Patient-specific three-dimensional printing for pre-surgical planning in hepatocellular carcinoma treatment
Introduction
Liver lesion resection is often a complex task requiring sound comprehension of patient specific anatomical and pathological characteristics to ensure optimal surgical outcomes (1-6). Diagnostic imaging plays a major role in the diagnosis and characterisation of liver lesions, with imaging appearance and radiological reporting outcomes being utilized to direct patient-specific treatment planning (7,8). However, confidence in the comprehension of anatomical, pathological and structural complexities which surgeons require to conduct hepatic resections, may not always be supported by two-dimensional (2D) imaging alone (2).
Multi-planar medical images and three-dimensional (3D) reconstructions have the ability to provide valuable information related to the lesion and its extent. However, complex anatomical networks and information pertaining to spatial relationships, structural depth, and topological characteristics may not be truly appreciated (2,5,9). Currently, 3D printed models are being used as clinical tools in the endeavour to bridge this gap, enhancing the viewer’s cognitive comprehension of anatomy and pathology in areas such as medical education, surgical training, surgical planning and operative simulation (2,3,9-20). The tactile experience is known to support a holistic evaluation of important anatomical and/or pathological features (10), with the manipulation and visualization of the physical 3D printed models providing the in-depth understanding required to plan accurate, safe, and effective surgical procedures (4,5,13,14,16,17-21).
3D printed liver models have been found to enhance the viewer’s understanding of highly variable and complex hepatic anatomy (13,14,16,18,22). There are a number of case studies that deem 3D printed liver models to be useful clinical tools in pre- and perioperative processes for the resection of liver lesions and in living donor liver transplant procedures (3,6,9,13,16,23). However, evidence-based research evaluating the feasibility of 3D liver models in hepatocellular carcinoma (HCC) treatment is lacking. HCC is known for its pathological complexity, often spreading to surrounding intrahepatic structures and thus contributing to challenges in clinical management (1). If surgical intervention is classified as a viable option for treatment, it is imperative that the procedure is verified as safe and effective to ensure optimal surgical outcomes (1-3). Further exploration in the application and clinical value of 3D printed models for pre-operative planning in HCC treatment is therefore warranted. It is also noted that there is currently no research investigating the application of 3D liver models in diagnostic reporting for HCC, which precedes and often directs the decision for surgical management by providing diagnosis and report of imaging characteristics (1,8).
A recent systematic review identifies the need for a large-scale study that implements qualitative and/or quantitative methods to holistically evaluate the clinical applications of 3D printed liver models, with the outcomes of existing case studies alone perceived to lack credibility due to the absence of qualitative and/or quantitative data (20). This research may be considered as a small-scale pilot study implementing an evidence-based, holistic methodological approach. The aim of this study is to investigate the feasibility of utilizing 3D printed liver models as clinical tools in pre-operative planning for resectable HCC lesions. Both qualitative and quantitative approaches to data collection and analysis are applied to assess the clinical value of a single patient-specific 3D liver model in two major pre-operative stages. This includes diagnostic reporting and pre-surgical planning. Details of the production process, and statistical analysis of 3D model accuracy are also included to evaluate whether the reported applications are feasible.
Methods
Ethics approval for this study was obtained from Curtin University’s Human Research Ethics Committee (HRE2017-0153). Due to the retrospective collection of de-identified computed tomography (CT) image data, ethics approval from the clinical center and patient consent was waived.
Generation of 3D printed liver model
A four-step production process was implemented to generate the patient-specific 3D liver model utilized for this study (Figure 1) (3,6,9,10,21,24,25).
