
- Sayed D, Hong C, Chen IY, et al. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res 2007;100:416-24. [Crossref] [PubMed]
- Chery J, Wong J, Huang S, et al. Regenerative Medicine Strategies for Hypoplastic Left Heart Syndrome. Tissue Eng Part B Rev 2016;22:459-69. [Crossref] [PubMed]