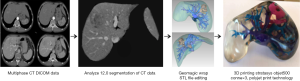
CT data acquisition
An anonymised, contrast-enhanced abdominal CT Digital Imaging and Communications in Medicine (DICOM) dataset complying with a set of specified image selection criteria (Supplement I) was obtained. Arterial and portal venous datasets at 1 mm slice thickness had been acquired using a 64-slice CT scanner (Siemens Definition, Siemens Healthcare, Forchheim, Germany). Image datasets for both arterial and portal venous enhancement phases were required to allow for adequate visualization of the tumour, normal liver parenchyma, portal and hepatic venous networks, and arterial supply (23).
Image segmentation
Medical imaging visualization, analysis, and segmentation was conducted using a commercially available biomedical image processing software, Analyze 12.0 (AnalyzeDirect, Inc., Lexana, KS, USA). Image visualization and processing tools within this software were used to (I) apply a median filter, reducing image noise and enhancing the anatomical boundaries for accurate segmentation; (II) segment structures of interest; and (III) export the segmented data in standard tessellation language (STL) file format as required for editing and 3D printing (10,16,18,22-25). Anatomical structures that were perceived likely to affect decisions made in surgical planning included the inferior vena cava, portal vein and associated branches, hepatic veins, hepatic artery, the tumour and associated arterial supply, and the liver body (2,3,9,23). These structures were segmented for inclusion within the final 3D liver model using a combination of manual and semi-automatic tools as defined in Supplement II.
Image data editing
Geomagic Wrap 2017 (3D Systems, Korea, Inc., Seoul, Korea) was used to refine segmented structures in preparation for 3D printing. This involved the utilization of manual, semi-automatic and automatic editing tools. All individual files were then decimated and exported in STL file format for 3D printing.
3D printing
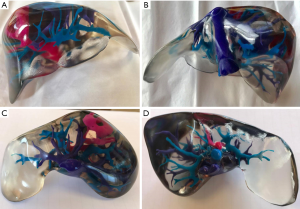
PolyJet print technology was identified as the most appropriate 3D printing method due to its ability to print highly accurate, multi-material models in a timely manner with little post-processing requirements (4,12). Stratasys’ Objet500 Connex3 multi-material printer, available through Objective 3D (Melbourne, Victoria, Australia), was selected for 3D printing. PolyJet VeroClear transparent and rigid opaque photopolymers were used to print the model (Figure 2), which was scaled down to 60% of its true size. Additionally, a full-size model depicting the arterial enhancement characteristics of the tumour and associated hepatic arterial supply was printed using the rigid opaque VeroMagenta photopolymer (Figure 3).

Data collection and analysis
Measuring model accuracy
Two independent observers completed measurements for five key anatomical landmarks including antero-posterior and medio-lateral diameters for the superior and inferior aspect of the inferior vena cava, the portal vein, and hepatic artery. Three measurements in each orientation at each landmark were performed and recorded in millimetres (24). This process was repeated for the three main stages of 3D model production, with measurements acquired from the original CT data using the measurement tool in RadiAnt DICOM viewer, the STL files using the measurement tool available in Geomagic Wrap, and the final 3D printed liver model using electronic callipers.
All measurements acquired from the 3D printed liver model were scaled up using the calculation depicted in Eq. [1], where MS represents the scaled-up measurement, and MO represents the raw measurement acquired from scaled down 3D printed liver model. This ensured consistency for accurate statistical analysis.
MS = (MO × 0.6) + MO [1]
Determining clinical value in radiology
Two independent radiologists were recruited through selective sampling to evaluate the usefulness of 3D printed liver models in the diagnostic reporting process for HCC cases (Supplement III).
Both participants were given the CT images and 3D printed liver model to complete a short nine-question survey (Supplement IV) involving questions related to the potential usefulness in the application of 3D printed models in radiology, and more specifically, as tools in diagnostic reporting for HCC. Participants answered multiple-choice questions, 5-point Likert scales, and open-ended questions for short answer responses. The level of agreement existing between the two professionals was assessed by a single observer, determined through comparative analysis of responses to each survey question (26).
Determining clinical value in surgical planning
Two independent abdominal surgeons were recruited through selective sampling to evaluate the potential usefulness of 3D printed liver models in surgical planning for resectable HCC lesions (Supplement III).
Both surgeons completed an interactive thirteen-question survey involving a combination of multiple choice and short answer questions (Supplement V). To complete the questionnaire, both participants were given the 3D printed liver and tumour models, in addition to a 3D reconstruction of the printed model. Questions were related to the 3D models provided, and how these models may be applied in practice.
The level of inter-observer agreement was determined and quantified by Cohen’s kappa statistics, and responses to short answer questions were discussed as appropriate.
Statistical analysis
Accuracy measurements recorded by two independent observers were entered into SPSS (SPSS 24.0, IBM Corporation, Armonk, NY, USA) for statistical analysis. A paired sample t-test was applied to statistically evaluate the magnitude and direction of difference in the mean measurements of specified anatomical landmarks acquired at each stage of data processing (5). Accuracy of the 3D liver model was achieved if the difference between in the mean of measurements obtained from the CT data and the 3D liver model was ≤1 mm (24). Paired samples statistics are presented as mean ± standard deviation, and paired differences are presented as the mean with the associated 95% confidence interval (CI) and t-statistic. T-statistics are presented as t(df)=t, p where t is the t-value, df is the degrees of freedom (N-1), and P is the P value. Statistical significance was defined where P<0.05, being used to accept or reject the null (HO) or alternative hypotheses (HA), where HO=µdifference and HA=µdifference≠0.
The surgeon’s responses to multiple-choice questions were assessed for inter-observer agreement through Kappa analysis. Crosstabulation results produced by SPSS were used to calculate the percentage agreement (P) as depicted by Eq. [2], where A represents the number of times both surgeons agreed in support of the use of 3D liver models, a represents the number of times both surgeons selected a multiple-choice question that did not support the use of 3D liver models, and T representing the total number of questions used for kappa analysis (27,28).

As the percentage agreement value does not account for the possibility for random agreement, Cohen’s kappa statistic (κ) is used to determine and quantify the level of inter-observer agreement. The calculated kappa statistic is interpreted using a standardised range of κ values. A κ value of 0 indicates poor inter-observer agreement; 0.01 to 0.20, slight agreement; 0.21 to 0.40, fair agreement; 0.41 to 0.60, moderate agreement; 0.61 to 0.80, good agreement; and 0.81 to 1.00, excellent agreement (27,28). The κ value was considered statistically significant if P<0.05.
Results
Cost and time required for 3D model production
The scaled down 3D liver model and the full-size tumour model were priced at AUD $1,250 and AUD $175 respectively. The total time required to produce the 3D liver model was 25.25 hours including 6 hours for segmentation, 4 hours for editing, 11 hours for printing, and 4.25 hours for post-print processing. Post-processing for the liver model involved cleaning, polishing and applying a clear coat to achieve transparency. The tumour model required 5.15 hours to print, and an additional 1 hour for post-processing which involved cleaning and drying.
3D printed liver model accuracy
The mean differences between paired variables, the t-values, and associated descriptive statistics were calculated using a paired sample t-test (Table 1). The Shapiro-Wilk’s test was utilized to assess normality, affirming normal distribution for measurement data acquired from the CT images (P=0.07), STL files (P=0.24), and 3D liver model (P=0.46). There were no outliers identified in the data collected for statistical analysis.

Full table
The average measurements acquired from the original CT data (18.28±9.31 mm) were greater than that of the STL file data (15.63±8.06 mm), with a difference of 2.65 mm (95% CI, −3.65 to 8.95), t [11] =0.93, P=0.37. A greater difference in the total average measurements is noted when comparing measurements from the original CT data to the physical 3D liver model (14.47±7.71 mm), with a variance of 3.81 mm (95% CI, −1.93 to 9.55), t [11] =1.46, P=0.17. The smallest difference in the means is demonstrated when pairing the measurements acquired from the STL files and 3D liver model, with a variance of 1.16 mm (95% CI, −0.31 to 2.63), t [11] =1.74, P=0.11.
Statistical significance was not achieved for any of the three-paired sample t-tests. As each of the mean differences were not statistically significant from zero, we fail to reject the null hypothesis (HO) and reject the alternative hypothesis (HA), where HO=µd=0 and HA=µd≠0.
Radiologist’s questionnaire response
When asked how useful they perceived the 3D printed liver model to be as a tool in diagnostic reporting for the given case, radiologist one (R1) deemed the model to be somewhat useful and radiologist two (R2) deemed it to be not useful at all. Both radiologists agreed that the 3D liver model did not improve their understanding of patient-specific pathological characteristics when compared to the CT images. R1 considered 3D liver models to be potentially useful supplementary tools in the diagnostic reporting process when viewing image datasets for patients with complex hepatic lesions, compared to R2 who indicated no use for 3D models in the diagnostic reporting process for hepatic lesions. Both participants considered the 3D models to be useful in assisting interprofessional communication between radiologists and associated health professionals involved in the treatment planning for patients with complex liver lesions. R1 did not believe that the 3D liver model would be useful in the education and training for junior radiologists or registrars, while R2 was unsure about its potential.
In response to short answer and feedback questions, one participant commented on the liver model’s ability to allow for a better appreciation of the tumour’s orientation in space within the liver. It was also suggested that the liver model might be a useful supplementary tool for surgical trainees when learning how to interpret multi-planar medical images.
Surgeon’s questionnaire response
Percentage agreement was calculated, quantifying the number of times both surgeons agreed that the 3D printed liver model was, or was not considered useful in the surgical management of HCC lesions. In this case, 80% agreement was achieved. A κ value of 0.38 (P=0.24) was obtained, indicating fair agreement.
Both surgeons believed that the 3D printed liver model allowed for a better perception of information related to structural depth and spatial relationships when compared to the corresponding 3D reconstructed image provided. Participants indicated that this information is particularly important in surgical planning for HCC. Both surgeons believed that the 3D liver model would be useful in the process of identifying a safe surgical pathway, in addition to intraoperative navigation and orientation. The separate tumour model was also deemed to be potentially useful in treatment planning, however it cannot be confirmed that it was able to provide additional information when compared to the 3D reconstructed image. Neither surgeon was able to confirm that the 3D models could reduce operating times by replacing the need for intraoperative visualization aids (such as Doppler ultrasound or cholangiography), although they both believed in the potential for the models to reduce the chance of intraoperative complications in complex HCC cases. The surgeons were in disagreement when asked if the scaled down model impacted on their ability to visualize the details of anatomical structures.
Discussion
3D model production
The cost and time required for 3D liver model production remains to be a major limitation (4,9,13,18). In aim of reducing print time and cost, the liver model was printed at 60% of its true size. While it is not a true representation of the patient’s anatomy, the scaled down model is relatively light, can be held in one hand, and is easy to manipulate. This finding is supported by recent research conducted by Igami et al. (9) and Oshiro et al. (23) who produced liver models scaled down to 70% and 50% of their true size respectively. The results of this study were not able to confirm whether the size of the printed model had an impact upon the viewer’s ability to identify or distinguish the details of intrahepatic anatomy and pathology. However, it can be confirmed that there is an element of structural distortion caused by light refraction from the transparent material inside the liver as reported by Oshiro et al. (23).
Both segmentation and editing processes are tedious and contribute significantly to the considerable time required for 3D liver model production. 3D model fabrication methods are well defined in existing literature (3,6,9,10,18,21,22,24,25), however there remains a need for efficient, accurate and cost effective automated programs to further streamline the liver model production process, consequently improving feasibility and encouraging the integration of 3D printed liver models within clinical practice (9,12,13,18,20-22).
Individuals creating the 3D liver model for pre-operative applications should attain a sound understanding of the software involved, as well as the anatomy and pathology of interest (20,29). Involvement of the reporting radiologist and surgeon (or surgeons) in 3D liver model production should also be considered to ensure accuracy of segmentation and identification of appropriate critical structures required for surgical planning (10,20). The importance of satisfying certain pre-requisites and implementing methods for quality control pertaining to software management and model production should be considered by future researchers. This may facilitate the acquisition of a patient-specific 3D printed liver model that is pathologically, anatomically, and structurally correct (20,29).
Model accuracy
The results for each of the three paired samples t-tests demonstrate a mean difference greater than 1 mm, indicating that undesirable variances exist in the measurements for specified anatomical landmarks at each stage of 3D model production. Although the segmentation, editing, and 3D printing processes are all considered as possible influential factors contributing to the observed variance, it is logical to surmise that the difference was largely influenced by the segmentation and editing process (10,22), with the mean difference in CT and STL file measurements being greater than that of the STL and 3D liver model measurements.
The most recent systematic review conducted by Witowski et al. (20) identifies the lack of studies implementing quantitative methods to statistically verify liver model accuracy. Model accuracy is important in the evaluation of feasibility, as it ensures accuracy and safety in the delineation of effective surgical pathways (20). While there are two studies that are identified to evaluate liver model accuracy through statistical analysis (5,13), this is the first study attempting to define the magnitude and direction of difference in accuracy measurements acquired at each stage of 3D liver model fabrication. Soon et al. (5) implemented a similar methodological approach in the quantitative evaluation of model accuracy, utilizing a paired samples t-test to compare measurements acquired from the original CT data and the corresponding 3D printed model. While the authors were able to confirm model accuracy with a mean difference of 0.1±0.06 mm (P<0.05), their results are not completely comparable to this study due to the fact that the full-size model utilized was printed with Fused Filament Fabrication technology (5).
Zein et al. (13) produced three 3D liver models, printed using PolyJet technology. Intraoperative measurements were acquired and compared to measurements taken from the corresponding liver model, which is regarded as the gold standard in the evaluation of 3D liver model accuracy (20). The models were deemed highly accurate, with mean dimensional errors of <1.3 mm for vascular structures, and <4.0 mm for the entire liver model (13). Differences existing between the quantitative methods and accuracy tolerances applied are identified to cause difficulties when comparing results. A universal threshold in the classification of 3D liver model accuracy should be defined and applied in future research to allow for consistency in quantitative evaluation and true comparison of results between studies.
Clinical value in radiology
In accordance with the results yielded from the radiologist’s questionnaire, it is apparent that 3D liver models are perceived to have minimal value in radiology, and more specifically as tools supplementing the diagnostic reporting process for HCC cases. These results alone deem the application of 3D printed liver models in the diagnostic reporting for HCC lesions as unfeasible. However, promising results were obtained with regards to the possibility of utilizing the patient-specific model to facilitate effective interprofessional communication between radiologists and other health professionals involved in pre-operative processes.
Based on the survey outcomes and participant feedback, it is evident that the real clinical value of the 3D printed liver models lies in applications beyond diagnostic reporting where individuals are likely to gain a more in-depth and holistic understanding of anatomy and pathology through 3D representations of 2D images. Physical 3D modelling has been identified to support efficient and effective perception of positional and structural information through the direct visualization of anatomy and pathology (2,3,9,13,14,16,22,25). This ultimately surpasses the need for mental 3D reconstruction involved when attempting to understand 2D images (29), which may be considered particularly valuable in the case of HCC due to the relative complexity of hepatic anatomy and pathological characteristics associated with certain HCC lesions (1,5).
Clinical value in surgical planning
It should be noted that this is the first study attempting to evaluate the clinical value of 3D liver models in surgical planning for HCC implementing both qualitative and quantitative approaches to data collection and analysis. Findings within case studies that utilized 3D printed liver models as tools in the surgical management for various liver lesions provided valuable information in the development of survey questions for this study (3,9,13,16,23). Such information allowed for the evaluation of whether the applications identified in current literature were transferrable and applicable to HCC treatment.
Encouraging results were obtained with regards to the perceived usefulness of both the 3D liver and tumour models as tools in surgical planning for complex resectable HCC lesions, verifying the feasibility of this application. Specifically, it was confirmed that the 3D liver model could assist in the identification of a safe surgical pathway through enhancing the viewer’s perception of structural and spatial information related to the case at hand (3,9,13,16,23). Contrary to the findings of a study conducted by Kong et al. (14), the 3D liver model used in this study was perceived to allow for better appreciation of spatial and structural characteristics when compared to the corresponding 3D reconstructed image provided. However, the relatively basic nature of the 3D visualization software employed could have influenced this result. Although the participants could not confirm the 3D liver model’s ability to replace the need for intraoperative visualization tools (such as Doppler ultrasound and cholangiography) as suggested by Igami et al. (9) and Zein et al. (13), they deemed the models to be potentially useful tools in intraoperative navigation and orientation (3,9,13,16).
Study limitations, recommendations and future research
There are a number of limitations associated with this small-scale study that must be noted. First, despite efforts to ensure consistency in the orientation and location of measurements for each anatomical landmark recorded by two independent observers, the chance for inconsistencies cannot be ruled out and must be considered as a possible factor influencing the observed variance in the means. Increasing the number of recorded measurements and number of observers involved in recording measurements may address this and help to reduce the chance for or effect of measurement error (24). Second, relying on medical images to acquire the baseline accuracy measurements has its disadvantages due to associated inaccuracies of 2D multi-planar imaging, consequently influencing the evaluation of model accuracy (13). Obtaining intra-operative measurements from the native liver as achieved by Zein et al. (13) would be considered as the gold standard in the evaluation of model accuracy (20). Third, while printing the 3D liver model at 60% of its true size allowed for a significant reduction in costs and improved model tangibility (9,23), the potential loss of visual or spatial information as a result of the scaling down process remains a possibility and is identified as an area for further exploration. Lastly, it must be noted that the principle researcher alone was responsible for quantitative comparative analysis of survey responses. Introducing a second objective observer into this process would assist in minimizing the potential for bias, thus improving credibility of results (26).
This study demonstrates how a mixed methods approach may be implemented to allow for a holistic evaluation of feasibility. Future research should implement a similar mixed methods approach with larger sample sizes to further assess the clinical value and application of patient-specific 3D liver models in surgical management of HCC. Applications other than surgical planning should also be explored, including the possibility for the 3D liver models to act as facilitators of patient-surgeon communication, interprofessional communication, as well as surgical education and training for HCC lesions (20). Future research in this area is warranted.
In conclusion, study outcomes indicate that there is minimal value in the application of 3D printed models in radiology, and more specifically in the diagnostic reporting process for HCC lesions. Encouraging results were achieved with regard to the potential clinical value of utilizing patient-specific 3D printed liver models in the pre-surgical management for resectable HCC lesions. The feasibility of this application is currently challenged by identified limitations including the cost and time of model production, and potential inaccuracies introduced with each stage of model fabrication.
Supplementary

Full table
Full table
Supplement III Selection criteria for participant recruitment
- Selection criteria for the recruitment of radiologists
Radiologists were approached to voluntarily participate in this study in accordance with the following selection criteria.
The participant was required to: - Currently specialise in diagnostic reporting for abdominal imaging, body imaging, and/or oncology;
- Be currently practicing within this specialisation;
- Be located within the Perth metropolitan area.
- Selection criteria for the recruitment of surgeons
Surgeons were approached to voluntarily participate in this study in accordance with the following selection criteria.
The participant was required to: - Currently specialise in abdominal surgery, hepatic surgery, and/or treatment of hepatic lesions;
- Be currently practicing within this specialisation;
- Be located within the Perth metropolitan area.
Supplement IV
Supplement V
Acknowledgements
We would like to thank Dr. Andrew Squelch from Curtin University for his guidance in image processing and segmentation; and Mark Walters, Senior Research Scientist from Princess Margaret Hospital Crani-Maxillo-Facial Unit’s for his advice and support throughout the pre-print editing process. Thanks also to Anne D’Arcy-Warmington for her support with statistical analysis.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: Ethics approval for this study was obtained from Curtin University’s Human Research Ethics Committee (HRE2017-0153). Due to the retrospective collection of de-identified computed tomography (CT) image data, patient consent was waived.
References
- Ertel AE, Shah SA. Surgical approaches to hepatocellular carcinoma. Semin Roentgenol 2016;51:88-94. [Crossref] [PubMed]
- Memeo R, De’Angelis N, De Blasi V, Cherkaoul Z, Brunetti O, Longo V, Piardi T, Sommacale D, Marescaux J, Mutter D, Pessaux P. Innovative surgical approaches for hepatocellular carcinoma. World J Hepatol 2016;8:591-6. [Crossref] [PubMed]
- Souzaki R, Kinoshita Y, Ieiri S, Hayashida M, Koga Y, Shirabe K, Hara T, Maehara Y, Hashizume M, Taguchi T. Three-dimensional liver model based on preoperative CT images as a tool to assist in surgical planning for hepatoblastoma in a child. Pediatr Surg Int 2015;31:593-6. [Crossref] [PubMed]
- Yao R, Xu G, Mao SS, Yang HY, Sang XT, Sun W, Mao YL. Three-dimensional printing: review of application in medicine and hepatic surgery. Cancer Biol Med 2016;13:443-51. [Crossref] [PubMed]
- Soon DS, Chae MP, Pilgrim CH, Rozen WM, Spychal RT, Hunter-Smith DJ. 3D haptic modelling for preoperative planning of hepatic resection: A systematic review. Ann Med Surg (Lond) 2016;10:1-7. [Crossref] [PubMed]
- Alkhouri N, Zein NN. Three-dimensional printing and pediatric liver disease. Curr Opin Pediatr 2016;28:626-30. [Crossref] [PubMed]
- Willatt J, Ruma JA, Azar SF, Dasika NL, Syed F. Imaging of hepatocellular carcinoma and image guided therapies - how we do it. Cancer Imaging 2017;17:9. [Crossref] [PubMed]
- Choi JY, Lee JM, Sirlin CB. CT. Radiology 2014;273:30-50. [Crossref] [PubMed]
- Igami T, Nakamura Y, Hirose T, Ebata T, Yukihiro Y, Sugawara G, Mizuno T, Mori K, Nagino M. Application of a three-dimensional print of a liver in hepatectomy for small tumours invisible by intraoperative ultrasonography: preliminary experience. World J Surg 2014;38:3163-6. [Crossref] [PubMed]
- Mitsouras D, Liacouras P, Imanzadeh A, Giannopolous AA, Cai T, Kumamaru KK, George E, Wake N, Caterson EJ, Pomahac B, Ho VB, Grant GT, Rybicki FJ. Medical 3D printing for the radiologist. Radiographics 2015;35:1965-88. [Crossref] [PubMed]
- Ventola CL. Medical applications for 3D printing: current and projected uses. P T 2014;39:704-11. [PubMed]
- Kim GB, Lee S, Kim H, Yang DH, Kim YH, Kyung YS, Kim CS, Choi SH, Kim BJ, Ha H, Kwon SU, Kim N. Three-dimensional printing: Basic principles and applications in medicine and radiology. Korean J Radiol 2016;17:182-97. [Crossref] [PubMed]
- Zein NN, Hanouneh IA, Bishop PD, Samaan M, Eghtesad B, Quintini C, Miller C, Yerian L, Klatte R. Three-dimensional print of a liver for preoperative planning in living donor liver transplantation. Liver Transpl 2013;19:1304-10. [Crossref] [PubMed]
- Kong X, Nie L, Zhang H, Wang Z, Ye Q, Tang L, Li J, Huang W. Do three-dimensional visualization and three-dimensional printing improve hepatic segment anatomy teaching? A randomized controlled study. J Surg Educ 2016;73:264-9. [Crossref] [PubMed]
- Preece D, Williams SB, Lam R, Weller R. “Let's get physical”: Advantages of a physical model over 3D computer models and textbooks in learning imaging anatomy. Anat Sci Educ 2013;6:216-24. [Crossref] [PubMed]
- Xiang N, Fang C, Fan Y, Yang J, Zeng N, Liu J, Zhu W. Application of liver three-dimensional printing in hepatectomy for complex massive hepatocarcinoma with rare variations of portal vein: preliminary experience. Int J Clin Exp Med 2015;8:18873-8. [PubMed]
- Naftulin JS, Kimchi EY, Cash SS. Streamlined, inexpensive 3D printing of the brain and skull. PLoS One 2015;10:e0136198. [Crossref] [PubMed]
- Witowski JS, Pędziwiatr M, Major P, Budzyński A. Cost-effective, personalized, 3D-printed liver model for preoperative planning before laparoscopic liver hemihepatectomy for colorectal cancer metastases. Int J Comput Assist Radiol Surg 2017;12:2047-54. [Crossref] [PubMed]
- Jones DB, Sung R, Weinberg C, Korelitz T, Andrews R. Three-dimensional modeling may improve surgical education and clinical practice. Surg Innov 2016;23:189-95. [Crossref] [PubMed]
- Witowski JS, Coles-Black J, Zuzak TZ, Pedziwiatr M, Chuen J, Major P, Budzynki A. 3D printing in liver surgery: A systematic review. Telemed J E Health 2017;23:1-5. [Crossref] [PubMed]
- Bücking TM, Hill ER, Robertson JL, Maneas E, Plumb AA, Nikitichev DI. From medical imaging data to 3D printed anatomical models. PLoS One 2017;12:e0178540. [Crossref] [PubMed]
- Madurska MJ, Poyade M, Eason D, Rea P, Watson AJ. Development of a patient-specific 3D-printed liver model for preoperative planning. Surg Innov 2017;24:145-50. [Crossref] [PubMed]
- Oshiro Y, Mitani J, Okada T, Ohkohchi N. A novel three-dimensional print of liver vessels and tumors in hepatectomy. Surgery Today 2017;47:521-4. [Crossref] [PubMed]
- Ho D, Squelch A, Sun Z. Modelling of aortic aneurysm and aortic dissection through 3D printing. J Med Radiat Sci 2017;64:10-17. [Crossref] [PubMed]
- Leng S, Chen B, Vrieze T, Kuhlmann J, Chen B, McCollough CH. Construction of realistic phantoms from patient images and a commercial three-dimensional printer. J Med Imaging (Bellingham) 2016;3:033501. [Crossref] [PubMed]
- Cho JY, Lee EH. Reducing confusion about grounded theory and qualitative content analysis: Similarities and differences. Qual Rep 2014;19:1-20.
- Gisev N, Bell JS, Chen TF. Interrater agreement and interrater reliability: key concepts, approaches, and applications. Res Social Adm Pharm 2013;9:330-8. [Crossref] [PubMed]
- McHugh ML. Interrater reliability: the kappa statistic. Biochem Med (Zagreb) 2012;22:276-82. [Crossref] [PubMed]
- Javan R, Herrin D, Tangestanipoor A. Understanding spatially complex segmental and branch anatomy using 3D printing. Acad Radiol 2016;23:1183-9. [Crossref] [PubMed]




